Bioinformatics Analysis and Characteristics of the giant panda Interferon-alpha
Автор: YueYi, Zhiwen Xu
Журнал: International Journal of Information Engineering and Electronic Business(IJIEEB) @ijieeb
Статья в выпуске: 1 vol.3, 2011 года.
Бесплатный доступ
In this report, the amino acid sequence of giant panda interferon-α (gpIFN-α) was determined and compared with 15 corresponding IFN-α sequences. Phylogenetic analysis showed that the 15 interferons fell into two large groups. The giant panda and ferret branched and were most closely related to fox and dog and evolved into a distinct phylogenetic lineage from that of eukaryotic mammalians which evolved into another lineage. After analyzing the encoded amino acid sequence of the gpIFN-α using bioinformatics, the results revealed that in the full amino acid sequence, there were no transmembrane domain, one N-glycosylation sites, eight O-glycosylation sites and nine antigenic determinants. Secondary structure analyzed showed that the Alpha helix, Extended strand, Beta turn and Random coil each occupied 60.37%(99aa), 4.88%(8aa), 9.76%, 25%(41aa) respectively. In conclusion, our results will give the opportunity to investigate more in detail function study in giant panda and add to studies on the evolution of the IFN system in vertebrates and avian more generally.
Ailuropoda melanoleuca, interferon alpha, clone, sequence analysis, Structure analysis
Короткий адрес: https://sciup.org/15013065
IDR: 15013065
Текст научной статьи Bioinformatics Analysis and Characteristics of the giant panda Interferon-alpha
Published Online February 2011 in MECS
Interferons (IFNs) are a multi-gene family of inducible cytokines which have a major role in immune defense
Corresponding author: Zhiwen Xu;
against virus infections but are also recognized for their antiproliferative and immunomodulatory activities [1, 2]. IFNs are produced in numerous cell types including lymphocytes (T and B cells), macrophages, fibroblasts, blood vessel endothelial cells and osteogenetic cells, and are known to be important elements in antivirus reply [3]. They were classified as Type I IFNs (virus-infected IFNs), Type II IFNs (immune IFNs) and type III [4]. The alphainterferons belonged to type I interferons, are mainly produced by virus-infected peripheral blood leukocytes, lymphobl- astoid and myeloblastoid cell lines[5]. It is considered to be a choice drug to treat virus disease and tumour in clinics.
The giant panda (Ailuropoda melanoleuca) is a much loved animal all over the world and is considered a symbol of China[6], However, it is also one of the world's most endangered species, as well as a flagship species for conservation. The latest molecular censusing research used fecal samples and nine microsatellite loci shows that the estimated number of wild pandas is 1596, and the number of captive pandas is about 161. In recent years, strenuous efforts have been made to protect this animal and considerable knowledge of its physiology, biochemistry, genetic diversity and ecology has been onrd gained, but death of disease are one of the major problems facing giant panda health. The research on the diseases of giant panda revealed that there were more than 40 kinds of diseases do exist. They are mainly viruses, bacteria and parasite. Therefore, studies of cytokine genes of the giant panda become important and necessary to facilitate the use of these cytokines in the immunotherapy of infectious diseases of the giant panda.
During the past decade, a variety of interferon genes have been cloned and sequenced from a number of species, including human, canine, porcine, chicken. And many studies had been performed on them. nonetheless, research on the giant panda Interferon-alpha is still rare. Although the gene has been reported since 2007 [7], very few information about the structures and functions of giant panda IFN-α has been published. In order to understand the bioinformatics and characteristics of the gp IFN-α at the molecular level and to extend the
-
II. Materials and methods
A.Culture of peripheral blood lymphocytes
Adult giant pandas were phlebotomized at the Ya’an Research Base of Giant Panda Breeding under required procedures. Peripheral blood lymphocytes were separated by gradient centrifugation using Lymphocyte Separation Medium (TBD, Tianjin, China), and the Cells (5×106cells/ml) were then resuspended and cultured in RPMI-1640 medium (Gibco, Grand Island,NY, USA) supplemented with 10% fetal calf serum, 100U/ml penicillin, 100 mg/ml streptomycin. Lymphocytes were stimulated with concanavalin A at a final concentration of 15 mg/ml and incubated for 16 h at 37 V in a humidified incubator with 5% CO2. After 16 h, the stimulated cells were washed twice with Hanks’balanced salt solution and collected by centrifugation.
-
B. RNA extraction
Total RNAs from the harvested cells were isolated using TRIzol reagent (TaKaRa, DaLian, Japan) according to the manufacturer’s instruction. RNA quality was evaluated on 1.0% agarose gels containing ethidium bromide (10 mg/mL). RNA samples were used subsequently for RT-PCR or stored at -70 C .
-
C. RT-PCR amplification of the IFN-a gene
Synthesis of the first strand of complementary DNA (cDNA) was performed using an RNA polymerase chain reaction (PCR) kit (TaKaRa, Dalian, Japan). The cDNA was synthesized at 37C from RNA by using oligo-dT (Promega, Madison, WI,USA). The following IFN-a specific primers were used in the amplification: IFN-a applications of recombinant giant panda IFN-α, we are trying to analyze and report some information about the structure and function of its encoded protein. Furthermore, to illustrate its evolutionary relationships among the eukaryotic mammalians, a phylogenetic tree was constructed. Our results will give the opportunity to investigate in detail on the evolution of the IFN system in avian and mammals more generally, and may provide some insights for further research the function and biological activities about the gpIFN-α.
forward, 5’- GGATCC TGTGACCTGCCTCAGAACCA TGGCCTG-3’ containing the BamHI site, and reverse, 5’- GTCGAC TCATTTCTCGCTCCTTAGTCTTTCTTG-3’containing the SalI site. The PCR was carried out in a 20μL total volume reaction mixture that consisted of 10μL 2×PCR Mixture (TaKaRa, Dalian, Japan), 0.5μL each primer (25 mM), 1.0μL template cDNA (about 300 ng) and 8μL ddH2O. The conditions of PCR were: 95 °C for 5 min, 35 repeated cycles of 95 C for 30 s, 58 C for 40 s and 72 C for 45 s, and then a final extension at 72 C for 10 min. The products of these PCR were electrophoresed on 1% agarose gelscontaining ethidium bromide and visualized under an ultraviolet light.
-
D. Cloning and Sequencing of the IFN-a Gene
The PCR products were purified by using a TIANprep Mini Plasmid Kit (TianGen) according to the manufacturer’s instructions. The purified PCR products were cloned into pMD19-T vector (TaKaRa), and then transformed into Escherichia coli (E.coli) DH5α competent cell, After that, the positive recombinant clone was selected by the Amp/IPTG/X-Gal agar plate. Plasmid DNA was identified by bacterial colony PCR with a forementioned conditions, and digested with restriction enzymes BamHI and SalI (TaKaRa, Dalian, Japan). Then it had to be fractionated in 1% agarose gels. Sequencing reactions was also performed by TaKaRa.
-
E. Analyzing nucleotide sequence of IFN-a the gene by bioinformatics
We incorporated IFN-α sequences described for giant panda as well as 14 other mammalian and avian species from NCBI, and their GenBank accession numbers are listed in Table 1. Multiple sequence alignments of deduced amino acid sequences of these 15 interferons were performed by using ClustalW multiple alignments. Phylogenetic trees were constructed using the neighbor -joining method in MEGA version 4.0. To estimate the transmembrane domain of the gpIFN-α, the amino acid sequence was analyzed by using online prediction tools. Prediction of the transmembrane segment were analyzed es/NetNGlyc/), NetPhos2.0 serv-ices/NetPhos/), and BepiPred 1.0 Server , respectively. The structure of Intron and exon was analyzed by using Genscan . Secondary structure was predicted by using SOPM method on web site with TMHMM -MM/). Using the DNAStar 7.0 and on-line predicted tools to analyze the structural properties of the gpIFN-α amino acids sequence. The glycosylation sites, phosphorylation site and epitope were analyzed using the programs of NetNGlyc1.0
-NPSA/. The similar three dimensional structure was obtained online by Swiss-Model server (http:// swiss -model. and was observed with RasMol 2.7 software. Rare condons content analysis of the gpIFN-α.gene by using codon usage data base on line.
TABLE I. Accession No. of interferon-a ftom 15 different species
|
Species |
GenBank Accession No. |
Animals' IFN-α |
Natural animal |
Length (bp) |
|
AY323972 |
Bubalus bubalis IFN-α |
Bubalus bubalis |
498 |
|
|
DQ520882 |
Canis familiaris IFN-α |
Canis familiaris |
495 |
|
|
FJ959074 |
Capra hircus IFN-α |
Capra hircus |
501 |
|
|
NM_001099441 |
Equus caballus IFN-α |
Equus caballus |
486 |
|
|
NM_001009851 |
Felis catus IFN-α |
Felis catus |
516 |
|
|
NM_001194384 |
Macaca mulatta IFN-α |
Macaca mulatta |
501 |
|
|
Mammal |
EU863618 |
Mustela putorius furo IFN-α |
Mustela putorius furo |
495 |
|
HM636502 |
Pteropus vampyrus IFN-α |
Pteropus vampyrus |
540 |
|
|
AY331298 |
Sus scrofa IFN-α |
Sus scrofa |
501 |
|
|
EF990625 |
Vulpes vulpes IFN-α |
Vulpes vulpes |
540 |
|
|
BC074928 |
Homo sapiens IFN-α |
Homo sapiens |
501 |
|
|
HQ378189 |
grint panda IFN-α |
grint panda |
495 |
|
|
Birds |
X84764 |
duck IFN-α |
duck |
492 |
|
AB021154 |
Gallus gallus IFN-α |
Gallus gallus |
492 |
|
|
EU022750 |
Shitou Goose IFN-α |
Shitou Goose |
492 |
Ш. RESULTS
A. Analysis the amino acids sequences of gpIFN- а
The gpIFN-α gene was composed of 495 nucleotides and was translated into a putative peptide of 164 amino acid residues(Fig. 1). Also it has been submitted to GenBank and the accession number HQ378189 was assigned for the IFN-a of the giant panda. Through on-line
analysis, it had a putative molecular mass of 18.52 kDa and a predicted isoelectric point (PI) of 6.10. In addition, the results showed that the formular of the gpIFN-α gene was C818H1281N225 O246S10, the total number of atoms was 2580, the instability index was 49.27, the aliphatic index was 79.37, and the grand average of hydropathicity (GRAVY) was 0.809. The total number of negative charged residues (Asp + Glu) and positive charged residues (Arg + Lys) was 18 and 16, respectively. The estimated half-life of gpIFN-α was: 7.2 hours (mammalian reticulocytes, in vitro), >20 hours (yeast, in vivo), >10 hours (Escherichia coli, in vivo). Through the analysis of DNAStar7.0, the polypeptide encoded by the gpIFN-α was composed of 58 hydrophobic amino acids, 51 hydrophilic amino acids,16 basic amino acids and 18
acidic amino acids. Furthermore, through the prediction of GENESCAN on line, the consequence indicated that there was no intros presenting in the sequence.
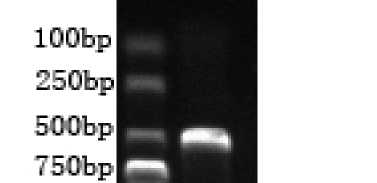
Figure 1. RT-PCR of giant panda IFN-a. The expected about 500bp fragment of giant panda IFN-a cDNA was amplified.
B. Functional sites analysis
By using different online web servers, we gained more informations about the gp IFN-α. Such as follows: Firstly, the mature amino acid of gp IFN-α had one potential N-glycosylation sites and eight potential O-glycosylation sites. The potential N-glycosylation sites located at 78aa, and the biggest potential value was 0.6653. The O-glycosylation sites located at 70aa, 73aa, 74aa, 109aa,
141aa, 147aa, 148aa, and 162aa respectively. These results revealed that like other mammalian type I IFNs, the gpIFN-α was also a glycoprotein. Secondly, the phosphorylation sites were predicted through on-line analysis software NetPhos2.0 services/NetPhos/). The result was shown in fig. 3. When taken the threshold as 0.5, the mature amino acid sequence had a total of eleven potential phosphorylation sites, including six serine sites, four threonine site and one tyrosine site. thirdly, one cAMP-and cGMP-dependent protein kinase phosphorylation site (at aa residue 22), two Protein kinase C phosphorylation sites (at aa residues 141, 162) and three casein kinase II phosphorylation site (at aa residue 27,80,94) can be clearly identified. And also contain a signature of interferon alpha, beta and delta family (at aa residue 121). Fourthly, there were six cysteines residue respectively locate at 1, 29, 69, 87, 100 and 145 amino acids. At last there were twenty-one protein-protein interaction sites that were likely to be hot spots for protein interaction.
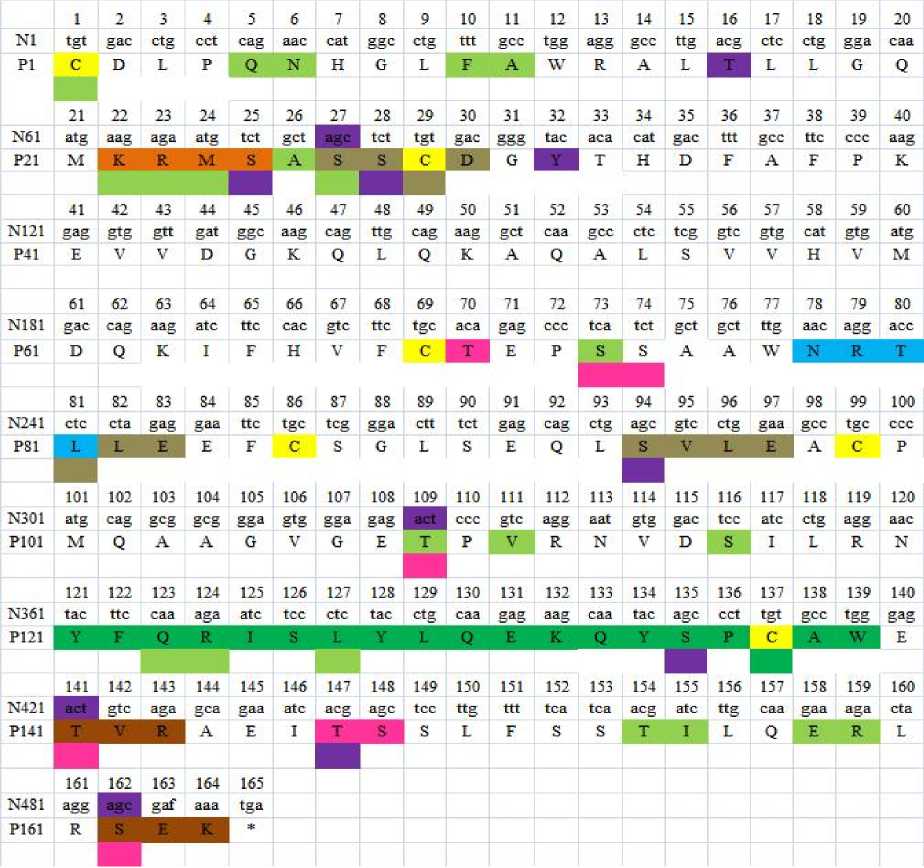
Figure 2. Nucleotide sequence of gpIFN-α gene and its deduced amino acid sequence. In the amino acid sequence, there are some repetitional functional sites, we use different colors to distinguish them. the boxed letter highlighted in blue indicates the putative N-glycosylation site; phosphorylation sites are identified by the letters highlighted in purple; N-glycosylation site is identified by the letters highlighted in pink; cAMP-and cGMP-dependent protein kinase phosphorylation site is identified by the letters highlighted in orange; Protein kinase C phosphorylation sites are identified by the letters highlighted in brown; casein kinase II phosphorylation site is identified by the letters highlighted in gray; cysteines residue marked with yellow protein-protein interaction sites are marked with reseda; the interferon alpha, beta
and delta family signature is identified by the letters highlighted in green; and also the asterisk indicates the stop codon.
-
C. Transmembrane domain and antigenic Determinants of the gpIFN-α protein
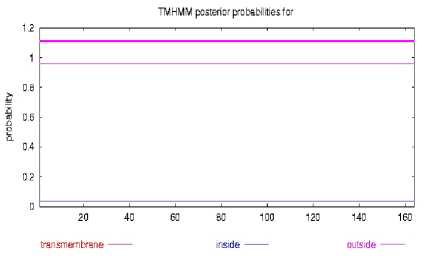
3. The prediction of transmembrane segments.
The prediction of transmembrane domain revealed that the gp IFN-α amino acid had non-transmembrane domain (Fig. 3). Through the prediction of online web servers, we found that gp IFN-α amino acid sequence contained nine antigenic determinants and was mainly positioned at 1-4aa, 26-33aa, 37aa, 41-49aa, 71-77aa, 101-113aa, 133-139aa, 161-162aa, 164aa (fig. 4).
Figure
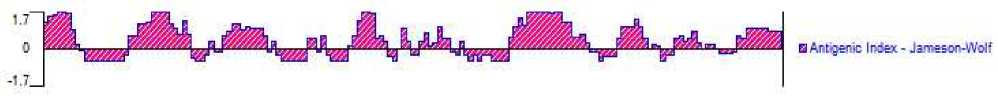
Figure 4. The predicted results of the antigenic determinants. It was performed with DNAStar 7.0.
-
D. The prediction of Secondary structure and tertiary
structure of IFN-a
The prediction of gp IFN-a’s secondary structure was shown in Figure 5(A) and Figure 5(B). The result indicated that the alpha helix ( H ) , extended strand ( E ) , Beta turn and random coil ( C ) each occupied 60.37%(99aa), 4.88%(8aa), 9.76% (16aa), 25%(41aa) respectively. The alpha helix of the protein have higher
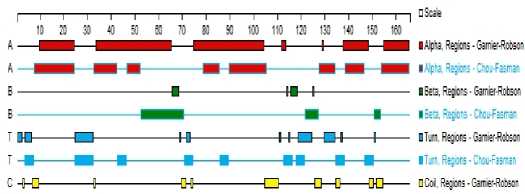
Figure 5(B). The gp IFN-α secondary structure is predicted with DNAStar 7.0
chemical bonding energy, which can firmly maintain proteinic higher structure, and mainly situated in aa 8-26, 40-56, 74-98, 114-130, 139-162. And the rest components are mainly at aa 1-7, 27-39, 44-49, 61-73, 99-113 and 131-138.
The similar three dimensional structure of the IFN-a by Swiss-Model automobile homology modeling database is showed in Fig. 6. The templet is 1itfA with 54.82% identity. Both of the two pictures were displayed by RasMol 2.7 software with Ribbons mode, the left picture with group colour pattern while the right picture with structure colour pattern.
10 20 30 40 50 50
I I I I I I
EP 5 SAAWNRTLLEEFCSGLSEQLSVLEACPMQAAGVGETPVRNVDSILRNYFQRISLYLQEKQYSPC hhhhhhhhhiihhhhhhhhhhhlihhh: ^ htt ^ hhhhhhhhhhhhhhhhhttt:
TVRAEITSSLF55TILQERLRSEK hhhhhhhhhhhhhhhhhhhhhhtt
Figure 5(A). The secondary structure of IFN-a is predicted by
SOPM method with the online web server
, h represents alpha helix, e represents extended strand, c represents random coil, and t represents beta turn.
-
E. Similarity and Phylogenetic analysis
Similarity comparison of gpIFN-α amino acid sequence with those of 14 other mammalian and avian
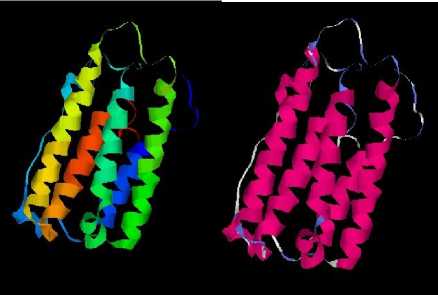
Figure 6. The 3D model of IFN-a. The left with group colour pattern: according to their location, that amino acids are colored from blue, green, yellow, orange to red, with the N’ terminal as blue, the C’ terminal as red.The right with structure colour pattern: according to their secondary structure, that amino acids are colored with the alpha helix as scarlet, the turn as light blue, other as white.
types of human, ferret, dog, cat, fox, chicken and duck IFN-a were approximately 58.2%, 73.9%, 70.3%, 70.9%, 70.3%, 26.2%, 21.3%, respectively (Fig. 7). This revealed the big similarity between the giant panda and ferret. This may provide some clues to explore the enigmatic relationship.
|
grintjianda ... 2D IP 2N B£fi FAW 8 6 i Г Ml^piEASS pGYTHDB1 37 ferret ... :nb;D9SLLHWm< IL12MRRDEASSI1NYTN0E5 37 dog ... 2^ jPCT a3L39SR7ЯзЯКВЬЕ AGS IDHYTNDI1 37 fox ...:DlpiAa;LR9SR7lclL12MRR[jEAGslMBbnDEA 37 cat ... IdIp^SMOSRSaIcIlISURROEASS I2KDHSDE1 37 human ... ID lEIiSlDliRRr L В A2M^HS|PS5 IliMDRHDEl 37 cattle ...:E«aTaSIA9RRv’LH;HLKp7S₽SEIL2DRNDEl 37 goat ...№*а1а51АЯ:-87Ыь52^В^5Р55|02ВКЕ0ЕЬ 37 house ... |ЯнТа513Ш?М1|32№М5₽К5 jDKDRNDES 37 monkey ... 2185139336 h »ЬЙ2М^315₽Е5 LkDRHDES 37 pteropus .. J^H;PaS179336 ls2URBI5PF5 IGKDBEDES 37 pig ... |Д;та5[АНТ161111А2№В15РК5 jOEH^RDES 37 duck FSCSPgRLHE^FAWES J2 ЯмфРЗРТСР |?CQBSPCS 40 chicken .ACNHHRFQIATFSHESn;|FRD|kPTLLQL IpgHNASCS 39 goose FSCSp|RI.H^FPWDSg 2^n[nPSPTCp|pBQBA₽CS 40 Consensus cdlp hsl n г 1 11 qnrr s sc dr df |
grint_panda ferret dog fox cat human cattle go at house monkey pteropus pig duck chicken goose Consensus |
EP FF EP EP EP EP EP EP EP EP FF ф FF F^ FF £p |
GE ф Ki i EE EE EE EE EE si 1 CI CI |
vv^c vfcc LFCC L?EC ‘^S 2 ETC algIc alg|c #БС 2 EEC А?й alg: llE ILDI LLDI fde |
№ FAL № QRL cEb: N^E s;l s;l n^f: n^ NPF № NET SNT NET q |
;ka$ IKACg Si м :pi® ;ka№ ;m$ 2МЖ .kM jig® № 2МЖ ;. ■ |kaqa |
,5V .FV ,5V .FV - 5V I5V :dV .FV :f| :gv a^ AL №1 ЖТ Mi SV |
vE Lr Л ;: л Lr Lr Lr VE Lr 7r VE AI AI |
IvMeQkI 7TNRKT vMiEjkv 7MIQKV VINQKI CLIQ2I EVIQHT CVIQHT Eiijci Ш$1 EVTRQT рмьБЕт HLLQHL ^ILQHL HLLQHL q |
1 2 f |
ivft i|lf: iLEZ :lf: if|ez £ei 2 les 2 LES ILFS ILES _ьф jbES mb (ILS )ILS Ifs |
EIep. ^A. PDI. PDT. Eeas |tkd. TEG. ICG. IDG. IKE. |lEA. IpG. Bps. 5P5. SP5. t |
SSi5 76 SW 76 SSAP 76 SSAP 76 SSAaI 77 SSaJ 76 S® 76 uEl 76 SSSA 76 SSAA 76 SS|^ 76 S06A 76 DEM 77 IF13 76 IE6i 77 ssaa |
|
grint panda |
ЩявтЕгБе е&ВЦзеЕхЬ уЯе5В|рмЕ В Я11 Lb EE LC 5 И 5E QI GP IE A l PL j ВыМ? LL3ELC5 FL 5EQL T_ JEA |P1; BnMCLL^ELCSGLFDGLDeIeAiFLG В ЯIT Lb EE pb Г ^L dr|q-i1 T P ba! vv j gDEDLbDKF:iE|bo|tE ISA IVMQ |
АА&'еЕтркй&’. |
114 |
grint_panda |
CS |
I |
L |
RN. . |
jQEI |
5 |
LY |
G |
EK |
S jCABi |
E |
r1 |
AE |
S |
|||||
|
ferret |
iAGVEETPLV^. |
114 |
ferret |
ES |
I |
1 |
RN. . |
ри |
5 |
LY |
Q |
EK |
S IcABi |
E |
I VE |
AEIM |
К |
||||||
|
dog |
iAGLAmLMHE. |
114 |
dog |
CS |
1 |
RT. . |
Iori |
5 |
LY |
Q |
DRNB |
S САЙ |
E |
5VE |
AE |
f |
|||||||
|
fox |
iaglaetIlmhs. |
114 |
fox |
CS |
L |
RS. . |
jSRl |
5 |
LY |
G |
Ep |
ЯН |
S САЙ |
E |
IVE |
AE |
iE |
||||||
|
human |
?фреф|РЕфЕр eervgetJlm^. |
116 114 |
human |
IHFE |
CS CS |
I I |
L |
RN. . AVKK |
i i |
ink in |
5 |
LY LY |
Q G |
EK EK |
kL ^F |
S jCAK S |сАй |
E E |
■VE jVE |
AEIMR AEIMR |
||||
|
cattle |
BsESLb DKL?AALD2Q17ElEb!LR; |
eeglrgaIilke. |
114 |
cattle |
CS |
5 |
AVRK |
-:rE |
LY |
EK |
K? |
S |сАй |
E |
jVE |
M |
г |
R |
||||||
|
coat |
gDQSLbDKL?AAL:2GirE О jLR2 |
^GLRGAfLL?;5 . |
114 |
frn at. |
]S |
5 |
AVRK |
1 |
ir^ |
LY |
t |
EK |
51 |
IS 8САЙ |
E |
AE |
|||||||
|
house monkey |
Вэезиск^|ут&^2сг:е Iej.IlsI gECNHEKESi:E|bt2G15cliAln; Ё1Е1ЬЬЗНЕк№ъ12С15В Hl Li; |
svg^eEtplmSe. iAGVGEllLMbE. |
114 114 |
house monkey |
CS CS |
L I |
AVRR AVRK |
: |
}RI 2RI |
A |
LY LY |
Q [ |
EK EK |
^F |
s дсдй S |сдй |
E E |
■VE gvE |
AEIMR AEIMR |
|||||
|
pteropus |
!!УСГЕ™1ЕЕЕ. |
114 |
pteropus |
CS |
- |
AVRK |
ЮМ |
LY |
Q |
EK |
KH |
S СДЙ |
E |
■ F |
AEIMR |
||||||||
|
pig |
gDES^bpQF ZfE^b C^QL^E A >VM£. g^G|LEG^PljLE E . BlhtarhdEln^ghhihh IeE !ffaeaarlhrrgpr5l . |
114 116 |
pig |
E5 IL |
I S |
I |
AVRK NK. . |
1 |
;cE |
F |
HF |
Q Q |
EK NH |
FL |
s |сай S |САЙ |
E D |
■VE |
AE № |
VS5 AHA |
||||
|
chicken |
BhDSQRQSLLNRIHRITQH Ie: iLDSRETRSRTRPiPRNL. |
115 |
chicken |
....HLTIKK.. |
3 |
5CLHIE |
0 |
END it |
S |САй |
E |
Ive |
L(ARA |
|||||||||||
|
goose |
g ЬНI ARB D L LN g|l|gHB IH I- IE 1 1 FF M АШ EHRR G FRflE,. |
116 |
goose |
....HLGINK.. |
^ |
3 |
IC^HF |
Q |
NH |
Tit |
s |сдй |
3 |
gVE |
^ |
AHA |
||||||||
|
Consensus |
w 11 1c gl qql leac q |
e g etpl ne |
Consensus |
ds |
1 |
yf |
lit |
lylqek |
yspcawe |
vraeimr |
|||||||||||||
grintjanda ferret dog fox cat human cattle go at house monkey pteropus pig duck chicken
^F.^S±EqMI5E
PLY.a|sTv'L^SLB5R
SEP.SSIlLQi SF^.SSTILQS sEyyssialqr 515.1|5СЧЩ?
is:
:n
I RR
IRK
ILS5E
IL8RK
AFS.SSIMQilSFhK
5f|s.|s SIMMS.
IRK
goose
Co ns e ns us
Cf’RIfiFbIRIMR. . .
aLbibbiignirt. .
CgpRIBRITRAHR...
sf sst Iq r I
Figure 7. Amino acid sequence alignment of giant panda IFN-α from other species. The sequence alignments were performed using CLUSTALX.
On the basis of the alignment of the IFN-α sequences from 15 distinct genes , a phylogenetic tree of the interferon family had been constructed by using neighbor-joining method. The phylogenetic tree told us that the 15 interferons fell into two large groups. And there was an excellent relationship between the giant panda and ferret IFNα. Further examination of the phylogenetic branch showed that the giant panda and ferret IFNα formed a monophyletic group distinct from dog and fox, confirming the similarity between giant panda and ferret IFN-α’s. The results also indicated that IFN-encoding regions of giant panda, ferret, dog and fox clustered together and evolved into a distinct phylogenetic lineage from that of mammals which evolved into another lineage. Phylogenetic analysis based on interferon sequences had also demonstrated that strains of the similar always clustered together.
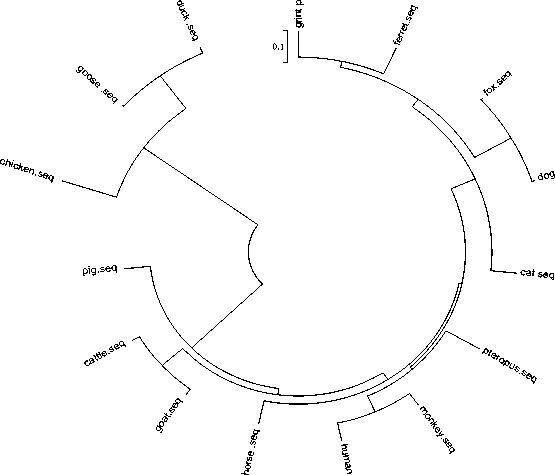
Figure 8. Phylogenetic relationships of IFN-a sequences from other species
-
F. Rare condons analysis of the gpIFN-α gene
Rare condons analysis showed that there were 15 rare condons (9.15%) in the giant panda IFN-α gene by using codon usage database on line ( http://nihserver. . Five AGG codons and four AGA codons were the rare Arg condons. Two CTA codons were the rare Leu condons. Four CCC codons were the rare Pro condons. And also containing one consecutive rare codons AGACTAAGG.
W. Discussion
The giant panda is one of the surviving ancient animals with high scientific value. It is considered as the fossil alived because of its represents of an important stage of evolution[8]. It has been reported that giant pandas are particularly susceptible to infectious disease and parasitic, which can result a 44% infection rate from pneumonia and tick-borne disease and a 66.67% mortality rate from ascariasis[9]. Therefore, it is very important to study the immune system of giant panda, especially immune function genes. In order to further understand the biological function of giant panda IFN-α at the molecular level and to extend the recombinant interferon-based applications in the precious animal, in this report, we describe the basic structure, function and characteristic of the gpIFN-α through bioinformatics softwares and tools.
Signal peptides consist of short stretches of amino acids which, after protein delivery to the correct subcellular compartment, are frequently removed by specialized signal peptidases [10]. Only after removal of the signal peptide sequence, the precursor protein may be allowed to entry into the secretory pathway and become a normal function of the mature protein[11]. We plan to express the IFN-a gene in Ecoli expression system in the near future. So we clone the interferon alpha gene without single peptide. In the mature sequences there are six cysteine residues located in amino acid residues 1, 29, 69, 87, 100 and 145. Four of these six cysteines (at positions 1, 29, 100, and145) are conserved among most of the mammalian IFN-a proteins. The first residue of the mature peptide is cysteine residue which is important to disulfide bond formation and structure stabilization. Through the prediction of GeneScan, we find that the gp IFN-α gene is intronless, Similar with the other mammalian’s IFN-a[12, 13]. A motif of 19 amino acids(YFQRISLYLQEKQYSPCAW) surrounding by
Cys-145 is highly conserved among reference IFN-α sequences, which is a typical characteristic of Interferon alpha, beta and delta family, and may play an important role in the function of IFN-a.
Glycosylation is a complex, coor post-translational protein modification that serves to expand the diversity of the proteome [14]. It can influence the physical properties of the protein, including altered solubility, hydrophobicity, electrical charge and the protein thermal stability [15]. In the mature amino acid sequence of the gpIFN-α, there are one potential N-glycosylation site, eight potential O-glycosylation sites , eleven potential phosphorylation sites and nine antigenic determinants. There are also twenty-one protein-protein interaction sites that are likely to be hot spots for protein interaction. That is, residues that are in the interface between two proteins and that are crucial for the stabilization of the complex. Analysis of the gpIFN-α protein through bioinformatics methods reveals that the protein may good immunogenicity and also undergochemical modifications like glycosylation and phosphorylation.
At the same time, there are 15 rare condons (9.15%) in the giant panda IFN-α gene by using rare condons analysis. And one consecutive rare codons AGACTAAG G also were predicted. All the data initially indicated the excessive rare codons would affect gene’s effective expression and the production of protein. Through analyzing the content of rare codons, it provided theoretical data not only for selecting the suitable host of expression bacterium , but also for improving the gene’s expressing production with reforming codons[16] .
Phylogenetic trees infer the evolutionary relationships of species and patterns of gene duplications in multigene families[17]. In this study evolutionary relationship is analyzed by constructing a phylogenetic tree (shown in Fig. 9) based on the alignment of the IFN-α sequences from 15 distinct genes. The phylogenetic tree demonstrated that strains of the similar always clustered together.
In conclusion, bioinformatics are used to predict and analyze the molecular characteristics and evolution relationship of giant panda’s IFN-a. All the consequences provide a basis data for futher research, We hope that the functional studies on the recombinant IFN-a in future could provide clues to new preventative and therapeutic treatments to the giant panda, which will be quite useful for the conservation of this species.
Acknowledgment
Список литературы Bioinformatics Analysis and Characteristics of the giant panda Interferon-alpha
- C. Samuel, “Antiviral actions of interferons,” Clinical Microbiology Reviews, vol. 14, no. 4, pp. 778, 2001.
- H. Nguyen, J. Hiscott, and P. Pitha, “The growing family of interferon regulatory factors,” Cytokine & growth factor reviews, vol. 8, no. 4, pp. 293-312, 1997.
- M. Colonna, A. Krug, and M. Cella, “Interferon-producing cells: on the front line in immune responses against pathogens,” Current opinion in immunology, vol. 14, no. 3, pp. 373-379, 2002.
- W. Stewart, and J. Vil ek, The interferon system: Springer-Verlag New York, 1979.
- P. Familletti, R. McCandliss, and S. Pestka, “Production of high levels of human leukocyte interferon from a continuous human myeloblast cell culture,” Antimicrobial agents and chemotherapy, vol. 20, no. 1, pp. 5, 1981.
- W. Feng, and G. Li, “The saving of giant panda,” Published by Sichuan Science and Technology Publishing Company, pp. 113-129, 2000.
- X. Tan, Y. Tang, Y. Yang et al., “Gene cloning, sequencing, expression and biological activity of giant panda (Ailuropoda melanoleuca) interferon-[alpha],” Molecular immunology, vol. 44, no. 11, pp. 3061-3069, 2007.
- L. Xu, B. Zeng, R. Peng et al., “Molecular cloning and sequence analysis of the gene encoding interleukin-6 of the giant panda (Ailuropoda melanoleuca),” Journal of Natural History, vol. 42, no. 39, pp. 2585-2591, 2008.
- W. Feng, R. Wang, S. Zhong et al., “Analysis on the dead cause of the anatomical carcass of giant panda (Ailuropoda melanoleuca),” A study on breeding and diseases of the giant panda, pp. 244-248.
- H. Tjalsma, A. Bolhuis, J. Jongbloed et al., “Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome,” Microbiology and Molecular Biology Reviews, vol. 64, no. 3, pp. 515, 2000.
- G. von Heijne, “The signal peptide,” Journal of Membrane Biology, vol. 115, no. 3, pp. 195-201, 1990.
- R. Wonderling, T. Powell, S. Baldwin et al., “Cloning, expression, purification, and biological activity of five feline type I interferons,” Veterinary immunology and immunopathology, vol. 89, no. 1-2, pp. 13-27, 2002.
- R. Devos, H. Cheroutre, Y. Taya et al., “Molecular cloning of human immune interferon cDNA and its expression in eukaryotic cells,” Nucleic acids research, vol. 10, no. 8, pp. 2487, 1982.
- E. Weerapana, and B. Imperiali, “Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems,” Glycobiology, vol. 16, no. 6, pp. 91R, 2006.
- R. Huby, R. Dearman, and I. Kimber, “Why are some proteins allergens?,” Toxicological Sciences, vol. 55, no. 2, pp. 235, 2000.
- S. Zhang, A. Cheng, M. Wang et al., "Molecular Cloning and Nucleotide Sequence Analysis of the Newly Identified UL53 Gene of Duck Enteritis Virus." pp. 1-6.
- S. Kumar, K. Tamura, and M. Nei, “MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment,” Briefings in bioinformatics, vol. 5, no. 2, pp. 150, 2004.


