An Efficient System for Medical Image Retrieval using Generalized Gamma Distribution
Автор: T.V. Madhusudhana Rao, S.Pallam Setty, Y.Srinivas
Журнал: International Journal of Image, Graphics and Signal Processing(IJIGSP) @ijigsp
Статья в выпуске: 6 vol.7, 2015 года.
Бесплатный доступ
Efficient diagnosis plays a crucial role for treatment. In many cases of criticalness, radiologists, doctors prefer to the usage of internet technologies in order to search for similar cases. Accordingly in this paper an effective mechanism of Content Based Image Retrieval (CBIR) is presented, which helps the radiologists/doctors in retrieving similar images from the medical dataset. The paper is presented by considering brain medical images from a medical dataset. Feature vectors are to be extracted efficiently so as to retrieve the images of interest. In this paper a two-way approach is adopted to retrieve the images of relevance from the dataset. In the first step the Probability Density Functions (PDF) are extracted and in the second step the relevant images are extracted using correlation coefficient. The accuracy of the model is tested on a database consisting of 1000 MR images related to brain. The effectiveness of the model is tested using Precision, Recall, Error rate and Retrieval efficiency. The performance of the proposed model is compared to Gaussian Mixture Model (GMM) using quality metrics such as Maximum distance, Mean Squared Error, Signal to Noise Ratio and Jaccard quotient.
Generalized Gamma Distribution, Content Based Image Retrieval, Relevancy, Correlation, Quality metrics
Короткий адрес: https://sciup.org/15013882
IDR: 15013882
Текст научной статьи An Efficient System for Medical Image Retrieval using Generalized Gamma Distribution
Published Online May 2015 in MECS DOI: 10.5815/ijigsp.2015.06.07
Brain disease is one of the most striking factors for the increase in mortality rates. In India this rates have been increasing continuously [24]. Most of the diseases causing brain tumors may be either Benign or Malignant. The classifications of these are subjected to the size of the tumors. Hence effective diagnosis and identification of these diseases will be of crucial importance. Most of the cases related the treatment of these diseases is subjected to MR imaging. MR imaging is mainly choosing because of its property of non-ionization. Along with the advantage, the grater disadvantage with these techniques is that the reports are generated by the radiologists using the visual perception of the MR images [22]. However as the number of cases of brain related diseases are increasing, the visual perception may lead to incorrect decisions. Another added factor to this is the proportionate increase of the radiologists versus the increase in the number of cases is lagging. Hence the advantage of using automated systems for effective brain image scanning and report generations are of crucial importance. This paper highlights a methodology which helps to retrieve the relevant images from the databases. This system may help in particular to the paramedics, radiologist residing in remote areas to suggest a basic treatment for patient in residing in rural areas. So that he/she can be shifted to nearby super specialized hospitals.
Many systems have been presented in the literature based on Picture Archive and Communication Systems (PACS) [18]. Also text based retrieval systems are listed in the literature wherein the relevant text/keywords from the lab reports are retrieved against a query. However these systems have their own drawbacks since text annotation is difficult [14], though, the usage of text will be useful to some of the patients eager to know about some related facts about their disease. Hence, in this paper we present a novel methodology of CBIR using Generalized Gamma Distribution which helps to retrieve similar images based on content as well as text. The updated equations of the model parameters are estimated using Expectation Maximization (EM) algorithm. The main advantage behind the usage of this distribution is of the fact that the human tissues are asymmetric in nature and it can handle the speckles more efficiently.
Assuming that the shape and texture features play a vital role in identifying the contents more effectively [20][21], in this model identification of Malignant and non-Malignant tissues is experimented. Other models based on auto regression [7], wavelets [19], semantic gap [10][13], relevance feedback [23], Gabor filter [4], K- means algorithm [3][16], Genetic algorithm [9], KD-Tree [8], binary splitting [2], quad trees [11] have been proposed in the literature. However, these models lack in efficiency while retrieving the images of relevance because of several disadvantages like K-means algorithm is sensitive to initial clusters which may not be efficient for handling medical images where the data is both continuous and discrete. Models like binary splitting, KD-Tree, Genetic algorithms are subjected to parametric dependence and complexity [5][15]. Hence efficient methods to retrieve the images of interest will be of a great advantage.
-
A. The Related Work
Models based on Gamma, Log-Normal and Nakagami distributions are also highlighted in the literature for handling asymmetric distributions. However, Nakagami distribution overrules the other distributions while characterizing the speckles in tissues [17]. Nevertheless the Nakagami distribution fails in handling larger impulse response of the speckles generated by human speckles compared to Generalized Gamma Distribution.
This paper presents a methodology using Generalized Gama Distribution. The organizing structure of the paper is as follows. The proposed methodology is presented in Section II. The experimentation, results together with performance analysis and evaluation are highlighted in Section III, the final Section IV concludes the paper.
-
II. Proposed Methodology Using Generalized Gamma Distribution
The proposed methodology is depicted in the block diagram shown in Figure 1, and it will be very much useful for medical image analysis for certain remote areas, where only the minimum first aid is available. Using the developed methodology, procedures can be evaluated and necessary treatments can be derived.
These methods are deployed mainly on the brain image data collected from University of Rennes1 database. Every brain image consists of vital information such as white matter (WM), Gray Matter (GM) and Cerebro Spinal Fluid (CSF). In order to assess the damage of the brain tissues, effective methodologies are to be developed to discriminate WM, GM, and CSF.
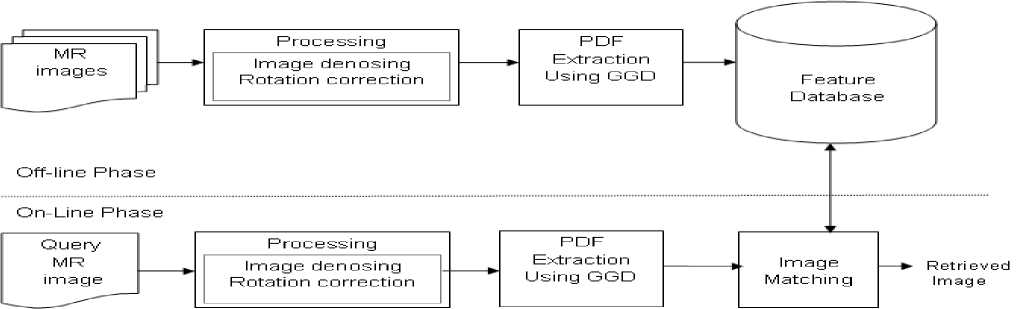
Fig. 1. Block diagram of the proposed model
A. Generalized Gamma Distribution
Generalized Gamma Distribution is proposed for the classification of the images in this paper. In general, in Content Based Medical Image Retrieval, the damaged tissues are to be recognized. These damaged pixels behave in a different manner compared to the nondamaged pixels and the intensity levels of these pixels will be more i.e. the number of non-damaged pixels are more than the damaged pixels. And these damaged pixels will have their peeks at the origin and hence the damaged medical images will have their distributions with long tails. In order to interpret these distributions, tail distributions will be more appropriate and to cater these distributions the best suited distribution is Generalized Gamma Distribution. Generalized Gamma Distribution includes several other distributions such as Weibull distribution, Gamma distribution, Log-normal distribution, Chi-square distribution and Exponential distribution as particular cases. The main advantage of using Generalized Gamma Distribution is that, it can
sensitize the medical image features in presence of noise and with minimum variance more effectively.
The Probability Density Function of the Generalized Gamma Distribution is of the form
f (x, k, c, a, b) =
c (x - a)ck 1
e
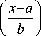
bck Г( k )
Where ‘a’, ‘b’ and ‘x’ are called gamma variants and ‘c’ and ‘k’ are called shape parameters. By varying the value of the shape parameters, the particular cases of gamma distribution can be modeled.
B. Expectation Maximization (EM) Algorithm for Generalized Gamma Distribution
In order to retrieve the relevant images more efficiently and effectively, the parameters of the medical images are to be estimated effectively. The parameters, Expectation Maximization algorithm is used for obtaining the final
estimates. The final updates derived in [12] and
are
presented here.
N
a
( l + 1) = уS E ( l ) [ t k ( Z s , 6 ( l ) )]
k
s = 1
c =
1
1 dr
c
—
Г дc
k logl
—
a
b
a = x
1 — ck
—
1 дГ
c
x
—
a
c — 1
—
b =
k = 1 +
Г дa
b
b
ck
^ (x — a ) c
1 дГ
—
Г д b
J_ e — t ( log e t У 1 dt 0
Г ( k — 1 ) c log]
x
—
a
—
1 дГ
b
Г дk
-
C. Image Database And Features Considerd
In order to test the validity of the images a database consists of 1000 images pertaining to brain tumor disease is used. The images collected include, the inhomogeneities of the brain tumor such as SEIZURES, MS-LESIONS and SCELEROSIS. Each image is of fixed size which is normalized and pre-processed. In order to have a effective recognition of the deformities, the features are to be extracted basing on position, color and texture, since any deformity in the brain image to be retrieved effectively needs to figure out the exact location for which the position of the (x, y) coordinates place a vital role where ‘x’ denotes the width and ‘y’ denotes the height.
The in-homogeneities in the brain can be identified and differentiate appropriately basing on the color also. Hence for each image the color components are also considered. Another feature which is of prime important for any retrieval and identification is the texture. In this methodology the texture of images are extracted and these features are given as input to the Generalized Gamma Distribution.
-
D. Image Matching Using Correlation
In this paper we present a novel methodology for identifying the relevancy between the images in the database and feature or a query image is considered for relevance. The main advantage of this methodology [1] is that it can be used to test whether a subpart of the image
belongs to the image in the database or not and it is given by
. . У n L * K
C ( L , K ) = ------- (7)
sum ( L j • L 2 ) * sum ( K • K 2)
Where L is the image from the database and K is the query image (Note: K and L should be of same dimension) and we say that the query image K is the part of the image L, if the correlation approaches to near 1.
-
E. CBIR Algorithm For The Proposed Methodology
-
• Step 1: Obtain histogram of all the images in the database, in order to model the various particular cases of Generalized Gamma distribution
-
• Step 2: Calculate PDFs of Generalized Gamma Distribution
-
• Step 3: Find the relevant image based on correlation using the formula
C ( L , K ) = , S i = 1 L *—, ------7 (8)
sum(L , • L^ ) * sum ( K • K )
Using the correlation value, most relevant images can be extracted.
-
• Step 4: Retrieve similar images based on correlation value of step 3
The brain images from the image dataset are given as inputs to the Generalized Gamma Distribution proposed in section II (A). The PDF of the images is retrieved and stored. The query image is processed using section II (B). The relevant images that are matched are compared against the PDF.
The input query image is processed using the methodology under-laid in Section II (C) and the relevant images are retrieved. The performance of the developed methodology is tested using metrics like Precision, Recall, Error rate and Retrieval efficiency. In order to present this paper we have considered an image dataset obtained from University of Rennes1. The relevant images from the data set against the query are matched based on Region of Interest (ROI) using the equation 25.
The methodology is tested by varying the images ranging count from 200, 300, 500 and 1000 images in the database. The next level of analysis is to identify the match between the ROI and the images considered with above sizes. Since it is mandatory to test the appropriateness of the developed methodology, we have tested the performance on different operating systems by varying image sizes as mentioned above. The time elapsed against each of the images under different operating system environments like Windows, Linux, UNIX is considered.
Personal Computer (PC) with Windows operating system with 200 images in the database is considered and the result was successful and the time elapsed is 1.8 seconds. The tests were conducted in the same environment on databases of sizes 300, 500 and 1000 and the time elapsed for retrieval is 2.5 seconds, 2.75 seconds and 2.9 seconds respectively. The results were tested under the other operating systems with the above dataset and time taken is as follows.
The table 1 presents the effectiveness of the model where it is tested in different Operating System environments by varying the image sizes. In both the cases of operating Systems the model performs better retrieval accuracy in minimum time.
Table 1. Performance of the system by varying the image sizes
|
Operating System |
Images Size |
Time elapsed in seconds |
|
Linux |
200 |
1.85 |
|
300 |
2.6 |
|
|
500 |
2.8 |
|
|
1000 |
2.85 |
|
|
UNIX |
200 |
1.8 |
|
300 |
2.65 |
|
|
500 |
2.75 |
|
|
1000 |
2.8 |
The query medical image is considered and is to be matched with the similar images in the database, to identify whether the patient is normal or suffering from disease. A local database is maintained and the query image considered is processed with these images in the local database for relevance. The Query image is processed based on Query based image technique presented in Section II (B) and the relevant images are obtained based on relevance method. These are presented in Figure 2 and Figure 3.
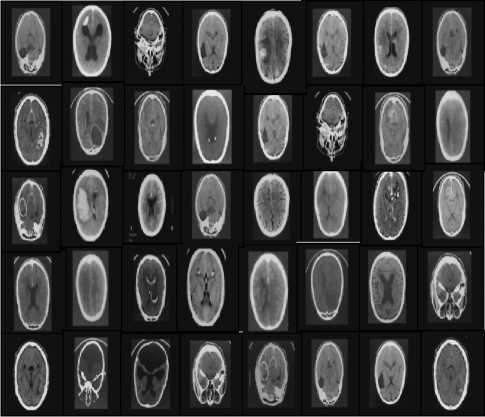
Fig. 2. Image Dataset
Disease
Brain tumor
Query image
Relevant images
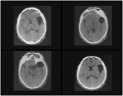
NonRelevant images
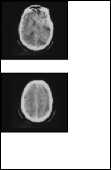
Fig. 3. Input Query Image versus Retrieved Relevant and Non- Relevant images
-
A. Evaluation
The outputs derived against the query processed are evaluated using metrics like Precision, Recall, Error rate and Retrieval efficiency.
-
• Precision
It is the ratio of the number of relevant images retrieved to the total number of irrelevant and relevant images retrieved. It is usually expressed as a percentage.
Precision = (A / (A + C))* 100;
-
A: Number of relevant images retrieved.
-
C: Number of irrelevant images retrieved.
A + C: Total number of irrelevant + relevant images retrieved.
-
• Recall
It is the ratio of the number of relevant images retrieved to the total number of relevant images in the database. It is usually expressed as a percentage.
Recall = (A / (A + B)) * 100
-
A: Number of relevant images retrieved
-
B: Number of relevant images not retrieved
A + B: The total number of relevant images
-
• Error rate
Error rate = Number of non-relevant images retrieved / Total number of images retrieved.
-
• Retrieval efficiency
Retrieval efficiency = Precision = Number of relevant images retrieved/ Total number of images retrieved (If number of retrieved images > number of relevant images), otherwise
Retrieval efficiency = Number of relevant images retrieved/ Total number of relevant images.
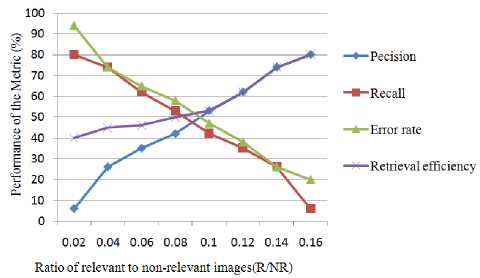
The outputs derived using the proposed metrics are presented in Table 2 and in Figure 4.
Table 2. Precision, Recall, Error rate and Retrieval efficiency based on relevant and non-relevant images
|
No. of relevant images in the database |
No. of nonrelevant images in the database |
Ratio of relevance to nonrelevance |
Precision |
Recall |
Error rate |
Retrieval efficiency |
|
120 |
750 |
0.16 |
80 |
6 |
94 |
40 |
|
98 |
700 |
0.14 |
74 |
26 |
74 |
45 |
|
78 |
650 |
0.12 |
62 |
35 |
65 |
46 |
|
60 |
600 |
0.1 |
53 |
42 |
58 |
50 |
|
44 |
550 |
0.08 |
42 |
53 |
47 |
53 |
|
30 |
500 |
0.06 |
35 |
62 |
38 |
62 |
|
18 |
450 |
0.04 |
26 |
74 |
26 |
74 |
|
8 |
400 |
0.02 |
6 |
80 |
20 |
80 |
-
B. Analysis Of The Methodology Against The Time Taken
Table 3 describes the recognition accuracy against the time taken. From this table it can be clearly seen that the relevant images are retrieved effectively.
Table 3. Showing the accuracy of Present model
|
Brain tumor |
Brain Hemorrhage |
Fractures |
|
|
Accuracy (%) |
98 |
96 |
97 |
|
Time taken in seconds |
1.8 |
2.0 |
2.2 |
-
C. Performance Evaluation
The retrieved efficiency, in case of medical images against the query image is evaluated using Image quality metrics. The various metrics considered for the work include, Maximum distance, Mean Squared Error, Signal to Noise Ratio and Jaccard quotient. The formulas for computing the above quality metrics are presented in Table 4.
Using these formulae the performance evaluation is carried out and the proposed model is compared to GMM and the results obtained are tabulated and presented in Table 5.
Fig. 4. Variation of Precision, Recall, Error rate and Retrieval efficiency values with relevant to non-relevant images ratio
Table 4. Formulas for Quality Metrics
|
Quality metric |
Formula to Evaluate |
|
Maximum Distance |
Max{ F ( j , k ) - F ( j , k )} |
|
Mean Squared error |
52 M , X N , [ O { F ( j , k )} - O { F ( j , k )}]2 1 52 M , У N , [ O { F ( j , k )}]2 MN Z—ij =1 Z—ik =1L 1 vy, < W, 7JJ у j^^j =1 Z—ik =1L 1 , 7^J Where M,N are image matrix rows and columns |
|
Signal to noise ratio |
( MAX, ) .og, 0 [ -Jmse J Where, MAX I is maximum possible pixel value of image, MSE is the Mean squared error |
|
Jaccard quotient |
| X n y| = a X u У a + b + c XY Where, a — |X П У | , b — -- , C — -- , d — |X U У | and X, Y are input and output image intensities |
Table 5. Evaluated Image Quality Metrics
|
Image |
Quality Metric |
GMM |
Generalized Gamma Distribution |
Standard Limits |
Standard Criteria |
|
Maximum Distance |
0.422 |
0.9325 |
-1 to 1 |
Closer to 1 |
|
|
Mean Squared error |
0.04 |
0.094 |
0 to 1 |
Closer to 0 |
|
|
Signal to noise ratio |
17.41 |
33.89 |
-∞ to ∞ |
As big as Possible |
|
|
Jaccard quotient |
0. 089 |
0.703 |
0 to 1 |
Closer to 1 |
|
|
Maximum Distance |
0. 221 |
0.912 |
-1 to 1 |
Closer to 1 |
|
|
Mean Squared error |
0 2404 |
0.2019 |
0 to 1 |
Closer to 0 |
|
|
■WB |
Signal to noise ratio |
14.45 |
39.85 |
-∞ to ∞ |
As big as Possible |
|
Jaccard quotient |
0.0677 |
0.7921 |
0 to 1 |
Closer to 1 |
|
|
Maximum Distance |
0.345 |
0.807 |
-1 to 1 |
Closer to 1 |
|
|
Mean Squared error |
0.22 |
0.2123 |
0 to 1 |
Closer to 0 |
|
|
Signal to noise ratio |
19.88 |
39.71 |
-∞ to ∞ |
As big as Possible |
|
|
Jaccard quotient |
0. 043 |
0.7143 |
0 to 1 |
Closer to 1 |
|
|
I ■*■! |
Maximum Distance |
0. 224 |
0.971 |
-1 to 1 |
Closer to 1 |
|
Mean Squared error |
0.24 |
0.1192 |
0 to 1 |
Closer to 0 |
|
|
Signal to noise ratio |
21.42 |
37.41 |
-∞ to ∞ |
As big as Possible |
|
|
Jaccard quotient |
0. 045 |
0.874 |
0 to 1 |
Closer to 1 |
-
IV. Conclusion
In this paper an efficient methodology for brain MRI image retrieval against the query image is proposed. This proposed system helps to retrieve the relevant images from the image dataset effectively. The proposed model exhibits good recognition rates of above 90%. The evaluation of the developed model is carried out by comparing with the existing models based on GMM, by using quality metrics. The results show that, this developed algorithm outperforms the existing algorithm. This methodology suits very well in applications wherein assistance is needed for the doctors at remote areas.
-
[1] Ahmad Fauzi M. F., and Ahmad W., “Efficient block based matching for content based retrieval of CT head images,” In Proceedings of the IEEE 10th workshop on multimedia signal processing, Cairns, Qld, pp. 142-147, 2008.
-
[2] Babu G. P., and Murty M. N., “A near-optimal initial seed value selection in k-means means algorithm using a genetic algorithm,” Pattern Recognition Letters, vol. 14, no. 10, pp. 763-769, 1993.
-
[3] Bishnu P. S., and Bhattacherjee V., “Software fault prediction using quad tree-based K-means clustering algorithm,” IEEE Transactions on Knowledge and Data Engineering, vol. 24, no. 6, pp. 1146-1150, 2012.
-
[4] Buciu I., and Gacsadi A., “Directional features for automatic tumor classification of mammogram images,” Biomedical Signal Processing and Control, vol. 6, no. 4, pp. 370-378, 2011.
-
[5] Hamarneh G., and Li X., “Watershed segmentation using prior shape and appearance knowledge,” Image and Vision Computing, vol. 27, no. 1, pp. 59-68, 2009.
-
[6] Ibrahim S. I. Abuhaiba,Ruba A. A. Salamah,"Efficient Global and Region Content Based Image Retrieval", IJIGSP,vol.4,no.5,pp.38-46,2012.
-
[7] Kassner A., and Thornhill R. E., “Texture analysis: a review of neurologic MR imaging applications,” American Journal of Neuroradiology, vol. 31, no. 5, pp. 809-816, 2010.
-
[8] Likas A., Vlassis N., and Verbeek J. J., “The global k-means clustering algorithm,” Pattern recognition, vol. 36, no. 2, pp. 451-461, 2003.
-
[9] Linde Y., Buzo A., and Gray R., “An algorithm for vector quantizer design,” IEEE Transactions on
Communications, vol. 28, no. 1, pp. 84-95, 1980.
“Medical image segmentation using geometric active contour model based on level set method,” In
Proceedings of the IEEE Pacific Rim Conference on communications, computers and signal processing,
Victoria, BC, pp. 577-580, 2007.
-
[16] Ng H. P., Ong S. H., Foong K. W. C., Goh P. S., and Nowinski W. L., “Medical image segmentation using k-means clustering and improved watershed algorithm,” In Proceedings of the IEEE Southwest symposium on image analysis and interpretation, Denver, CO, pp. 61-65, 2006.
-
[17] Prasad B. G., and Krishna A. N., “Statistical texture feature-based retrieval and performance evaluation of CT brain images,” In Proceedings of the 3rd international conference on electronics computer technology, Kanyakumari, India, pp. 289-293, 2011.
-
[18] Quellec G., Lamard M., Cazuguel G., Roux C., and Cochener B., “Case retrieval in medical databases by
fusing heterogeneous information,” IEEE Transactions on Medical Imaging, vol. 30, no. 1, pp. 108-118, 2011.
-
[19] Rajkumar K., and Muttan S., “Medical image retrieval using energy efficient wavelet transform,” In Proceedings of the 2010 international conference on computing, communication and network technologies (ICCCNT), Karur, pp. 1-5, 2010.
-
[20] Sanjay S., and Trimbak S., “An Effective Mechanism to Neutralize the Semantic Gap in Content Based Image Retrieval (CBIR),” The International Arab Journal of Information Technology, vol. 11, no. 2, pp. 124-133, 2014
-
[21] Shahabi C., and Safar M., “An experimental study of alternative shape-based image retrieval techniques,” Multimedia Tools and Applications, vol. 32, no. 1, pp. 2948, 2007.
-
[22] Stoitsis J., Valavanis I., Mougiakakou S. G., Golemati S., Nikita A., and Nikita K.S., “Computer aided diagnosis based on medical image processing and artificial intelligence methods,” Nuclear Instruments and Methods in Physics Research Section A: Accelerators,
Spectrometers, Detectors and Associated Equipment, vol. 569, no. 2, pp. 591-595, 2006.
-
[23] Trpovski Z., “Content based image retrieval: from pixels to semantics,” In Proceedings of the 9th symposium on neural network applications in electrical engineering, NEUREL, Belgrade, pp. 7-12, 2008.
-
[24] World Health Organization Cancer Fact Sheets, Available at http://www.who.int/mediacentre/factsheets/fs297/en/ind ex.html, Accessed April 15, 2014.
Список литературы An Efficient System for Medical Image Retrieval using Generalized Gamma Distribution
- Ahmad Fauzi M. F., and Ahmad W., "Efficient block based matching for content based retrieval of CT head images," In Proceedings of the IEEE 10th workshop on multimedia signal processing, Cairns, Qld, pp. 142-147, 2008.
- Babu G. P., and Murty M. N., "A near-optimal initial seed value selection in k-means means algorithm using a genetic algorithm," Pattern Recognition Letters, vol. 14, no. 10, pp. 763-769, 1993.
- Bishnu P. S., and Bhattacherjee V., "Software fault prediction using quad tree-based K-means clustering algorithm," IEEE Transactions on Knowledge and Data Engineering, vol. 24, no. 6, pp. 1146-1150, 2012.
- Buciu I., and Gacsadi A., "Directional features for automatic tumor classification of mammogram images," Biomedical Signal Processing and Control, vol. 6, no. 4, pp. 370-378, 2011.
- Hamarneh G., and Li X., "Watershed segmentation using prior shape and appearance knowledge," Image and Vision Computing, vol. 27, no. 1, pp. 59-68, 2009.
- Ibrahim S. I. Abuhaiba,Ruba A. A. Salamah,"Efficient Global and Region Content Based Image Retrieval", IJIGSP,vol.4,no.5,pp.38-46,2012.
- Kassner A., and Thornhill R. E., "Texture analysis: a review of neurologic MR imaging applications," American Journal of Neuroradiology, vol. 31, no. 5, pp. 809-816, 2010.
- Likas A., Vlassis N., and Verbeek J. J., "The global k-means clustering algorithm," Pattern recognition, vol. 36, no. 2, pp. 451-461, 2003.
- Linde Y., Buzo A., and Gray R., "An algorithm for vector quantizer design," IEEE Transactions on Communications, vol. 28, no. 1, pp. 84-95, 1980.
- Li-xin S., Rui-feng C., and Qian W., "Image retrieval of calcification clusters in mammogram using feature fusion and relevance feedback," In Proceedings of the international forum on strategic technology, pp 15-18, 2010.
- Lu G., "Techniques and data structures for efficient multimedia retrieval based on similarity," IEEE Transactions on Multimedia, vol. 4, no. 3, pp. 372-384, 2002.
- Madhusudhana Rao T. V., Pallam Setty S., and Srinivas Y., "Content based Image Retrievals for Brain Related Diseases," International Journal of Computer Applications, vol. 85, no. 11, pp. 0975-8887, 2014.
- Mohamed M. Fouad,"Content-based Search for Image Retrieval", IJIGSP, vol.5, no.11, pp.46-52, 2013.DOI: 10.5815/ijigsp.2013.11.05
- Muller H., Michoux N., Bandon D., and Geissbuhler, A., "A review of content-based image retrieval systems in medical applications clinical benefits and future directions," International journal of medical informatics, vol. 73, no. 1, pp. 1-23, 2004.
- MyungEun L., SoonYoung P., and WanHyun C., "Medical image segmentation using geometric active contour model based on level set method," In Proceedings of the IEEE Pacific Rim Conference on communications, computers and signal processing, Victoria, BC, pp. 577-580, 2007.
- Ng H. P., Ong S. H., Foong K. W. C., Goh P. S., and Nowinski W. L., "Medical image segmentation using k-means clustering and improved watershed algorithm," In Proceedings of the IEEE Southwest symposium on image analysis and interpretation, Denver, CO, pp. 61-65, 2006.
- Prasad B. G., and Krishna A. N., "Statistical texture feature-based retrieval and performance evaluation of CT brain images," In Proceedings of the 3rd international conference on electronics computer technology, Kanyakumari, India, pp. 289-293, 2011.
- Quellec G., Lamard M., Cazuguel G., Roux C., and Cochener B., "Case retrieval in medical databases by fusing heterogeneous information," IEEE Transactions on Medical Imaging, vol. 30, no. 1, pp. 108-118, 2011.
- Rajkumar K., and Muttan S., "Medical image retrieval using energy efficient wavelet transform," In Proceedings of the 2010 international conference on computing, communication and network technologies (ICCCNT), Karur, pp. 1-5, 2010.
- Sanjay S., and Trimbak S., "An Effective Mechanism to Neutralize the Semantic Gap in Content Based Image Retrieval (CBIR)," The International Arab Journal of Information Technology, vol. 11, no. 2, pp. 124-133, 2014
- Shahabi C., and Safar M., "An experimental study of alternative shape-based image retrieval techniques," Multimedia Tools and Applications, vol. 32, no. 1, pp. 29-48, 2007.
- Stoitsis J., Valavanis I., Mougiakakou S. G., Golemati S., Nikita A., and Nikita K.S., "Computer aided diagnosis based on medical image processing and artificial intelligence methods," Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment, vol. 569, no. 2, pp. 591-595, 2006.
- Trpovski Z., "Content based image retrieval: from pixels to semantics," In Proceedings of the 9th symposium on neural network applications in electrical engineering, NEUREL, Belgrade, pp. 7-12, 2008.
- World Health Organization Cancer Fact Sheets, Available athttp://www.who.int/mediacentre/factsheets/fs297/en/index.html, Accessed April 15, 2014.


