Application of proteomic method in analysis of profile of protein expression in CSFV-infected PK-15 cells
Автор: Jinfu Sun, Li Geng
Журнал: International Journal of Image, Graphics and Signal Processing(IJIGSP) @ijigsp
Статья в выпуске: 5 vol.3, 2011 года.
Бесплатный доступ
Proteomic analysis is a powerful technology to enhance our insight into the pathogenesis, biomarkers and prevention of disease. Two-dimensional polyacrylamide gel electrophoresis (2-DE) is an important proteomics tool, where thousands of protein spots can be visualized, resulting in a global view of the state of a proteome. Viral infection will modify the patterns of host cell protein expression, which can affect the normal physiological function of host cell and determine viral pathogenic progress and consequence. To uncover host cellular responses in the early stage of classical swine fever virus infection, a proteomic analysis was conducted using 2DE followed by MALDI-TOF-TOF identification. Altered expression of 21 protein spots in infected pk-15 cells at 24 h p.i. were identified in 2D gels, with 13 of these being characterized by MALDI-TOF-MS/MS. These proteins function in cytoskeletal, energy metabolism, nucleic acid/processing, and cellular stress. The expression alteration of these proteins presents the changes in physiological functions of host cells and provides a clue for further understanding of the mechanisms of CSFV infection and pathogenesis.
Classical swine fever virus, 2-DE, Proteome, viral pathogenesis
Короткий адрес: https://sciup.org/15012172
IDR: 15012172
Текст научной статьи Application of proteomic method in analysis of profile of protein expression in CSFV-infected PK-15 cells
Published Online August 2011 in MECS
I . INTRDUCTION
Proteomic analysis is a powerful technology to enhance our insight into the pathogenesis, biomarkers and prevention of disease[1]. Two-dimensional polyacrylamide gel electrophoresis (2-DE) is an important proteomics tool, where thousands of protein spots can be visualized, resulting in a global view of the state of a proteome. By comparing the 2-D spot patterns from different samples, changes in individual proteins can be detected and quantified. This allows identification of protein markers that are characteristic of a specific physiological or pathological state of a cell or tissue [2].
Viral invasion will modify the patterns of host cell protein expression, which may affect the normal physiological function of host cell and determines viral pathogenic progress and consequence. Therefor, studies on viral infections proteomics contributes to uncover the mechanism of interaction between virus and host and viral molecular pathogenesis. In this study, a proteomic approach of 2-dimensional polyacrylamide gel electrophoresis (2-DE) was used to comparing protein expression patterns of normal and Classical swine fever virus (CSFV) infected cells, and obtained exclusive information about how host cells respond to CSFV infection.
CSFV is a small, enveloped virus belonging to the genus Pestivirus within the family Flaviviridae [3]. CSFV genome is a single positive-stranded RNA about 12.5 kb in length, with a single large open reading frame encoding a polyprotein that is cleaved into into 12 known proteins by cellular and viral proteases[4].
CSFV causes classical swine fever (CSF) that is a highly contagious swine disease characterized by fever, leukopenia, and hemorrhage. CSF is a notifiable one of the World Organization for Animal Health (OIE), usually results in high morbidity and mortality of infected swine and leads to substantial economic losses to the pig industry worldwide. Diseased animals show fever, anorexia, diarrhoea and haemorrhages in the skin, mucosa and internal organs [5,6]. During acute CSF, pigs show general immunosuppression featuring a dramatic decrease of peripheral B- and T-cells [7,8]. Moreover,
∗ This work is supported by Ph.D Initiatory Fund of Liaoning Province (grant No.20091028 ) and Fundamental Research Funds for the Central Universities (grant No.100405013 ).
granulocytopenia and induction of apoptosis of bone marrow haematopoietic cells were observed [9].
Some studies have attributed immune cell apoptosis or death to cytokines released from monocytes/macrophages activated by CSFV infection[10,11]. Bensaude et al. have shown that proinflammatory induced by CSFV infection disrupt the haemostatic balance and lead to the coagulation and thrombosis seen in acute disease[12]. Apart from these, other factor that contribute to the pathogenesis and progression of CSF is little known.
To furthly uncover the pathogenesis of CSFV, we analysed proteomic alteration in CSFV-infected PK-15 cells at 24 h p.i.. A total of 13 differentially expressed proteins have been identified. These differentially expressed proteins reflect changes in physiological functions of host cells and posses rich information concerning CSFV pathogenesis.
-
II. MATERIAL AND METHODS
-
A. CSFV Infection and Detection of Infected PK-15 Cells
Monolayers of PK-15 cells were infected with blood stock of the CSFV strain Shimen (106.9 TCID50/mL) at an input multiplicity of about 2 TCID 50 /cell. At 24 h p.i., cells were washed with PBS, fixed with 80% cold acetone in PBS at -20 ºC for 30 min and washed again with PBS. The cells were then incubated with pig anti-CSFV hyperimmune serum at 37 ºC for 1 h, washed four times with PBST (0.5% Tween-20 in PBS, pH7.4), and further incubated with rabbit anti-pig IgG conjugated with fluorescein isothocyanate (FITC; Sigma) at 37 ºC for 1 h in the dark. After four washes with PBST, CSFV infection of PK-15 cells were confirmed by fluorescence microscopy.
-
B. Sample Preparation for 2-DE
The culture media were removed and infected cell monolayers were harvested by addition of lysing solution (8 M urea, 4% CHAPS, 40mM Tris, 0.5% IPG buffer).Uninfected control PK-15 cells were treated in parallel in the same way. Protein concentration was measured with the Coomassie Plus-The Better BradfordTM Assay Kit (Amersham Biosciences, USA).
-
C. 2-DE
2-DE was performed using precast IPG strips [pH 3–10 NL (non-linear), 17 cm length; Bio-Rad, USA] in the first dimension (IEF). The IPG strips were rehydrated with 350 µL rehydration buffer (8 M urea, 2% CHAPS, 18 mM DTT, 0.5% IPG buffer pH 3–11 NL, and minute amounts of bromophenol blue) containing of protein samples 150 µg for silver stain, 700 µg for CBB stain. Active rehydration was achieved by applying 50 V for 16 h. IEF was carried out at 20 °C in the Protean IEF Cell (Bio-Rad, USA), with the current limited to 50 mA/strip and voltage program as: 250 V constant for 1 h,500 V constant for 1 h, 1000 V constant for 1 h, linear ramp to 8000 V over 1 h, then 8000 V constant for a total of 80 000 Vh. After IEF, the IPG strips were equilibrated by soaking first for 15 min in 50 mM Tris-HCl, pH 8.8, 6 M urea, 2% SDS, 30% glycerol, 2% w/v DTT and a trace of bromophenol blue, then for 15 min in the same solution, but containing 2.5% w/v iodoacetamide instead of DTT. The second dimensional separation was performed using 12.5% homogeneous SDS polyacrylamide gels.
-
D. Gel Staining
The gels were fixed in 50% methanol and 5% acetic acid in water for 30 min, and then washed in water for 1 hour and incubated in 0.02% sodium thiosulfate solution for 2 min. After washing twice in water for 10 sec each, the gels were stained in 0.1% silver nitrate solution for 30 min. Color development was allowed for 15 min in 2% sodium carbonate/ 0.04% formaldehyde. EDTA (1.46%) was used to stop the development and the stained gels were washed 3 times in water for 3 min each. CBB (G250, sigma, USA) staining was followed the procedure reported by Mathesius et al .[13].
-
E. Image Acquisition And Analysis
The silver stained gels were scanned in an ImageScanner (Amersham Biosciences, USA) operated by the LabScan 3.00 software (Amersham Biosciences, USA). Image analysis was carried out using PDQuestTM software 7.1(Bio-Rad, USA). Image spots were initially detected, matched and then manually edited. Intensity levels of the spots were normalized by expressing the intensity of each protein spot in a 2- DE gel as a proportion of the total protein intensity detected for the entire gel. Normalized protein spot volume (area x stain intensity) was calculated for each gel and used for statistical calculation of protein expression level, with the resulting spot volume percentage being used for comparison. Student’s t -test was performed for relative comparison of intensities between CSFV-infected and control groups at 24 h p.i. Only spots with infected/uninfected expression intensity ratios of >1.5 or <0.67 were selected as showing significant alteration (p ≤ 0.05) for analysis by mass spectrometry.
-
F. Protein Identification
Protein spots of interest were excised and transferred into 1.5 mL Eppendorf tubes. Spots were destained twice for 30 min at 37 °C with 200 mM NH 4 HCO3/40% acetonitrile (ACN) and dehydrated for 5 min in pure ACN. Then, spots were rehydrated with 25 mM NH 4 HCO 3 for 5 min and dehydrated again for 10 min in pure ACN. The last rehydration and dehydration step was repeated 3 times, following which the gel plugs were dried in a SpeedVac, then rehydrated in a minimal volume of trypsin solution (10 µg/mL in 25 mM NH4HCO3) and incubated at 37ºC overnight. Each supernatant was processed for MALDI-TOF-MS/MS analysis by direct application onto the sample plate with equal amounts of matrix. If necessary, the in-gel digests were extracted subsequently with 50% and 80% ACN, and then concentrated and desalted by ZipTips (Millipore, Bedford, MA, USA) prior to application on the sample plate. Peptides of each sample were analyzed in a 4700 Proteomics Station (Applied Biosystems, USA) in automatic mode. Samples were analyzed in the m/z range 800 to 4000, with an accelerating voltage of 20 kV. The parent ions with high intensity were selected and fragmented using CID. Spectra were internally calibrated with peptides from trypsin autolysis. Proteins were identified by peptide mass fingerprint (PMF) and confirmed by MS/MS analysis of two peptides in each sample. GPS Explore software (Applied Biosystems, USA) based on MASCOT searching engine (Matrixscience, UK) was used for protein identification in the NCBI non-redundant mammalian database. A maximum of one missed cleavage per peptide was allowed, a mass tolerance of 0.3 Da, and MS/MS tolerance of 0.4 Da were used. Protein identifications with a score value over 60 were positively assigned.
Ш. RESULT
-
A. CSFV Infection in PK-15 Cells
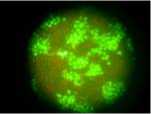
Since CSFV does not produce cytopathologic effects (CPE) in PK-15 cells, viral infection was confirmed by detection of CSFV antigen using IFA at 24 h p.i.. The results showed intense green staining in CSFV-infected PK-15 cells (see figure 1), while the mock controls showed a negative result, which indicated that CSFV infection in PK-15 cells was successful. Thereby, these cells were used for proteomic analysis.
-
B. Protein Expression Profile of CSFV-infected And Uninfected PK-15 cell
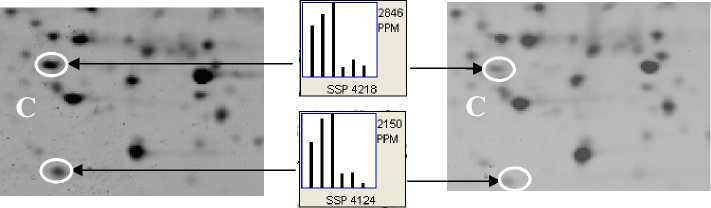
Protein profiles of CSFV-infected and uninfected PK-15 cells were analyzed by 2-DE with 150 μg protein loaded on each gel strip. To minimize inter-gel and inter-samples variation, 2 repeats of independent sample preparations and 2 repeats of independent 2-DE were performed under identical conditions. All gels provided high resolution in the separation of proteins, with each showing more than 1300 protein spots (see figure 2). On the stained gels, protein spot volume comparisons were made between infected and uninfected cell samples with the assistance of PDQuestTM software. A total 21 protein spots with at least 1.5-fold quantitative alteration (t test p <0.05) in CSFV-infected and uninfected PK-15 cells were identified on 2D gels at 24 h p.i.. A magnified comparison of the differentially expressed protein pattern of two spots ( ssp 4218 and ssp 4124) is shown in Fig. 2C.
-
C. Identification of Differentially Expressed Proteins
Protein spots with at least a 1.5-fold alteration (increase or decrease) were cut out and subjected to MALDI-TOF-MS/MS analysis. Of 21 differentially expressed protein spots identified above, 13 were unambiguously identified by their PMF and peptide sequences in the NCBInr database. The remaining 8 proteins spots were not identified successfully, either because the quantity was too low to produce a good spectrum or because the confidence of database searching through PMF or the data from MS/MS were insufficient to yield unambiguous results.
W. DISCUSSION
Proteomic analysis has provided a key approach to understand the molecular mechanisms of how the host reaction to viral invasion is associated with viral pathogenesis. Proteomic methods has revolutionized our ability to assess protein interactions and cellular changes on a global scale, allowing the discovery of previously unknown connections. Virus-induced changes in the cellular proteome may be assessed by comparing protein profiles before and after viral infection by two general approaches: 2D gel electrophoresis and quantitative mass spectrometry techniques. 2D gel methods also enable the detection of altered forms of a protein, including posttranslational modifications, which could have major effects on the function of the cellular protein. 2D gel electrophoresis has been used to determine changes in the cellular proteome upon infection by some different viruses, where protein spots that differ before and after infection are excised and identified by mass spectrometry [14]
пгю
Figrue 1. Detection of CSFV-infected PK-15 cells with indirect immune fluorescent staining at 24 h p.i.. Monolayers of PK-15 cells were infected with blood stock of the CSFV strain Shimen (106.9 TCID 50 /mL) at an input multiplicity of about 2 TCID 50 /cell , after incubated for 24 h at 37 °C, the CSFV infection was confirmed by detection of CSFV antigen using indirected immunofluorescence method. Each image is a representative of one of three independent experiments. Images were taken with an original magnification of 200 x .
PI 3 IEF 10
-------------------------►
PI 3 IEF 10
>
MM (kDa)


control CSFV
Figrue 2. Representative high resolution 2-DE images showing protein spots expressed in control (A) and CSFV-infected (B) PK-15 cells at 24 h p.i. .More than 1300 spots were detected in each gel. Equal amounts of total protein (150 µg) from infected and uninfected PK-15 cell lysates were resolved by 2DE. The protein spots were visualized by silver staining. (C) Magnified comparison maps of protein spots(ssp 4218 and ssp 4124) with arrow in above 2-DE gels. (MM, molecular mass; kDa, kilodalton)
TABLE 1. PROTEINS SHOWING AT LEAST 1.5-FOLD QUANTITATIVE DIFFERENCES IN EXPRESSION INCSFV-INFECTED
PK-15 CELLS BY 2-DE ANALYSIS
|
Protein description |
GI no. |
Pep. hitsa |
Unique Pep.b |
Functionc |
Ratio valued |
|
Cytoskeletal protein |
|||||
|
Beta-actin |
gi|14250401 |
16 |
12 |
involved in various types of cell motility |
0.39 |
|
Keratin,type II cytoskeletal 1 |
gi|1346343 |
17 |
13 |
Intermediate filament family |
0.46 |
|
Metabolic enzyme |
|||||
|
Bovine heart cytochrome c oxidase subunit |
gi|40889866 |
8 |
4 |
energy metabolism |
0.38 |
|
Isocitrate dehydrogenase [NAD+] subunit alpha |
gi|18250284 |
21 |
13 |
energy metabolism |
1.92 |
|
Stress Protein |
|||||
|
Thioredoxin 1 |
gi|6755911 |
9 |
5 |
disulfide reductase activity |
1.79 |
|
Putative 42-9-9 protein |
gi|15216177 |
7 |
4 |
disulfide reductase activity |
3.94 |
|
Peroxiredoxin 3 |
gi|2507170 |
8 |
4 |
redox regulation |
0.64 |
|
Heat shock protein 27 |
gi|55926209 |
13 |
7 |
molecular chaperoning, stress response |
0.2 |
|
nucleic acid /protein processing |
|||||
|
Splicing factor, arginine/serine-rich 3 |
gi|28629815 |
7 |
4 |
RNA processing |
0.35 |
|
Translational elongation factor 1 delta |
gi|33333168 |
26 |
15 |
translational elongation |
5.32 |
|
CIP29 protein |
gi|62202489 |
11 |
6 |
nucleic acids metabolism, cell growth |
0.19 |
|
ERp57 |
gi|62549200 |
8 |
4 |
protein disulfide isomerase family |
4.67 |
|
Peptidylprolyl isomerase |
gi|108463 |
7 |
4 |
accelerate protein folding |
0.4 |
a) The total number of peptides identified for the listed protein based on the MS/MS data acquired and the MASCOT searching criteria. b) The number of peptides for the given protein that have a unique amino acid sequence and is a subset of the total number of peptides identified. c) Protein function was retrieved from the Swiss-Prot and GenBank databases or relevant references. d) Levels of protein expression in infected cells to that of uninfected cells.
Mass spectrometric methods and automation of a large part of the process including robotics application have continued to improve dramatically in recent years, allowing both increased sensitivity and higher throughput. Improved software and databases containing different species genes—known or putative—are also now available, allowing automated data processing of the large volume of acquired mass spectra[15].
In present study, we have provided proteomic profile of host cells (PK-15) in early stage of CSFV infection. To identify unambiguous differences in all infected PK-15 cells and to ensure reliability of the proteomic data, two independent experiment including sample preparation and 2-DE were conducted. Each gel was loaded with a sample from a bottle of independent infected cells or control cells, and only spots that were: (i) consistently up- or down-expressed, or consistently present or absent in all 6 gels of infected or control cells; and (ii) with an alteration (infected/control) of ≥ 1.5 fold, with a value in Student’s t-test of p ≤ 0.05, were selected for further MALDI-TOF-MS/MS analysis. In this manner, of 21 protein spots, 13 differentially expressed proteins were identified by MALDI-TOF-TOF. These identified proteins play crucial roles involving in cytoskeletal, cellular metabolism, cellular stress and nucleic acid /protein processing (Table 1). Their function in response to CSFV infection may be as follows.
In the infected PK-15 cells, 2 cytoskeletal proteins of beta-actin and keratin are remarkably decreased (Table 1), which may result cytoskeletal remodeling. Studies demonstrated that various viruses manipulated and utilized the host cytoskeleton to achieve its infectious life cycle [16,17]. Thereby, change of two cytoskeletal proteins implicated that CSFV may also manipulated cytoskeletal network of target cells to facilitate its replication cycles.
Among of two differentially expressed energy metabolic enzymes, Cytochrome c oxidase is the rate-limiting step of the mitochondrial electron transport chain and represents a molecular switch that induces apoptosis under energy stress conditions [18]. Decreased cytochrome c oxidase subunit I expression correlated with apoptosis resistance in the nonneoplastic mucosa of patients with colon cancer [19]. Downregulation of bovine heart cytochrome c oxidase subunit I in CSFV-infected PK-15 cells may contribute to improving cell apoptosis resistance. The other metabolic enzyme of isocitrate dehydrogenase [NAD+] subunit alpha catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the citric acid cycle, its upregulation may be the result of host cells energy stress induced by CSFV infection.
In infected samples, four stess proteins were alterd in expression level. Two thioredoxins of putative 42-9-9 protein and thioredoxin 1 were increased. Thioredoxins can scaveng reactive oxygen species (ROS) both in vivo and in vitro . It has been reported that Thioredoxin 1reduces disulfide bonds and thus regulates the activity of transcriptional factors, such as AP-1, NF-κB, and p53 and overexpression of Trx-1 inhibits apoptosis and enhances cell growth[20]. Therefor, upregulation of two thioredoxins may contribute to PK-15 cell persistent growth, other than apoptosis. Contrary to which, the other two stress proteins of peroxiredoxin 3(Prdx3) and heat shock protein 27 (HSP27) were downregulated. Prdx3 is a thioredoxin-dependent peroxide reductase, which was shown to be able to protect against both oxidative and nitrosative stress. Its overexpression contributes to resistance to the increase in H2O2 and apoptosis caused by hypoxia [21]. HSP27 is a stress-inducible ubiquitous cellular protein and is involved in thermotolerance, cellular proliferation and apoptosis, and molecular chaperoning. It has been found to be up-regulated during certain viral infections. Downregulation of these two stress proteins and upregulation of two thioredoxins may be a specific mechanism of CSFV directing host cells counteracting oxidative stress.
Significantly, translational elongation factor 1 delta (EF-1 δ ) was remarkable upregulated in infected samples. Reports have shown that host eukaryotic translation elongation factors interact with several viral protein or RNA and played critical role in viral replication and pathogenesis, such as West Nile virus[22], dengue 4 virus [23], hepatitis C virus[24] and bovine viral diarrhea virus (BVDV) [25]. These viruses belong family Flaviviridae , specifically, CSFV and BVDV belong to the genus Pestivirus, having extremely similar genomic organization and patterns of replication and assembly as well as cross-antigenicity. Therefore, up-regulation of
EF-1 δ in earlier period of CSFV infection implicates that it may be recruited by the viral replication machinery for CSFV mRNA translation and protein synthesis in a similar way as BVDV does.
CIP29 (Hcc-1), a apoptosis-relevant proteins, is involved in nucleic acid metabolism, cell growth regulation and induction of apoptosis. CIP29 has multiple functions in regulation of cell fate and appears to inhibit cell proliferation by inducing G2/M arrest or by inducing apoptosis without arresting cells in G2/M [26,27]. A large decrease in CIP29 expression in CSFV-infected cells (5.2-fold downregulation) may contribute to the survival of infected cells and to viral persistence, which is in agreement with the observation that CSFV does not cause cytopathic effect or cell death of PK-15 cells during infection.
ERp57 is a member of endoplasmic reticulum chaperone proteins, catalyzes the rearrangement of -S-S-bonds in proteins and functions in endoplasmic reticulum quality control of newly synthesized proteins. It has been also report that ERp57 facilitates polyomavirus infection [28]. Upregulation of this protein in CSFV-infected samples might be related to an increase in protein synthesis or to a mechanism to refold/renature the misfolded/denatured proteins resulting by oxidative and cellular stress.
In summary, the present study provides proteomic profile of host cell in the early stage of CSFV infection by using proteomic approach of 2-DE and MS identification. Although the roles of altered host protein expression in viral replication and pathogenesis are not given and there are no data showing specific proteins associated with a viral infection, with the help of functional analysis of host proteins the proteomic analysis will be able to identify those proteins or specific biomarkers associated with viral replication and pathogenesis. This work has provided an essential basis for further understanding of the pathogenesis of CSFV and the development of new methods for the diagnosis and therapy of CSF infection.
Список литературы Application of proteomic method in analysis of profile of protein expression in CSFV-infected PK-15 cells
- S. Hanash, Disease proteomics. Nature, 422 (6928): 226-232, 2003.
- A. Alban, David S. O., L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, I. Currie, A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 3(1): 36-44, 2003.
- F. X. Heinz, M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, R. J. M. Houghton, Moormann, C. M. Rice, H. J. Thiel, Family Flaviridae. In: C. M. Fauquet, M. Mayo, J. Maniloff, U. Desselberger, L. A. Ball, (Eds.), Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses, Academic Press, San Diego, 2004, 981-998.
- T. Rumenapf, G. Unger, J. H. Strauss, H. J. Thiel, Processing of the envelope glycoproteins of pestiviruses. J. Virol. 67, 3288-3294, 1993.
- H..J. Thiel, P.G.W. Plagemann, V. Moennig, Pestiviruses. In Fields, Virology, 3rd edn. D. M. Fields, P. M. Knipe & Howley. Philadelphia: Lippincott–Raven. 1996.
- V. Moennig, P.G.W. Plagemann, The pestiviruses. Adv. Virus Res , 41: 53–98, 1992.
- M. Susa, M. König, A. Saalmüller, M. J. Reddehase, H. J. Thiel, Pathogenesis of classical swine fever: B-lymphocyte deficiency caused by hog cholera virus. J Virol. 66: 1171-1176, 1992.
- A. Summerfield, S. M. Knötig, K. C. McCullough,Lymphocyte apoptosis during classical swine fever: implication of activation-induced cell death. J Virol. 72: 1853–1861, 1998a.
- A. Summerfield, M. A. Hofmann, K. C. McCullough, Low density blood granulocytic cells induced during classical swine fever are targets for virus infection. Vet Immunol Immunopathol 63, 289–301, 1998b.
- C. Choi, K.K. Hwang, C. Chae, Classical swine fever virus induces tumor necrosis factor-alpha and lymphocyte apoptosis. Arch Virol, 149, 875-889, 2004.
- P.J. Sánchez-Cordón, Núñez A, F.J. Salguero, M. Pedrera, M. Fernández de Marco, J.C. Gómez-Villamandos, Lymphocyte apoptosis and thrombocytopenia in spleen during classical swine fever: role of macrophages and cytokines. Vet Pathol, 42, 477-488, 2005.
- E. Bensaude, J.L. Turner, P.R. Wakeley, D.A. Sweetman, C. Pardieu, T.W. Drew, et al., Classical swine fever virus induces proinflammatory cytokines and tissue factor expression and inhibits apoptosis and interferon synthesis during the establishment of long-term infection of porcine vascular endothelial cells. J Gen Virol, 85,1029-1037, 2004.
- U. Mathesius, G. Keijzers, S. H. Natera, J. J. Weinman, M. A. Diordjevic, B. G. Rolfe, Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics, 1: 1424–1440, 2001.
- K. L. Maxwell, L. Frappier, Viral proteomics. Microbiol Mol Biol Rev. 71(2): 398-411, 2007.
- X. S. Jiang, J. Dai, Q. H. Sheng, L. Zhang, Q. C. Xia, J. R. Wu, R. Zeng, A comparative proteomic strategy for subcellular proteome research: ICAT approach coupled with bioinformatics prediction to ascertain rat liver mitochondrial proteins and indication of mitochondrial localization for catalase. Mol Cell Proteomics, (1): 12-34, 2005.
- G. A. Smith, L.W. Enquist, Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol., 18: 136-61, 2002.
- K. Döhner, B.Sodeik, The role of the cytoskeleton during viral infection. Curr. Top Microbiol. Immunol., 285: 67-108, 2005.
- B. Adenbach, S. Arnold, I. Lee, et al. The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochim. Biophys. Acta, 1655: 400–8, 2004.
- C.M. Payne, H. Holubec, C. Bernstein, H. Bernstein, K. Dvorak, S.B. Green, M. Wilson, M. Dall'Agnol, B. Dvorakova, J. Warneke, H. Garewal. Crypt-restricted loss and decreased protein expression of cytochrome C oxidase subunit I as potential hypothesis-driven biomarkers of colon cancer risk. Cancer Epidemiol Biomarkers Prrev. 14(9): 2066-2075, 2005.
- G. Powis, D.L. Kirkpatrick, Thioredoxin signaling as a target for cancer therapy. Curr Opin Pharmacol., 7(4): 392–397, 2007.
- L. Nonn, M. Berggren, G. Powis, Increased expression of mitochondrial peroxiredoxin-3 (thioredoxin peroxidase-2) protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis. Mol. Cancer Res., 1(9): 682-689, 2003.
- Davis, W. G; Blackwell, J. L; Shi, P. Y; Brinton, M. A. Interaction between the cellular protein eEF1A and the 3'-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81 (18): 10172-10187, 2007.
- M. De Nova-Ocampo, N. Villegas-Sepúlveda, R. M. del Angel, Translation elongation factor-1α, La, and PTB interact with the 3' untranslated region of dengue 4 virus RNA. Virology, 295 (2): 337-347, 2002.
- Y. H. Kou, S. M. Chou, Y. M. Wang, Y. T. Chang, S. Y. Huang, M. Y. Jung, Y. H. Huang, M. R. Chen, M. F. Chang, S. C. Chang, Hepatitis C virus NS4A inhibits cap-dependent and the viral IRES-mediated translation through interacting with eukaryotic elongation factor 1A. J. Biomed. Sci., 13 (6): 861-874, 2006.
- C. M. Johnson, D. R. Perez, R. French, W. C. Merrick, R. O.Donis, The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J Gen Virol, 82: 2935–2943, 2001.
- C. L. Leaw, Ren, M. L. Choong, Hcc-1 is a novel component of the nuclear matrix with growth inhibitory function. Cell. Mol. Life Sci. 61: 2264-73, 2004.
- S. Fukuda, L.M. Pelus, Growth inhibitory effect of Hcc-1/CIP29 is associated with induction of apoptosis, not just with G2/M arrest. Cell Mol. Life Sci., 62: 1526-1527, 2006.
- C. P. Walczak, B. Tsai, A PDI family network acts distinctly and coordinately with ERp29 to facilitate polyomavirus infection. J. Virol. 85(5): 2386-2396, 2011.


