A Frame Work for Classification of Multi Class Medical Data based on Deep Learning and Naive Bayes Classification Model
Автор: N. Ramesh, G. Lavanya Devi, K. Srinivasa Rao
Журнал: International Journal of Information Engineering and Electronic Business @ijieeb
Статья в выпуске: 1 vol.12, 2020 года.
Бесплатный доступ
From the past decade there has been drastic development and deployment of digital data stored in electronic health record (EHR). Initially, it is designed for getting patient general information and performing health care tasks like billing, but researchers focused on secondary and most important use of these data for various clinical applications. In this paper we used deep learning based clinical note multi-label multi class approach using GloVe model for feature extraction from text notes, Auto-Encoder for training based on model and Navie basian classification and we map those classes for multi- classes. And we perform experiments with python and we used libraries of keras, tensor flow, numpy, matplotlib and we use MIMIC-III data set. And we made comparison with existing works CNN, skip-gram, n-gram and bag-of words. The performance results shows that proposed frame work performed good while classifying the text notes.
Health records, MIMIC-III, Deep learning, auto-encoder
Короткий адрес: https://sciup.org/15017079
IDR: 15017079 | DOI: 10.5815/ijieeb.2020.01.05
Текст научной статьи A Frame Work for Classification of Multi Class Medical Data based on Deep Learning and Naive Bayes Classification Model
Published Online February 2020 in MECS
To distinguish whether a patient experiences a specific illness, a regulated methodology requires the accompanying info: guides to become familiar with the examples for the malady (occasions speaking to patients) and the portrayal wellbeing status of the patient that is examined. Given that a suitable number of precedents are given (both positive and negative), this basic classification performs well. However, in the therapeutic area, this straightforward case isn't experienced that frequently. Surveying the wellbeing status of a patient does not generally prompt a solitary, singular therapeutic condition. Numerous wellbeing conditions are corresponded and impact one another, in this way suggesting comorbidity in a patient[1,2,3,4,5]. There is a level of uncertainty around the phrasing concerning comorbidity. The term characterizes a patient's wellbeing status when at least 2 conditions coincide. Various related builds are utilized to allude to comorbidity: multi disease, dreariness weight, and patient multifaceted nature. Previously, authors propose a top to bottom investigation of the comorbidity in patients which incorporates term definition, the nature, significance, and intricacy of this wellbeing condition. The nearness of various conditions in a patient can be a consequence of possibility, determination or causal affiliations. A typical and straightforward case of co-happening conditions is interminable obstructive aspiratory and liver ailment thinking about that they are brought about by propensities that are connected: smoking and liquor utilization.
In this research we concentrate on clinical text classification. Classification of text is the problem of assigning documents to different classes. Particularly, medical text or clinical notes plays key role in patient rick prediction. Clinical note analysis is key and significant importance in medical field. Because these are free text narratives generated by doctors and nurses, through patient make a diagnosis when he is clinic. They are regularly joined by a lot of metadata codes from the International Classification of Diseases (ICD), which extant an institutionalized method for demonstrating judgments and systems that were performed amid the experience.These ICD codes having verity of advantages, which varies from admission of hospital to billing and predictive analysis of patient condition. The major problem from traditional approach (manual) is more time consuming and error pruning. Automatic coding will be more accurate and very speedy approach. But mapping of medical text notes with ICD is a difficult task because of two major reasons. The space of the label is high dimensional, due to ICD-9 having 15K medical codes. Second, clinical content incorporates superfluous data, incorrect spellings and non-standard condensing, and a huge restorative vocabulary. These highlights consolidate to make the expectation of ICD codes from clinical notes a particularly troublesome errand, for PCs and human coders alike. In this paper we are presenting a multi class labelling model which is mapped to ICD codes. To assign automatic ICD codes based on ICU discharge summaries which are text format in better adapt to the multi label classification, we employ a per-label attention mechanism, which allows our model to learn distinct document representations for each label. This will be achieved using deep learning based auto-encoder and decoder for text feature extraction of different kinds of deceases, then these extracted features will be given to skip gram model for labelling then this will be given to navie-basian classification model. The classification is achieved for mapping for different ICD codes.
Research Objective is in view of the urge to analyse complex medical data to observe useful insights from the abundantly available medical data, this research aims to achieve the following objectives by employing multilayered networks. The main objectives is to develop a frame work for classification of multi class medical text data based on deep learning and naive bayes classification model.
The rest of the article is organized as follows, section-2 describes the details of existing literature, section-3 presents proposed framework, section-4 shows experimental setup and results and finally section five concludes the paper.
-
II. Literature Work
Tran et. al [8] determine tolerant vectors with their adjusted RBM engineering, at that point train a strategic relapse classifier for suicide hazard stratification. They explored different avenues regarding utilizing the full EHR information versus just utilizing determination codes, and found that the classifier utilizing the total EHR information with the eNRBM engineering for idea embedding performed best.
Essentially, Deep Patient created patient vectors with a 3-layer auto encoder, at that point utilized these vectors with calculated relapse classifiers to anticipate a wide assortment of ICD9-based ailment analyse inside an expectation window [9]. Their system indicated upgrades over crude highlights, with prevalent precision @k measurements for all estimations of k. In a theoretically comparable style, Z. Che et al. [10] additionally created patient vectors for use with straight classifiers, however decided on layer-wise preparing of a Deep Belief Network (DBN) trailed by a help vector machine (SVM) for grouping general malady analyse. Since preferably clinical notes related with a patient experience contain rich data about the aggregate of the confirmation, numerous examinations have analysed result forecast from the content alone. Jacobson et al. [11] looked at profound unsupervised portrayal of clinical notes for anticipating social insurance related contaminations (HAI), using stacked scanty AEs and stacked RBMs alongside a word2vec-based implanting approach. They found that a stacked RBM with term recurrence converse archive recurrence (tf-idf) pre-preparing yielded the best normal F1 score, and that applying word2vec prepreparing worked preferred with the AEs over the RBMs. At long last, utilized a two-layer DBN for recognizing osteoporosis. Their system utilized a discriminative learning organize where top hazard factors were recognized dependent on DBN remaking mistakes, and found the model utilizing all distinguished risk factors resulted in the best performance over baselines.
Clinical notes regularly incorporate unequivocal individual wellbeing data (PHI), which makes it hard to openly discharge numerous helpful clinical datasets [12]. As per the rules of the Health Information Portability and Accountability Act (HIPAA), all the clinical notes discharged must be free of delicate data, for example, names of patients and their intermediaries, recognizable proof numbers, emergency clinic names and areas, geographic areas and dates. Dernoncourt et al. [13] made a framework for the programmed de-recognizable proof of clinical content, which replaces a generally relentless manual de-identification process for sharing confined information. Their structure comprises of a bidirectional LSTM arrange (Bi-LSTM) also, both character and wordlevel embedding’s. The creators observed their technique to be best in class, with a gathering approach with restrictive irregular field’s additionally faring great. In a comparative undertaking, Shweta et al. [14] investigate different RNN models and word installing methods for distinguishing conceivably recognizable named substances in clinical content. The creators show that all RNN variations beat conventional.
Choi et al's. Med2Vec structure [15,16] for learning idea and patient visit portrayals utilizes a non-cynicism imperative upheld upon the scholarly code portrayals. The creators take the k biggest estimations of every section of the coming about code weight grid as a particular illness assemble that is interpretable upon subjective investigation. They likewise perform a similar procedure on the subsequent visit installing lattice for examining the kinds of visits every neuron figures out how to distinguish. Additionally, eNRBM design likewise upholds non-pessimism in the loads of the RBM. The creators guarantee that the subsequent weight sparsity means that which kinds of information sources enact the scanty neurons and can be utilized as a system to gauge the inherent dimensionality of the information. They additionally build up a novel regularization system for advancing basic smoothness dependent on the structure of restorative ontologies, by encoding the progressive systems into an element diagram with edge loads dependent on the cosmology separations. Both of these imperatives are added to the general target work for producing vector portrayals of therapeutic ideas.
This section gives the theoretical background of the literature survey carried out for this research work. It delineates the state-of-art data mining, machine learning and deep learning methodologies for handling medical text data. It gives the required inputs necessary to achieve the research objectives.
-
III. Proposed Model
Clinical note analysis having significant importance in medical field. Because these are free text narratives generated by doctors and nurses, during patient encounters when he is clinic. To analyse clinical text is a complex process [6,7]. Traditional data mining methods are failed to get the significant outcomes form the clinical text. To analysis the clinical text in this paper we proposed a frame work, this frame works on multi class classification of clinical text data. Which is composed using Glove model for feature extraction form text data, after words those features are given to a deep earning based auto-encoder for train the features after words naive-basin classification is applied for making multi classes. The proposed model is as follows:
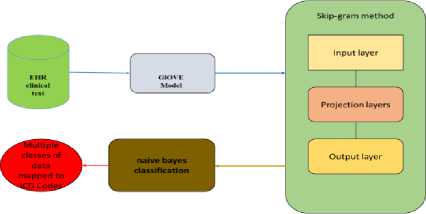
Fig.1. Multi-label multi class classification architecture for EHR data
-
A. The GloVe Model
The GloVe demonstrate represents Global Vectors which is an unsupervised learning model which can be utilized to acquire thick word vectors like Word2Vec. Anyway the method is extraordinary and preparing is performed on an amassed worldwide word-word co-event network, giving us a vector space with significant substructures. This technique was imagined in Stanford by Pennington et al. what's more, I prescribe you to peruse the first paper on GloVe, 'GloVe: Global Vectors for Word Representation' by Pennington et al. which is a magnificent perused to get some point of view on how this model functions[17].
We won't cover the execution of the model without any preparation in an excessive amount of detail here however on the off chance that you are keen on the genuine code, you can look at the official GloVe page. We will keep things basic here and endeavour to comprehend the essential ideas driving the GloVe show. We have discussed tally based grid factorization techniques like LSA and prescient strategies like Word2Vec. The paper guarantees that right now, the two families endure huge downsides. Techniques like LSA proficiently influence measurable data however they do generally inadequately on the word relationship errand like how we discovered semantically comparable words. Techniques like skip-gram may improve the situation on the relationship assignment, yet they ineffectively use the measurements of the corpus on a worldwide dimension.
The fundamental approach of the GloVe display is to initially make a colossal word-setting co-occurrence network comprising of (word, setting) combines to such an extent that every component in this framework speaks to how frequently a word happens with the unique situation (which can be a succession of words).
Considering the Word-Context (WC) lattice, WordFeature (WF) grid and Feature-Context (FC) network, we attempt to factorize WC = WF x FC, to such an extent that we mean to reproduce WC from WF and FC by duplicating them. For this, we ordinarily instate WF and FC with some irregular loads and endeavour to increase them to get WC' (an estimation of WC) and measure that it is so near WC. We do this on numerous occasions utilizing Stochastic Gradient Descent (SGD) to limit the mistake. At long last, the Word-Feature lattice (WF) gives us the word embedding’s for each word where F can be present to a particular number of measurements. A vital point to recollect is that both Word2Vec and GloVe models are fundamentally the same as by the way they work. The two expect to construct a vector space where the situation of each word is affected by its neighbouring words dependent on their specific circumstance and semantics. Word2Vec begins with neighbourhood singular instances of word co-event sets and GloVe begins with worldwide amassed co-event insights over all words in the corpus.
-
B. Learning Features with Auto-encoder
The Auto-encoder is a feed forward neural system with a balanced structure and a peculiar integer of layers by limiting the remaking blunder between the info information at the encoding layer and its recreation at the deciphering layer, in the mapping weight vectors. Autoencoder has a special preparing procedure to change the element of the information, e.g., from abnormal state to bring down one, or the other way around. It ensures that each neuron in info/yield layers can have a balanced correspondence to the component. For the least difficult case with a shrouded layer littler than the info/yield layer and straight actuations just, the auto-encoder executes a pressure conspire and performs similarly as PCA. Late examinations found that nonlinear auto-encoders are fit for ordering particular kinds of multimodal and nonlinear areas precisely, thus uncover a lot further associations between factors. To upgrade the execution of an autoencoder, the info vector should be component astute standardized to yJ-yJ n j
Where yj the normal estimation of each is highlight and oj is the difference. We accept the standardized vector Y as the contribution of the auto-encoder and characterize U as the conduct preparing set which is prepared to sustain into the auto-encoder with a size of N.
Where m is the measure of the dataset. The model is planned as a different layer structure (≥3), intending to pack and concentrate essential dormant highlights for further investigation, for example, grouping and relapse. The primary layer is the info layer, the last one is the yield layer, and others are shrouded layers.
Auto-encoders offer a method for characterizing a nonlinear type of M D, e (y) with parameters D, e that can fit to the dataset. In addition, the objective of repeating the auto-encoder is to gain proficiency with a personality function
л
M D ,e( y) = У ] ~ У ] (3)
Where y is the yield vector comparing to the input vector y j . We let m k signify the quantity of layers in our system, , i.e., mk = 5;^(k) indicates the parameter (or weight) related with the association between unit I in layer k, and unit j in layer k + 1; e ( k) is the inclination term related with unit j in layer k+1. The loads in each layer begin with an arbitrary esteem and repeat when endeavouring to limit the misfortune between the first info and its yield. We use digression work tanh(.) as r y -r -y initiation work between layers as. o(y) = —----
Г У + Г -У
Utilizing the chain guideline to back proliferate blunder subordinates through more profound layers to shallower ones, the angle strategy can without much of a stretch deal with the procedure. Accepting the third layer for instance (k=3), with an inadequate requirement on the concealed units, as it were for the greater part of its components to be zero, the normal value 9 of initiation level of shrouded neuron I on the preparation set is:
91=^1=! Ь*(у«Ъ (4)
Where b(3) indicates the enactment level of shrouded neuron I. Parameter ρ indicates the ideal dimension of sparsity whose esteem is near zero (e.g., 0.01). Presently we can characterize the general cost capacity to be
C(O,e)= 1 z 7 =1 W-y)z + PYL klog£ +
" L pt
(l-^log^] (5)
1-ptl
Where f is the quantity of neurons in the concealed layer. As appeared in Equation (5), we apply Kullback-Leibler difference as the punishment term, β is the heaviness of the punishment term. In the BP (Back
Propagation) preparing process, the refresh of loads is determined as:
^ [- ' ■ si c;) ^ <4) +^(^ p+^-p )].y ' (^v C3) ) (6)
In this examination, among the concealed layers, it is a sort of layer-wise-pre-preparing process, in other Words, the shallow layers (k=2) are utilized to get familiar with the genuinely basic and direct controls from units, and afterward the more profound concealed layers (k=3) are connected to learn idle directions or multimodal areas.
-
C. Naive bayes (NB) Classification
Naive Bayes model is definitely not hard to create and particularly important for sweeping instructive accumulations. Close by ease, Naive Bayes is known to beat even exceedingly refined gathering systems. Bayes hypothesis gives a method for ascertaining back likelihood P(c|x) from P(c), P(x) and P(x|c). Take a gander at the condition beneath:
p(c/x) =
p(c/x)p(c) P(x)
-
IV. Experimental Setup
For to perform experimental analysis we use python libraries of keras, tensor flow, numpy, matplot lib…etc. And we used 16GB RAM and 500 GB HDD with 2 GB graphic card and Intel I5 processor. As a platform we used UBUNTU 16.04 LTE. And the data set we used here is MIMIC-III [18].
MIMIC III ('Medical Information Mart for Intensive Care') is a substantial, single-focus database including data identifying with patients admitted to basic consideration units at an extensive tertiary consideration emergency clinic. Information incorporates imperative signs, meds, research facility estimations, perceptions and notes diagrammed via care suppliers, liquid parity, system codes, and analytic codes, imaging reports, clinic length of remain, survival information, and that's only the tip of the iceberg. The database bolsters applications including scholastic and modern research, quality enhancement activities, and advanced education coursework.
MIMIC III contains information related with 53,423 particular medical clinic confirmations for grown-up patients (matured 16 years or above) admitted to basic consideration units somewhere in the range of 2001 and 2012. Moreover, it contains information for 7870 neonates conceded somewhere in the range of 2001 and 2008. The information covers 38,597 unmistakable grown-up patients and 49,785 medical clinic confirmations. Mainly here we used 3 tables data those are discharge summaries, clinical notes which was written by doctors and nurses who are in ICU and other notes related to ICU. From that we performed the experimental evaluation with our proposed model to identify the class of the data which belongs to which class of ICD-9 codes.
Table 1. Clinical notes classification with respect to ICD-9 codes
|
S.no |
ICD-9 codes |
Discharge summaries |
Clinical notes |
Others Notes |
|
1 |
001 - 139 |
0.32 |
0.3488 |
0.31616 |
|
2 |
140 - 239 |
0.23 |
0.2507 |
0.22724 |
|
3 |
240 - 279 |
0.43 |
0.4687 |
0.42484 |
|
4 |
280 - 289 |
0.24 |
0.2616 |
0.23712 |
|
5 |
290 - 319 |
0.71 |
0.7739 |
0.70148 |
|
6 |
320 - 389 |
0.56 |
0.6104 |
0.55328 |
|
7 |
390 - 459 |
0.43 |
0.4687 |
0.42484 |
|
8 |
460 - 519 |
0.87 |
0.9483 |
0.85956 |
|
9 |
520 - 579 |
0.76 |
0.8284 |
0.75088 |
|
10 |
580 - 629 |
0.21 |
0.2289 |
0.20748 |
|
11 |
630 - 679 |
0.03 |
0.0327 |
0.02964 |
|
12 |
680 - 709 |
0.21 |
0.2289 |
0.20748 |
|
13 |
710 - 739 |
0.52 |
0.5668 |
0.51376 |
|
14 |
740 - 759 |
0.12 |
0.1308 |
0.11856 |
|
15 |
780 - 789 |
0.65 |
0.7085 |
0.6422 |
|
16 |
790 - 796 |
0.71 |
0.7739 |
0.70148 |
|
17 |
797 - 799 |
0.56 |
0.6104 |
0.55328 |
|
18 |
800 - 999 |
0.43 |
0.4687 |
0.42484 |
|
19 |
V CODES |
0.32 |
0.3488 |
0.31616 |
|
20 |
E Codes |
0.23 |
0.2507 |
0.22724 |
Table 2. Classes of deceases
|
proposed work |
Skipgram |
n-gram |
CNN |
Bag of Words |
||
|
Class1 |
Pr |
89 |
46 |
41 |
42 |
87 |
|
Re |
66 |
80 |
55 |
56 |
57 |
|
|
F 1-score |
68 |
58 |
47 |
48 |
64 |
|
|
Ac |
96 |
93 |
88 |
90 |
91 |
|
|
Class2 |
Pr |
81 |
73 |
78 |
80 |
72 |
|
Re |
72 |
33 |
42 |
43 |
60 |
|
|
F 1-score |
81 |
46 |
55 |
56 |
62 |
|
|
Ac |
93 |
88 |
85 |
87 |
88 |
|
|
Class3 |
Pr |
68 |
22 |
27 |
28 |
45 |
|
Re |
72 |
30 |
39 |
40 |
38 |
|
|
F 1-score |
68 |
25 |
32 |
33 |
41 |
|
|
Ac |
90 |
79 |
79 |
81 |
90 |
|
|
Class4 |
Pr |
68 |
49 |
49 |
50 |
81 |
|
Re |
71 |
48 |
54 |
55 |
67 |
|
|
F 1-score |
72 |
48 |
51 |
52 |
71 |
|
|
Ac |
82 |
75 |
71 |
73 |
88 |
|
|
Class5 |
Pr |
77 |
34 |
42 |
43 |
68 |
|
Re |
47 |
56 |
46 |
47 |
49 |
|
|
F 1-score |
61 |
43 |
44 |
45 |
51 |
|
|
Ac |
72 |
71 |
67 |
69 |
88 |
|
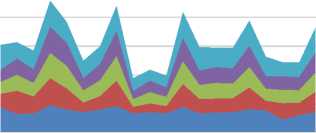
The table shows the percentage of patient belongs to which kind of decease and those will be evaluated using precession, recall, F1-score and accuracy of that model. And here we presented top-5 class of decease which are obtained form results section. The below table represents the five classes and here we made comparative analysis with respect to existing models of skip-gram, n-gram, CNN and bag of words approach. The deceases are chronic, heart, brain, liver and lungs.
500 d>
5 400 га си 300
5 200 го
|
£ 2 о |
£ 2 о |
£ 2 о |
5: 2 о |
5: 2 о |
|
Class1 |
Class2 |
Class3 |
Class4 |
class5 |
Class type
-
■ proposed work ■ Skip-gram ■ n-gram
-
■ CNN ■ Bag of Words
Fig. 2. Different Classes of Deceases Comparison
Here we saw that the proposed technique has the best execution on practically the majority of the assessed qualities. The n-gram and pack of-words based techniques are reliably more fragile than the proposed, validating the discoveries in writing that word installing's enhance execution of clinical NLP assignments. We moreover examine in the case of considering longer expressions enhances display execution. Here we demonstrate the distinction in F1-score between models with expressions length changing from 1st to 5th classes. Whenever the data size increase proposed method does not show much variance, but other models were not show the considerable performance loss for longer phrases. Proposed model shows significant performance than compared to existing work.
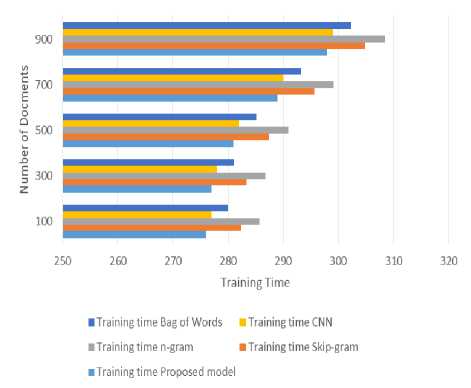
Fig. 3. Computation time for Training
Here fig.3 shows the computation time for training of different mechanisms with respect to number of documents here proposed model gives lower computation time for training because it gets features from GloVe model it done features very quickly and accurately. CNN gives better than remaining but other models are not performed up to the mark due to their constraints of not handling medical text. And when the size of the set increases it does not show much deviation by proposed model, but existing works are considerable shows decrease in training performance.
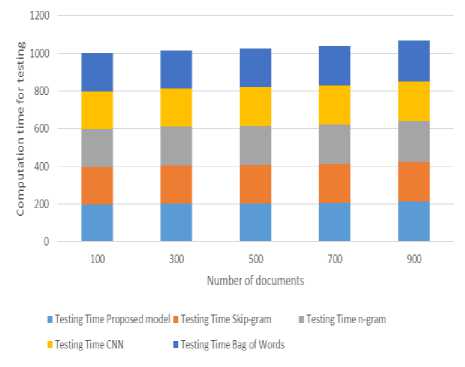
Fig. 4. Computation time for testing
Here fig.4 shows the computation time for testing the different mechanisms with respect to number of documents here proposed model gives lower computation time for testing because it gets features from GloVe model it done features very quickly and accurately. CNN gives better than remaining but other models are not performed up to the mark due to their constraints of not handling medical text. And when the size of the set increases it does not show much deviation by proposed model, but existing works are considerable shows decrease in training performance.
Medical text classification is a complex task because of its nature. In this research work we used deep learning based clinical note multi-label multi class approach using GloVe model for feature extraction from text notes, AutoEncoder for training based on model and Navie basian classification and we map those classes for multi- classes. And we perform experiments with python and we used libraries of keras, tensor flow, numpy, matplotlib and we use MIMIC-III data set. And we made comparison with existing works CNN, skip-gram, n-gram and bag-of words. The performance results shows that proposed frame work performed good while classifying the text notes.
-
V. Conclusion
Text classification is a complex task with regular methods but it gives significant impact to the socity. Medical text classification is a complex task because of its nature. In this paper we used In this paper we used deep learning based clinical note multi-label multi class approach using GloVe model for feature extraction from text notes, Auto-Encoder for training based on model and navie basian classification and we map those classes for multi- classes. And we perform experiments with python and we used libraries of keras, tensor flow, numpy, matplotlib and we use MIMIC-III data set. And we made comparison with existing works CNN, skip-gram, n-gram and bag-of words. The performance results shows that proposed frame work performed good while classifying the text notes.
Hybridization is widely used to overcome problems of different individual techniques in various domains. One of the directions for future work is to investigate the hybridization of Navie Bayes with other approaches such as fuzzy theory that may give more promising outcomes.
Список литературы A Frame Work for Classification of Multi Class Medical Data based on Deep Learning and Naive Bayes Classification Model
- Hai, J., et al. (2019). "Multi-level features combined end-to-end learning for automated pathological grading of breast cancer on digital mammograms." Computerized Medical Imaging and Graphics 71: 58-66.
- Kocbek S, Cavedon L, Martinez D, Bain C, Mac Manus C, Haffari G, et al. Text mining electronic hospital records to automatically classify admissions against disease: measuring the impact of linking data sources. J Biomed Inform. 2016; 64:158–67.
- Yetisgen-Yildiz M, Pratt W. The effect of feature representation on Medline document classification. AMIA Annu Symp Proc. 2005;2005:849–53.
- Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, et al. 2016. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 35:1160–69.
- Beaulieu-Jones, B.K., Greene, C.S., et al.: Semi-supervised learning of the electronic health record for phenotype strati cation. Journal of biomedical informatics 64, 168 -178 (2016).
- Ferr´andez O, South BR, Shen S, Meystre SM (2012) A hybrid stepwise approach for de- identifying person names in clinical documents. In: Proceedings of the 2012 Workshop on Biomedical Natural Language Processing, Association for Computational Linguistics, pp 65–72.
- Lin YK, Chen H, Brown RA , MedTime: A temporal information extraction system for clinical narratives. Journal of biomedical informatics 46:S20–S28 (2013).
- T. Tran, T. D. Nguyen, D. Phung, and S. Venkatesh, “Learning vector representation of medical objects via EMR-driven nonnegative restricted Boltzmann machines (eNRBM),” Journal of Biomedical Informatics, vol. 54, pp. 96–105, 2015.
- R. Miotto, L. Li, B. A. Kidd, and J. T. Dudley, “Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records.” Scientific reports, vol. 6, no. April, p. 26094, 2016.
- Z. Che, D. Kale, W. Li, M. Taha Bahadori, and Y. Liu, “Deep Computational Phenotyping,” in Proceedings of the 21st ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2015, pp. 507–516.
- J. C. Denny, T. A. Lasko, and M. A. Levy, “Computational Phenotype Discovery Using Unsupervised Feature Learning over Noisy, Sparse, and Irregular Clinical Data,” PLoS ONE, vol. 8, no. 6, 2013.
- C. Angermueller, T. P¨arnamaa, L. Parts, and O. Stegle, “Deep learning for computational biology,” Molecular systems biology, vol. 12, no. 7, p. 878, 2016.
- Franck Dernoncourt, Ji Young Lee, Peter Szolovits, “De-identification of Patient Notes with Recurrent Neural Networks,” arXiv:1606.03475v1 [cs.CL] 10 Jun 2016.
- A. E. Shweta, S. Saha, and P. Bhattacharyya, “Deep learning architecture for patient data de-identification in clinical records,” ClinicalNLP 2016, p. 32, 2016.
- E. Choi, M. T. Bahadori, and J. Sun, “Doctor AI: Predicting Clinical Events via Recurrent Neural Networks,” arXiv, pp. 1–12, 2015.
- E. Choi, M. T. Bahadori, E. Searles, C. Coffey, and J. Sun, “Multi-layer Representation Learning for Medical Concepts,” arXiv, pp. 1–20, 2016.
- Pennington, Richard Socher, Christopher D. Manning " GloVe: Global Vectors for Word Representation" Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages 1532–1543 (2014).
- http://dx.doi.org/10.13026/C2XW26


