Application of Sparse Coded SIFT Features for Classification of Plant Images
Автор: Suchit Purohit, Savita R. Gandhi
Журнал: International Journal of Image, Graphics and Signal Processing(IJIGSP) @ijigsp
Статья в выпуске: 10 vol.9, 2017 года.
Бесплатный доступ
Automated system for plant species recognition is need of today since manual taxonomy is cumbersome, tedious, time consuming, expensive and suffers from perceptual biasness as well as taxonomic impediment. Availability of digitized databases with high resolution plant images annotated with metadata like date and time, lat long information has increased the interest in development of automated systems for plant taxonomy. Most of the approaches work only on a particular organ of the plant like leaf, bark or flowers and utilize only contextual information stored in the image which is time dependent whereas other metadata associated should also be considered. Motivated from the need of automation of plant species recognition and availability of digital databases of plants, we propose an image based identification of species of plant when the image may belong to different plant parts such as leaf, stem or flower, fruit , scanned leaf, branch and the entire plant. Besides using image content, our system also uses metadata associated with images like latitude, longitude and date of capturing to ease the identification process and obtain more accurate results. For a given image of plant and associated metadata, the system recognizes the species of the given plant image and produces an output that contains the Family, Genus, and Species name. Different methods for recognition of the species are used according to the part of the plant to which the image belongs to. For flower category, fusion of shape, color and texture features are used. For other categories like stem, fruit, leaf and leafscan, sparsely coded SIFT features pooled with Spatial pyramid matching approach is used. The proposed framework is implemented and tested on ImageClef data with 50 different classes of species. Maximum accuracy of 98% is attained in leaf scan sub-category whereas minimum accuracy is achieved in fruit sub-category which is 67.3 %.
SIFT, Sparse Coding, Plant Species, Content based retrievel, Spatial Pyramid matching, HSV color space, Texture fetaures extraction
Короткий адрес: https://sciup.org/15014237
IDR: 15014237
Текст научной статьи Application of Sparse Coded SIFT Features for Classification of Plant Images
Published Online October 2017 in MECS
Plant Taxonomy is a science to separate plants into similar groups based on the characteristics like color of the flower, shape of the flower, leaf shape and form, fruits, bark of the stem etc. Plant recognition when done manually by specialized taxonomists, suffers from perceptual biasness, cost of hiring of experts and shortage of experts (‘‘taxonomic impediment’’) [1]. The process of manual taxonomy becomes time consuming and tedious when more and more images are added to the database. Automation of the recognition process can improve the time, efficiency, accuracy and cost associated with the recognition process. This has given rise to the demand of automatic tools for plant species recognition and classification. Major support system for the automation is availability of digitized databases with high resolution plant images annotated with species names and metadata like date and time, lat long information aided by high resolution cameras available on handheld devices. Royal Botanic Gardens, Kew provides a digital catalogue of over 200,000 high-resolution images, with more being added continuously as part of an ongoing digitization project. Motivated from availability of pictures of plants in digital format and need of automation, we propose an automatic plant classification system using techniques from image processing, computer vision and machine learning fields. The current approaches for plant species identification utilize either only leaf form and shape [2]-[12 or combination of leaf and bark [13] or color, shape and texture of the flowers [14]-[19] to identify the species of the plant. This paper presents a system that identifies the species of the plant based on the information provided by different parts of the plants like leaf, flower, fruit and bark of the stem. The major contributions to this paper are:
-
1. An automatic plant species recognition framework which considers different organs of the plant like leaf, flower, fruit, and bark to identify the species while including seasonal and topographic
-
2. Existing leaf detection approaches utilize global features of leaf like diameter, length, width, area, perimeter, aspect ratio, eccentricity etc. which require domain knowledge. In contrast our proposed approach based on sparsely coded SIFT features automatically generates features and classify the images without knowledge of domain that too with an exceptional accuracy of 98%.
-
3. Sparsely coded SIFT representation approach makes the classification robust in cluttered environment like flower amongst leaves, images with objects of different orientation like horizontal and vertical stem images, images corrupted with noise.
parameters in classification. The uniqueness of the framework is that for different parts of the plants different techniques are used. The system along with the test image identifies the part of the plant through meta data provided with the image and then dynamically selects the technique according to the part of the plant. The appropriate technique according to the particular organ of the plant has been decided after careful and rigorous testing.
-
II. Related Work
Most of the work done on plant species recognition works on leaf images of plant. A number of projectsystems namely Leafsnap in America [2], CLOVER [4] in Asia, Pl@ntNet[5], ReVes[6] and ENVIROFI [7] in Europe are based on leaf identification to recognize the species. These approaches utilize global features of leaves like color ,texture (entropy, energy, contrast) , shape (eccentricity, circularity , aspect ratio, rectangularity) and applied them to different classification approaches to identify the species. The problem with these approaches is that leaves of many plants like decidous trees are not available throughout the year. Moreover the leaves may be too young or may be too distorted or less informative as large banana leaves, needles in pine trees etc. To refine the leaf approach, Zuolin ZHAO proposed the method for recognizing plants by combining leaf and bark features together. The characteristic parameters of leaves and bark were extracted based on background segmentation and filtering method, and plants were recognized using SVM [13]. Zisserman proposed flower classification approach based on combined vocabulary of shape, texture color features. The system was tested on 103 classes [14]-[15]. Wenjing Qi, et al. suggested the idea of flower classification based on local and spatial cues with help of SIFT feature descriptors [16]. Yong Pei and Weiqun Cao provided the application of neural network for performing digital image processing for understanding the regional features based on shape of a flower [17]. Salahuddin et al. proposed an efficient segmentation method which combines color clustering and domain knowledge for extracting flower regions from flower images [18]. D S Guru et al. developed an algorithmic model for automatic flowers classification using KNN as the classifier [19]. But flowers have limitation that it is difficult to analyze shapes and structures of flowers since they have complex 3D structures. Naiara, Javier presented an automatic system for the identification of plants based on the content of images and metadata associated to them. The classification has been defined as a classification plus fusion solution [20].The authors of [21] have combined different views of plant organs (such as flowers, bark, leaves) using a late fusion process for efficient plant classification process.
-
III. Data Set
In this study, ImageCLEF 2015 [40] data is used
which focuses on 1000 herb, tree and fern species centered on France and neighbouring countries. Seven types of image content are considered: scan and scanlike pictures(free from background) of leaf, and 6 kinds of images of different organs of the plant taken from different views like Flower, fruit, stem and bark, branch, leaf and entire view. The data set is built through a crowd sourcing initiative conducted by T e l a Botanica and covers 1000 species [22]. The dataset contains 1,13,204 images provided for training. Each image is associated with an xml file containing contextual meta data described as below:
“ObservationId: Plant observation ID from which several pictures can be associated.
o FileName: Name of the image file with which it is associated.
o Content: Branch, Entire, Flower, Fruit, Leaf,
LeafScan, Stem.
o ClassId: Class number ID that must be used as ground-truth. It is a numerical taxonomical number used by Tela Botanica.
o Species: Species names (containing 3 parts: the Genus name, the Species name, and the author who discovered or revised the name of the species).
o Genus: Name of the Genus, one level above the Speciesin the taxonomical hierarchy used by Tela Botanica.
o Family: Name of the Family, two levels above the Speciesin the taxonomical hierarchy used by Tela Botanica.
o Date: (if available) Date when the plant was observed.
o Vote: (round up) Average of the user ratings on image quality.
o Locality: (if available) Locality name, most of the time a town.
Fig. 1 shows a sample picture of plant. The xml file of the same is as follows:

Fig.1. Sample Image
Due to unavailability of hardware resources, we have used 10 classes for classification purpose with five organs of plant in each class. Since supervised learning has been employed in our proposed framework, more and more images are required for training to achieve better results. So we have chosen top 10 species with maximum number of images provided in training. We have taken 75 images per species for flower, leaf and leafscan category whereas 50 images per class are taken for fruit and stem category. 25 images per class have been used for testing.
-
IV. Methodology
The species recognition process is implemented as a classic Image Classification problem. Image classification algorithms classify the objects based on their visual/semantic content described by features. The features can be global like colour, shape, texture and describe the image as a whole whereas local features describes local patches. Initial researches in Image classification were based on pixel based global features like colour, texture, shape, histogram etc. The global features are found to be affected significantly by various illumination effects, viewing angles, noise and other distortions and it is found that classifiers based on global features lack in accuracy. Hence recent researches are focusing on classification based on local features for quantification of visual information present in the image [24].We have used both global and local features subjective to the category of the plant image belongs to . The choice of features had been inferred through extensive testing of both the approaches for different categories. It was concluded that sparsely coded SIFT features approach gives best accuracy for leaf and leaf scan like images, images of stem/bark and fruits categories whereas colour and texture are additional discriminative features required for flowers. The framework is described in Fig. 2. The next two subsections explain both the approaches in detail.
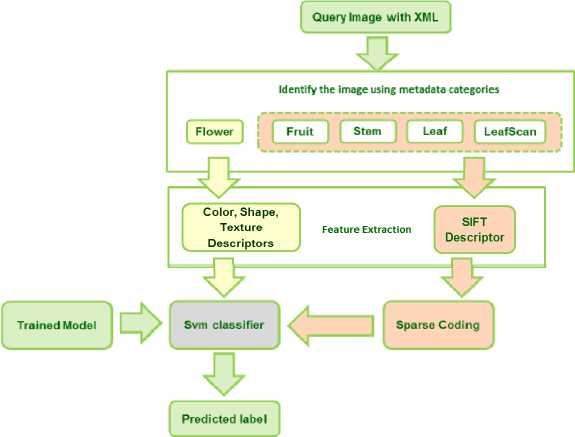
Fig.2. Framework of recognition system
-
A. Sparse coded SIFT feature representation
A good local feature should be easy to extract, distinctive, repetitive, invariant and robust to noise occlusion and clutter. A survey on local features has proved the superiority of a local feature detector and descriptor founded by David Lowe popularly known as known as SIFT or Scale Invariant Feature Transform. SIFT was founded in 1999 and summarized in 2004. “Scale Invariant Feature Transform is a method for extracting distinctive features from images which are invariant to image scale and rotation and provide robust matching in the presence of some affine transformations, change in viewpoint, addition of noise and change in illumination”[23]. SIFT descriptor calculates value of 4X4 grid around feature points from eight directions, which is 128-bit feature vector and many features are detected from each image (Fig 3(b)). It is evident from the picture that the SIFT algorithm rejects keypoints from the low contrast areas like background which is helpful when the object of interest is present in the cluttered environment. Due to rotational invariance, SIFT also reduces the ambiguity in case of stem images which can be vertical or horizontal.


(b)
Fig.3. (a) Original Image. (b) SIFT features extracted from the image while leaving the background environment
The classifiers expect the image to be described as a single vector, hence the SIFT vectors need to be quantized. One of the popular approaches is Bag-of-visual words in which the features are quantized using flat k-means or hierarchical k-means and then computes the histogram for semantic classification [27] .The discriminatory power of Bag-of- words suffers due to quantization errors and loss of spatial order of descriptors. An extension of Bag-of-features model was proposed in [28] called Spatial pyramid matching. It segments image into 2lX2l segments in different scale l=0,1, 2 and computes BoF histograms in each segment, and finally concatenates them to form a vector representation of the image. This algorithm is widely used in many computer vision applications, but problem of one or more vocabularies still exists, hence we have used Sparse coding to compress and quantize SIFT vectors.
Sparse coding is a representation of data as a linear combination of atoms (patterns) learned from the data itself. Such a collection of atoms (code words) is called dictionary or codebooks [29].
For an input X,
D = {d1, d2, d3… dp } (1)
Dictionary D is set of normalized basis column vectors of size p such that there exists a vector α known as sparse coefficient vector α such that utilize sparse coding survey of which is presented in [29]. Success of sparse coding depends on selection of dictionary. One approach to choose Dictionary D is to choose from known transform (steerable wavelet, coverlet, contourlet, bandlets) These off-the-shelf dictionaries fails for specific images like face, digits, etc. [39] therefore current researches are focusing on learning the dictionary from a set of given input sample. The approach is known as Dictionary learning. Given a set of SIFT features , K random features are selected to train the dictionary using following optimization.
V y e Y ,min a^y — Da\ 2 + Я О ( ak ) (3)
A lot of research has been noticed in recent years in development of dictionary learning algorithms some of which are K-SVD of Aharon [38], Olshausen and field [18], SPAMS of Mairal [33] and others [34], [35], [36], [39]. Yang in 2011 proposed Fisher Discriminative Dictionary learning which instead of learning a common dictionary to all classes, learns a structured dictionary D as [D1, D2, D3… DC] where c is the number of classes hence increasing the discriminative power. The performance was reported to be highest using this learning method in terms of high recognition rate and low error rate. We have used Mairal’s online dictionary which is available in SPAMS a Sparse modeling software containing an optimization toolbox for various sparse estimation problems. Once the dictionary is learnt from eq. (1), it is applied to code all SIFT vectors in all images and sparse representation is generated via following optimization.
min D , ak
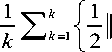
y — Da\ 2 + A Q ( ak ) l (4)
Therefore, for every SIFT vector, a sparse vector is learnt which are pooled using Spatial Pyramid matching before feeding to the classifier.
X=Dα
Where α should be as sparse as possible, i.e. most of the entries in α should be zero.
Sparse representation is more compact and high level representation of the image. As compared with vector quantization, sparse coding has a low reconstruction error, more separable in high dimensional spaces making them suitable for classification purposes. It can be seen in Fig. 4 that SIFT features before sparse coding occupied memory of 20GB whereas the memory reduced to 1 GB after sparse coding. The last years have witnessed an increase in computer vision algorithms that
Fig.4. Memory occupied by SIFT features has reduced by 20 times after applying sparse coding.
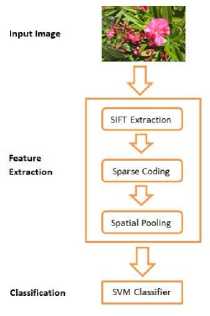
Fig.5. Framework for identification of species using leaf, bark, fruit images
-
B. Fusion of shape,color and texture features
As already mentioned, the most distinguishing characteristics of a flower image are the shape, color and texture. Using only one of these features do not provide accurate results since there can be more than specieshaving same colour, shape and texture. So we have used combination of all three features to represent flower images. To compact the features, we have used Bag-of-features approach to create visual vocabularies which are combined and fed into classifier as shown in Fig. 6.
Fig.6. Identification of species using flower image
The steps are summarized as:
-
• Segmentation is done to extract the flower image
from the background(Fig 9)
-
• Color features are extracted from HSV (Hue,
Saturation and Value) color model since it is less sensitive to illumination variations. Color visual words are created by clustering the HSV value of each pixel.
-
• Shape features are extracted by SIFT
representation and then clustered using bag-of-words approach. SIFT feature representation makes the approach robust against noise and occlusion as well reduces effects of rotation and scaling variances (Fig 3(b)).
-
• To find textures on the petals of the flowers, texture features are extracted by convolving the images with rotational invariant filters from an MR8 (Maximum Response) filter bank (Fig 8).
-
• All the vocabularies are combined into single vocabulary and fed to SVM classifier.
Fig.7. Textures extracted from flower image
Fig.8. Flower image before and after segmentation
V. Experiments for Parameter Setting
To implement the above techniques, experiments have been conducted for making the following decisions:
Choice of classifier (SVM/knn)
-
• Fixing upon particular size of image to maintain uniformity since all the images are with different sizes
-
• Experiments to decide upon number of sample
features to train the dictionary for sparse coding
-
• Number of iterations in sparse coding
-
• Choice of approach to be followed for each
category
The results and analysis of these experiments are as follows:
A. Experiments for choice of classifier
Table 1. Performance comparison of SVM and knn
|
Training Dataset per class |
Test Set (5 Images/class ) No. of classes |
SVM |
KNN |
||
|
Accuracy (%) |
Time (Seconds) |
Accuracy (%) |
Time (Seconds) |
||
|
50 |
4 |
30 |
3.51 |
20 |
2.45 |
|
100 |
4 |
85 |
8.13 |
80 |
4.54 |
|
150 |
4 |
90 |
11.5 |
95 |
6.2 |
|
200 |
4 |
90 |
16.41 |
90 |
8.13 |
|
250 |
3 |
100 |
12.87 |
86.67 |
7.96 |
|
300 |
3 |
100 |
16.16 |
93.33 |
9.38 |
|
350 |
3 |
100 |
23.24 |
93.33 |
10.86 |
|
400 |
3 |
100 |
27.2 |
93 |
12.17 |
From table 1, we can observe that on increasing the number of training images the accuracy increases using both of the classifier SVM and KNN. After the number of images for training reached to 250 per class the accuracy of SVM classifier reached to 100%. Another observation derived was that though SVM gives higher accuracy than that of knn but it ALSO takes more time than knn.
Table 2. Performance comparison of SVM and knn
|
Training Dataset |
Test Set (10 Images/ class) |
SVM |
KNN |
||
|
images/class |
No. of classes |
Accuracy (%) |
Time (Seconds) |
Accuracy (%) |
Time (Seconds) |
|
30 |
10 |
42 |
10.49 |
42 |
4.77 |
|
30 |
20 |
30 |
28.61 |
28 |
8.43 |
|
30 |
30 |
25 |
42.37 |
18 |
14.9 |
|
30 |
40 |
21.25 |
69.55 |
18 |
21.72 |
|
30 |
50 |
20 |
80.44 |
16.4 |
43.09 |
|
30 |
60 |
19.83 |
133.18 |
17.67 |
59.13 |
|
30 |
70 |
20.71 |
176.26 |
15.86 |
58.53 |
|
30 |
80 |
20.25 |
210.71 |
15.63 |
57.48 |
|
30 |
90 |
21.82 |
248.43 |
15.45 |
59.52 |
Table 3. Effect of image size on accuracy
|
Max size of image |
Accuracy (%) |
|
300 |
59.43 |
|
400 |
52.83 |
|
500 |
54.71 |
|
800 |
50.94 |
From the above table we observed that decreasing size of image does not reduce the accuracy. Infact on reducing the size from 400 to 300, accuracy is showing increase. Time taken for smaller size image is less because for smaller images less features are extracted resulting in less processing time during testing and training. Smaller size of image, less the number features extracted, reducing memory requirements.
-
C. Experiments for number of random samples for training the dictionary
Table 4. Number of random samples Vs accuracy
|
No. of Sample |
Random Samples (%) |
Accuracy (%) |
|
5 |
0.35 |
90.47 |
|
10 |
0.7 |
85.71 |
|
15 |
1.05 |
85.71 |
|
20 |
1.4 |
92.85 |
|
25 |
1.75 |
97.62 |
|
30 |
2.1 |
88.09 |
|
35 |
2.45 |
95.24 |
|
40 |
2.8 |
85.71 |
|
45 |
3.15 |
92.85 |
|
50 |
3.5 |
85.71 |
The dictionary learning takes random number of samples from the large dataset of the features extracted. From the above table and the graph we can observe that keeping the random number of samples higher does not increase the accuracy. The results of the experiment are fluctuating yet a stable increase in performance is observed around 1.75 to 2 %. So, we decided to keep the number of random samples as less as 2% of the total number of features extracted from the Images.
-
D. Experiments on number of iterations in dictionary learning
From above table 2 , we conclude that that the accuracy for both SVM and knn reduces as number of classes increase but it is yet relatively more while using SVM.
Looking at the accuracy efficiency of SVM from above observations, we set SVM as choice of classifier.
B. Experiment for size of image
Table 5. Number of Iterations Vs accuracy
|
No. of iteration |
Accuracy (%) |
|
10 |
41.79 |
|
25 |
41.79 |
|
50 |
43.28 |
This experiment is for the number of iterations while learning dictionary. Dictionary is updated in each iteration of dictionary learning. The more the number of iteration, the more time it takes for dictionary learning. So, to maintain tradeoff between accuracy and time, we set number of iterations to 10.
-
E. Experiments to decide on choice of approach
The above table shows the experimental results that compares the different approaches for classification, i.e. Color, Shape, Texture Based classification or Pixel based classification or sparsely coded SIFT based classification on each sub-category. We have tested different approaches on 10 classes of species with five sub-categories.
-
1) Flower: We observed that for flower sub
category, the species recognition accuracy was only 33.62% when we used intensity values as features i.e. the pixel based approach. When using sparse coded SIFT features pooled by SPM, the accuracy achieved was 69.54 whereas classification using combined vocabulary of color, shape and texture features gave maximum accuracy of 73.18.
-
2) Fruit: It was observed that intensity based approach gave very poor accuracy of 18.9% which further increased to 57% when combined vocabulary was used but maximum accuracy achieved was only 67.3% with Sparse coded SIFT feature representation.
-
3) Leaf: This category consists of leaf images on the plants with a cluttered background. With pixel representation, the accuracy is 22.9 which
improved to 45% with color, shape and feature based approach but again the maximum accuracy attained is 69.17 with SIFT and Sparse coding.
-
4) Leaf Scan: This type of category contains images of leaves totally free from background. Either they are scanned images or leaf of the plant is photographed. The accuracy for pixel based is 72.77 which jumps to 82.67 on application of combined vocabulary of shape, color and texture features but on using Sparse coded SIFT features approach, excellent accuracy of 98% was obtained.
-
5) Stem: Stem is the part of the plant which is characterized by different textures therefore a texture feature based approach should ideally be suitable for identification of plant species on the basis of stem image. But to our surprise, the SIFT based approach also outperforms here with a superior accuracy of 76.57% as compared to accuracy of 58.76% while using combined vocabulary and a very poor performance of 20.75% while using intensity representation.
-
VI. Results
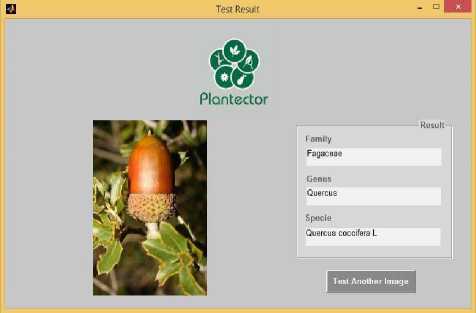
The framework for automatic plant species recognition is implemented in MATLAB in the form of an interactive software named Plantector snapshots shown in figure 11 and 12. The software accepts a plant image and its corresponding metada xml file. The software returns the family, genus and species as output. The software is tested on 10 classes of plant species under five categories namely flower, fruit, leaf, leafscan and stem. Each class of species has images from these five categories which have been used for training. To train the algorithm, 75 images per class has been taken from flower, leaf and leafscan whereas 50 images for fruit and stem classes. In total 3250 images have been available for testing. 25 images from each class have been taken for testing. So in all 250 images have been used for testing. We have implemented three approaches namely Color, shape and texture based approach,SIFT based approach and pixel based approach. From the observations summarized in Table 1 we concluded that when a flower image of the plant is provided for species recognition, the combined vocabulary approach outperforms the other two approaches whereas for all the other sub- categories namely stem, fruit, leaf and leafscan, Sparsely coded SIFT features pooled with SPM approach gives best accuracy.
Therefore to dynamically choose the approach, our Plant species recognition system first identifies the subcategory by reading the xml file provided with the test image and then chooses the approach dynamically. We also concluded that maximum accuracy is achieved in the sub-category leaf-scan which is 98% then in the stem sub-category which is 76.57, next in order is flower sub-category with 73.18 % accuracy and the two subcategories with lowest accuracies are leaf and fruit with 69.17% and 67.33 % respectively. The results can be directly analysed from the graph shown in Fig. 9
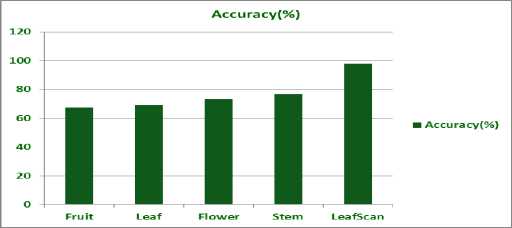
Fig.9. Accuracy comparison of various approaches
Table 6. Performance comparison of different classification approaches
|
Subcategory |
Approach / Classifier |
Classes |
Training |
Accuracy (%) |
|
Flower |
Color, shape, Texture Based |
10 |
75 |
73.18 |
|
Pixel based |
10 |
75 |
33.62 |
|
|
SIFT based |
10 |
75 |
69.54 |
|
|
Fruit |
Color, shape, Texture Based |
10 |
50 |
56 |
|
Pixel based |
10 |
50 |
18.9 |
|
|
SIFT based |
10 |
50 |
67.33 |
|
|
Leaf |
Color, shape, Texture Based |
10 |
75 |
45 |
|
Pixel based |
10 |
75 |
22.95 |
|
|
SIFT based |
10 |
75 |
69.177 |
|
|
Leaf Scan |
Color, shape, Texture Based |
10 |
75 |
82.67 |
|
Pixel based |
10 |
75 |
72.77 |
|
|
SIFT based |
10 |
75 |
98 |
|
|
Stem |
Color, shape, Texture Based |
10 |
50 |
58.76 |
|
Pixel based |
10 |
50 |
20.75 |
|
|
SIFT based |
10 |
50 |
76.57 |
-
VII. Conclusion
An automatic plant speciesrecognition application has been presented. Currently the application has been tested on 10 classes of speciesunder five organs due to hardware constraint. Motivated from success rate for 10 classes, best being 98% accuracy for leafscan images our future work will be to test on all 1000 classes. The future work aims at testing on more number of classes. We also aim to deploy the software as an Android mobile application which captures image of plant from the plant and performs recognition task through the application.
Список литературы Application of Sparse Coded SIFT Features for Classification of Plant Images
- James S. Cope, David P. A. Corney, Jonathan Y. Clark, Paolo Remagnino, and Paul Wilkin. Plant speciesidentification using digital morphometrics: A review. Expert Syst. Appl., 39(8):7562{7573, 2012.
- Kumar, N., Belhumeur, P.N., Biswas, A., Jacobs, D.W., Kress, W.J., Lopez, I.C., Soares, J.V.B.: Leafsnap: A computer vision system for automatic plant speciesidentification. European Conference on Computer Vision. pp. 502{516 (2012)
- Backes, A.R., Casanova, D., Bruno, O.M. Plant leaf identification based on volumetric fractal dimension. International Journal of Pattern Recognition and Artificial Intelligence 23(6), 1145{1160 (2009)
- Y. Nam, E. Hwang, and D. Kim. Clover: A mobile content-based leaf image retrieval system. In Digital Libraries: Implementing Strategies and Sharing Experiences, Lecture Notes in Computer Science, pages139{148. 2005.
- D. Barthelemy. The pl@ntnet project: A computational plant identification and collaborative information system. Technical report, XIII World Forestry Congress,2009.
- G. Cerutti, V. Antoine, L. Tougne, J. Mille, L. Valet, D. Coquin, and A.Vacavant.Reves participation -tree speciesclassification using random forests and botanical features. In Conference and Labs of the Evaluation Forum, 2012.
- Envirofi. http://www.envirofi.eu/.
- J.-X. Du, X.-F. Wang and G.-J. Zhang, “Leaf shape based plant species recognition,” Applied Mathematics and Computation, vol. 185, 2007.
- A. H. Kulkarni, H. M. Rai, K. A. Jahagirdar and P. S. Upparamani, (2013). A Leaf Recognition Technique for Plant Classification Using RBPNN and Zernike Moments, International Journal of Advanced Research in Computer and Communication Engineering, Vol. 2, Issue 1, pp. 984-988.
- Krishna Singh, Indra Gupta, Sangeeta Gupta, SVM-BDT PNN and Fourier Moment Technique for classification of Leaf, Internatio Guru, D. S., Y. H. Sharath, and S. Manjunath. Texture features and KNN in classification of flower images.IJCA, Special Issue on RTIPPR (1) (2010): 21-29, 2010.nal Journal of Signal Processing, Image Processing and Pattern Recognition Vol. 3, No. 4, December, 2010.
- Gopal, S. P. Reddyn and V. Gayatri, (2012). Classification of Selected Medicinal Plants Leaf Using Image Processing, IEEE International Conference on Machine Vision and Image Processing(MVIP), Taipei, pp. 5-8.
- O. Mzoughi, I. Yahiaoui, N. Boujemaa, and E. Zagrouba. Advanced tree speciesidentification using multiple leaf parts image queries. In IEEE International Conference on Image Processing (ICIP), 2013.
- Zuolin ZHAO, Gang YANG, Xinyuan HUANG. Plant Recognition Based on Leaf and Bark Images, School of Information Science and Technology, Beijing Forestry University, Beijing 100024, China.
- Nilsback and Andrew Zisserman. A Visual Vocabulary for Flower Classification.Computer Vision and Pattern Recognition, IEEE Computer Society Conference on. Vol.2, 2006.
- Nilsback, M.E., Zisserman, A. Automated flower classification over a large num-ber of classes. Indian Conference on Computer Vision, Graphics and Image Processing. pp. 722{729 (2008)
- Qi, Wenjing, Xue Liu, and Jing Zhao. Flower classification based on local and spatial visual cues. Computer Science and Automation Engineering (CSAE), Vol. 3,2012.
- Pei, Yong, and Weiqun Cao. A method for regional feature extraction of flower images.Intelligent Control and Information Processing (ICICIP), IEEE, 2010.
- Siraj, Fadzilah, Muhammad Ashraq Salahuddin, and Shahrul Azmi Mohd Yusof.Digital Image Classification for Malaysian Blooming Flower. Computational Intelligence, Modelling and Simulation (CIMSiM), IEEE, 2010.
- Guru, D. S., Y. H. Sharath, and S. Manjunath. Texture features and KNN in classification of flower images.IJCA, Special Issue on RTIPPR (1) (2010): 21-29, 2010.
- Naiara Aginako, Javier Lozano, Marco Quartulli, Basilio Sierra, Igor G.Olaizola.Identification of plant species on large botanical image datasets, Artificial Intelligence Department,University of the Basque Country,and Paseo Mikeletegi,57, 20009 Donostia-San Sebastián, 2014.
- H. Goeau, P. Bonnet, J. Barbe, V. Bakic, A. Joly, J.-F. Molino, D.Barthelemy, and N.Boujemaa. Multi-organ plant identification. In Proceedings of the 1st ACM International Workshop on Multimedia Analysis for Ecological Data, MAED '12,2012.
- http://www.tela-botanica.org
- Lowe, David G. "Distinctive image features from scale-invariant keypoints." International journal of computer vision (Springer Netherlands) 60, no. 2 (November 2004): 91-110.
- Lisin, Dimitri A, Marwan A Mattar, Matthew B Blaschko, Erik G
- Learned-Miller, and Mark C Benfield. "Combining local and global image features for object class recognition." Computer Vision and Pattern Recognition-Workshops. IEEE, 2005. 47-47.
- K, Mikolajczyk, and Schmid C. "A performance evaluation of local descriptors." IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE) 27, no. 10 (October 2005 ): 1615 - 1630.
- Lindeberg, T. Journal of Applied Statistics, 21(2):224-270.
- J, Sivic, and Zisserman A. "Video Google: a text retrieval approach to object matching in videos." Ninth IEEE International Conference on Computer Vision, 2003. Proceedings. Nice, France : IEEE, 2003. 1470 -1477.
- Lazebnik, Svetlana, Cordelia Schmid, and Jean Ponce. "Beyond bags of features: Spatial pyramid matching for recognizing natural scene categories." IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE, 2006. 2169-2178.
- J Wright, Y Ma, J Mairal, G Sapiro, TS Huang, S Yan - Sparse representation for computer vision and pattern recognition,Proceedings of the IEEE, 2010.
- B. Olshausen, D. Field(2004). Current Opinion in Neurobiology Current Opinion in Neurobiology 2004, 14(4):481–487
- Yang J-C, Yu K, Gong Y-H, et al.J OURNAL OF MULTIMEDIA, VOL. 9, NO. 1, Proceedings of IEEE Conference on Computer Vision and Pattern Recognition, pp. 1794-1801, 2009.
- Mairal, F. Bach, J. Ponce, G. Sapiro, and A. Zisserman. Supervised dictionary learning. In NIPS, 2009.
- Ramirez, P. Sprechmann, a nd G. Sapiro. Classification and clustering via dictionary learning with structured incoherence and shared features. In CVPR, 2010.
- J.C. Yang, K. Yu, and T. Huang. Supervised Translation-Invariant Sparse coding. In CVPR, 2010.
- M. Yang, L. Zhang, J. Yang and D. Zhang. Metaface learning for sparse representation based face recognition. In ICIP, 2010.
- J. Mairal, F. Bach, J. Ponce, G. Sapiro, and A. Zissserman Learning discriminative dictionaries for local image analysis. In CVPR, 2008.
- F. Rodriguez and G.Sapiro. Sparse representation for image classification: Learning discriminative and reconstructive non- parametric dictionaries . IMA Preprint 2213, 2007.
- Pham and S. Venkatesh. Joint learning and dictionary construction for pattern recognition. In CVPR, 2008.
- Meng Yang,Zhang, D.Xiangchu Feng,Zhang.Fisher Discriminative Dictionary Learning for Sparse Represenatation.ICCV,2011
- http://www.imageclef.org/lifeclef/2015/plant


