Comparative Analysis of Stress Tolerance in Millets
Автор: Hosita Gupta, Sugandha Tiwari
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 4 т.21, 2025 года.
Бесплатный доступ
Background: In the current scenario of a changing environment, climate resilient crops like Millets are promising. Various types of stress limit agricultural crop productivity and threaten food security. A comparative analysis of stress tolerance examines the ability of different millet varieties to withstand various environmental stresses like drought, cold, heat, flooding, rainfall, humidity, wind, light, radiation, elevated carbon-di-oxide, salinity, heavy metal, nutrient stress, etc. There are various morphological, physiological, biochemical and genetic characters (genes, transcription factors & proteins) present in millets contributing to their abiotic tolerance. Results: Millet crops possess diverse adaptive mechanisms to combat abiotic stresses like elevated carbohydrate level, low Na+/K+ ratio, decreased relative water content (RWC), decreased chlorophyll levels, increased proline, betaine, increase in the phytohormones like ABA & Jasmonic acid. Millets follow the C4 type carbon fixation pathway. Key genes involved in climate resilience are DREB, LEA, HSP, SOD, ABF, CBF. Key proteins like catalases, peroxidases, aquaporins, Zinc finger, PR-proteins & transcription factors like MYB, NAM, NAC, WRKY, bHLH are involved in stress tolerance. The paper reports the study on evolutionary relationship of ABA responsive bZIP, WRKY, DREB and HSP transcription factors in millets (Eleusine, Setaria, Panicum, Cenchrus, Sorghum) and cereal grasses (Maize and Rice). Conclusions: Millets (Eleusine, Setaria, Panicum) share strong evolutionary ties across all major stress related TF families. Cenchrus aligns with Panicum/ Setaria, but shows independent divergence in some TFs. Sorghum and Maize form a distinct cluster. Rice appears as a distant relative but useful as a reference but closely related to stress resilient millets. Foxtail millet (Setaria italica) has excellent abiotic stress tolerance due to its efficient ROS scavenging mechanism. In Pearl millet (Cenchrus americanus/Pennisetum glaucum) drought responsive genes belong to 8 functional groups -ABA signaling, hormone signaling, ion and osmotic homeostasis, TF mediated regulation, molecular adaptation, signal transduction, physiological adaptations, detoxification. Finger millet (Eleusine coracana) is an important climate smart crop with nutraceutical potential grown for creating sustainability in feed and fodder.
ABA signaling, climate resilience, DREB, ROS scavenging, RWC, signal transduction
Короткий адрес: https://sciup.org/143185124
IDR: 143185124
Текст научной статьи Comparative Analysis of Stress Tolerance in Millets
Millets are C4 respiratory resource & climate smart highly nutritious small grained cereal food, feed and fodder crops. They are eco-friendly requiring lower water, chemicals and human interventions, so a future crop for sustained agriculture and food security. They are on the basis of area grown and size of grain grouped into major and minor millets. Major millets include Sorghum (jowar) and Pearl millet (bajra). Minor millets include Finger millet (ragi), foxtail millet (kangani), little millet (kutki), kodo millet, barnyard millet (Sawan), proso millet (cheena) and brown top millet (korale) are minor millets. The millets contain 7-12% protein,2-5% fat ,6575% carbohydrates and 15-20% dietary fibre. Finger millet is the richest source of calcium (300-350 mg/100g). Millets show antioxidant activity due to the presence of phytates, polyphenols, tannins anthocyanins, phytosterols, and pinacosanols, thus preventing aging and metabolic diseases. Millets possess various morphological, physiological biochemical and molecular mechanisms imparting them climate resilience. So, millets are suitable for the Earth’s changing climate, for the cultivator and the consumer, enriching global biodiversity and our food grain basket. (Bhat, et al. , 2018.).
MATERIALS AND METHODS
For in silico comparative analysis of stress tolerance in millets firstly stress responsive proteins of Oryza sativa retrieved from the UNIPORT database (The UniProt Consortium, 2025) in FASTA format. Each of these retrieved proteins (query sequence) were blasted against various species of millets likewise Eleusine coracana, Setaria italica, Sorghum bicolor, Paspalum scrobiculatum, Cenchrus americanus and grasses like Zea mays via NCBI protein BLAST using non-redundant protein sequences with a threshold 0.05. BLAST (Basic Local Alignment Search Tool) helped to align & identify homologous sequences to query sequences. Ran the multiple sequence alignment (MSA) algorithm and saved the resulting file. MSA identified conserved regions, variation and phylogenetic relationships. A high percent identity with low E-value suggested a strong evolutionary relationship. COBALT (CONSTRAINT
BASED MULTIPLE ALIGNMENT TOOL) was used for constructing phylogenetic trees. For analysing a phylogenetic tree using BLAST results, homologous sequences were aligned using the Neighbor Joining evolutionary model. Also corresponding literature was correlated.
RESULTS
Foxtail millet (Setaria italica L.) has long been used as a model to understand stress responses in plants. The physiological analysis demonstrated that drought stress reduced the RWC and chlorophyll contents of leaves, but increased the REL, while the lengths and surface areas of root systems increased. The comparative proteomic analysis confirmed that the proteins were primarily associated with photosynthesis and leaf development. In contrast, responsive proteins in roots were primarily associated with the metabolic regulation of secondary metabolites and root cell development. The regulatory signaling pathways between leaves and roots in response to drought primarily involved secondary metabolite pathways, hormone regulation, and cell wall synthesis. (Gao, et al., 2023). The transcriptomic studies in Foxtail millet confirmed the upregulation of chloroplastic APX in response to salt stress and selenium supplementation. So, Se act as a mitigant at lower concentrations can alleviate NaCl stress in Foxtail millet. (Saleem, et al., 2023). Foxtail millet has a short growing cycle, lower amount of repetitive DNA, inbreeding nature, small diploid genome, and outstanding abiotic stress-tolerance characteristics The qRT-PCR revealed the post-transcriptional modification has crucial roles in regulating gene expression. Stress stimulates a regulatory network of different metabolic pathways (Pan, et al., 2018). S. italica and S. viridis reported stress-specific upregulation of SiαCaH1, SiβCaH5, SiPEPC2, SiPPDK2, SiMDH8, and SiNADP-ME5. Pearl millet (Pennisetum glaucum L.) is widely grown in arid and semi-arid regions, being tolerant to drought, due to certain molecular and genetic mechanisms operating within the crop. (Chakraborty, et al., 2022). Eleusine coracana (L.) revealed the activation of various genes involved in response to stress, specifically heat, oxidation-reduction process, water deprivation, and changes in heat shock protein (HSP) and transcription factors, calcium signaling, and kinase signaling. The heat stress activates genes such as bZIP involved in basal regulatory processes. Thus, the candidate genes, such as HSPs, are involved in tolerance towards heat stress in ragi (Goyal, et al. 2024).
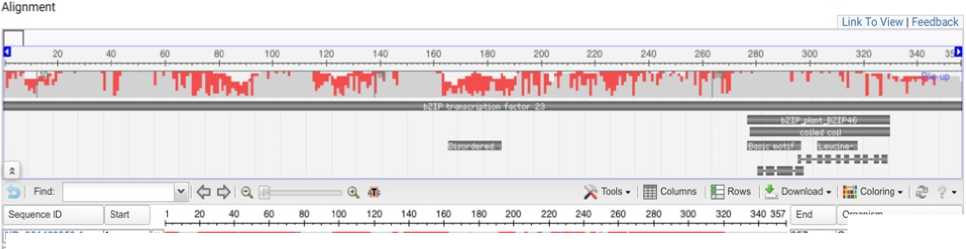
Multiple sequence alignment of bZIP transcription factors is shown on Figure 1. BLAST result showing description of stress responsive bZip transcription factors. Red blocks signify strong sequence similarity and functional conservation of bZIP family among millets and grasses. The sequences are orthologous, evolved from a common ancestor and involved in ABA mediated drought or stress signalling.
Phylogenetic tree showing the evolutionary relationship of ABA responsive bZIP transcription factors is shown on Figure 2. bZIP protein (PR202_ga02861) of Eleusine coracana (finger millet) closely clusters with known ABA-responsive transcription factors such as OsbZIP23 ( Oryza sativa ), ZmABF2 ( Zea mays ), and ABRE-binding factor 2 ( Cynodon dactylon ). This strongly indicates that the Eleusine bZIP protein is likely involved in ABA-mediated drought or stress signaling. Sorghum bicolor contains bZIP proteins that fall into a separate clade alongside maize’s bZIP46 indicating functional diversification. bZIP of Setaria italica shows more diversification.
Multiple sequence alignment shows that WRKY transcription factor proteins are involved in stress responses (Figure 3). WRKY sequences of Oryza sativa show highly conserved regions (red) among aligned sequences with Setaria, Paspalum, Eleusine, Sorghum, Zea mays and Panicum. Highly conserved regions of WRKY transcription factors show shared function across species and that they have evolved from a common ancestor with similar stress regulation pathways.
Phylogenetic tree (Figure 4). WRKY showing common ancestor, collinear relationships and evolutionary conservation. Proso millet (Panicum miliaceum), foxtail millet (Setaria italica) and Eleusine (Eleusine coracana) are placed in the same clade, suggesting a closer evolutionary relationship. Sorghum (Sorghum bicolor) is closer to maize and Rice (Oryza sativa) is a distant outgroup in the phylogenetic tree.
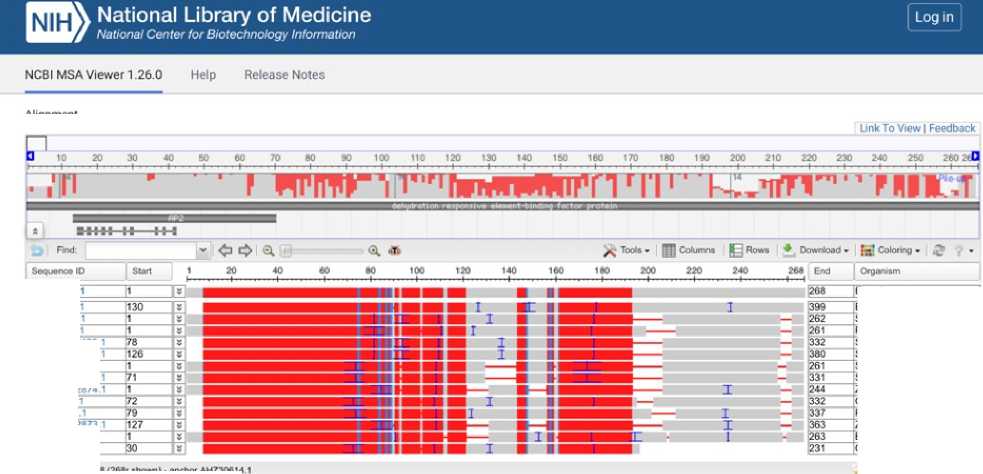
Multiple alignment: DREB transcription factor proteins shown on Figure 5. Eleusine coracana, Setaria italica, Sorghum bicolor, Cenchrus americanus, and Zea mays show very close alignment, indicating orthologous relationship. The evolutionary conservation in AP2 domain highlights the importance of DREB protein in drought stress response in millets, maize and other grasses.
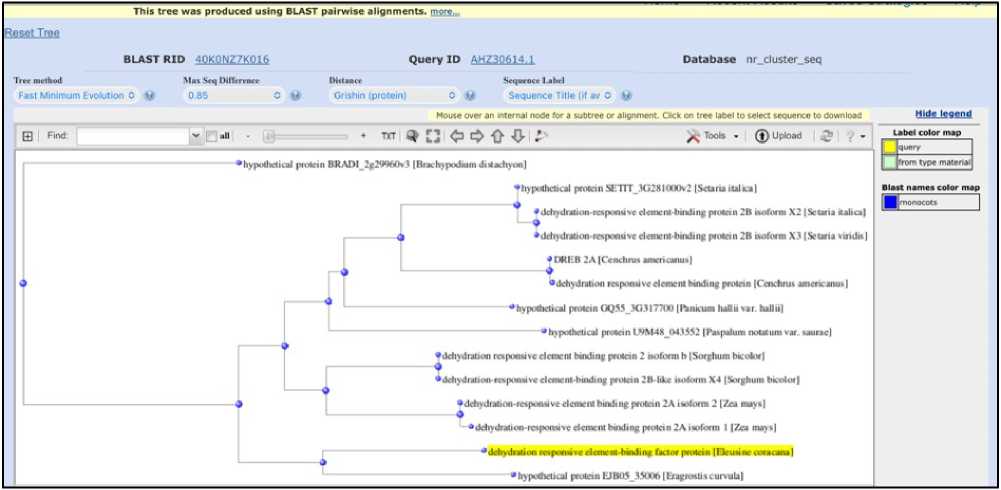
Phylogenetic tree of DREB shown on Figure 6. DREB transcription factors play a very important role in abiotic stress responses. Phylogenetic tree of DREB protein of Eleusine coracana shows a high sequence similarity with Eragrostis, Maize and Sorghum. Cenchrus (Pearl millet), Paspalum and Panicum show moderate evolutionary relationship with the DREB transcription factor from Eleusine coracana . Setaria and Brachypodium show early divergence and fast evolutionary rate of its DREB protein.
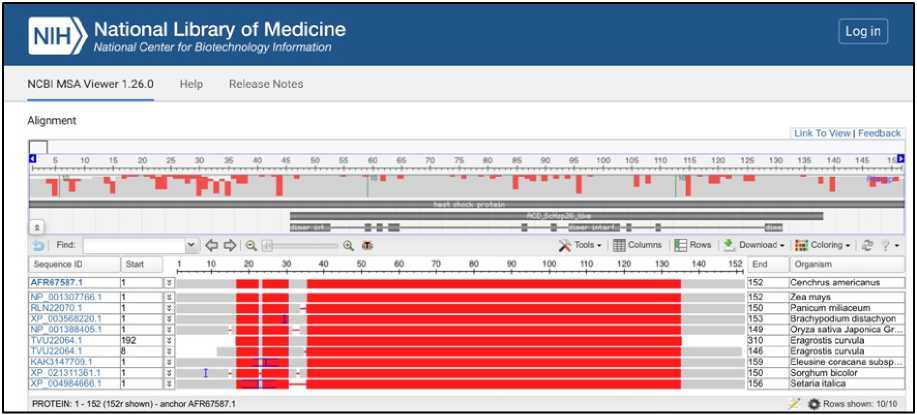
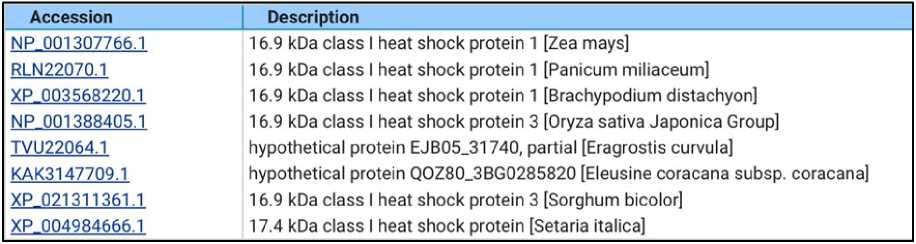
Multiple sequence alignment of Heat Shock Proteins is shown on Figure 7. Heat shock domain is highly conserved in Cenchrus, Maize, Brachypodium, Oryza sativa japonica group, Eragrostis, Eleusine, Sorghum and Setaria. The high sequence identity supports its functional conservation in stress responses among monocots.
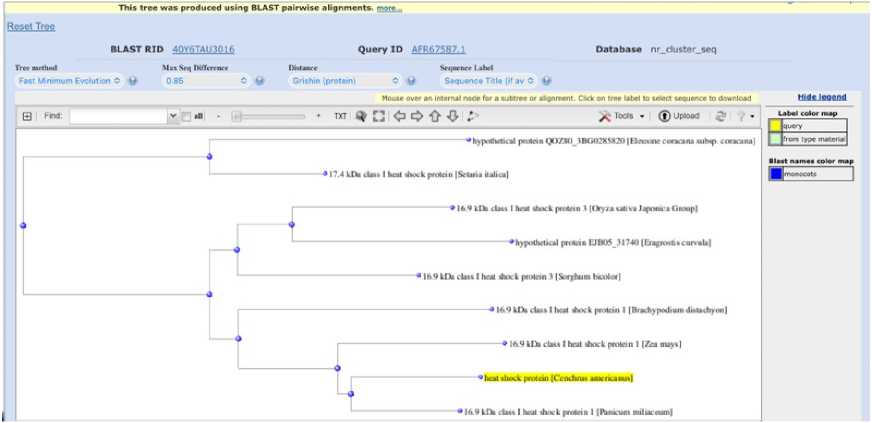
Phylogenetic tree showing the evolutionary relationship of HSP is shown on Figure 8. Cenchrus americanus, Eleusine coracana, Setaria italica, Zea mays, Sorghum bicolor , and others share a common evolutionary origin for Heat Shock Protein gene. The conservation of the heat shock protein across these species suggests it has an essential role in stress tolerance (heat, drought). The evolutionary closeness is especially strong between Eleusine and Setaria.
Table 1. Showing rainfall requirements & climate resilient traits in different millet crops. (Source: B Venkatesh Bhat, B Dayakar Rao & Vilas A. Tonapi. (2018) The Story of Millets, Karnataka state Department of Agriculture, Bengaluru, India with ICAR -IIMR, Hyderabad, India.)
|
MILLET CROP |
RAINFALL (Cms.) |
TRAITS |
|
PEARL MILLET |
80-95 |
Highly resilient to heat and drought, come up in very poor soils, but responsive to high input management |
|
FINGER MILLET |
90-130 |
Moderately resistant to heat, drought and humidity, adapted to wide altitude range |
|
FOXTAIL MILLET |
70-120 |
Adapted to low rainfall, high altitude |
|
KODO MILLET |
100-140 |
Long duration, but very hardy, needs little rainfall, comes up in very poor soils, good response to improved management |
|
SORGHUM |
100-125 |
Drought tolerant, excellent recovery mechanism from stresses, highly adapted to wide range of soils, altitudes and temperatures, responsive to high input management |
|
BARNYARD MILLET |
45-60 |
Very short duration, not limited by moisture, high altitude adapted |
|
LITTLE MILLET |
70-110 |
Adapted to low rainfall and poor soils- famine food; withstand waterlogging to some extent |
|
FONIO |
75-120 |
Shorter duration, Adapted to poorly fertile sandy and stony soils, low rainfall |
|
TEFF |
60-120 |
Short duration, drought and flood tolerant, high altitude adapted, fit in diverse cropping systems |
|
BROWN TOP MILLET |
60-80 |
Short duration, adapted to poor soils with less rainfall |
|
PROSO MILLET |
60-90 |
Short duration, low rainfall, high altitude adapted |
Table 2. Showing varied stress tolerance in millets
|
MILLET CROP |
STRESS TOLE RANCE |
MORPHOLOGICAL/ PHYSIOLOGICAL/ BIOCHEMICAL RESPONSE |
GENOMIC RESPONSE |
TRANSCRIPTO-MIC RESPONSE |
PROTEOMIC RESPONSE |
REFERENCE |
|
Setaria italica. L (foxtail millet) |
Drought |
Root length increased, decrease in RWC, decrease in chlorophyll content, increase in REL. |
MPK6, PAL, PER12, CA2, GAPA-2, ADF11 |
MAPK6, PAL1, PAL2, LOS4, s-APX, SIP2, BGL2, FUC1 |
Gao, et al. , 2023 |
|
|
Drought |
Arginine decarboxylase,spermidine synthase, polyamine oxidase, shikimate o-hydroxycinnamoyl transferase,caffeic acid 3-o-methyl transferase, cinnamoyl -co-A reductase, purple acid phosphatase, GPP,Callose synthase, tubulin α-1 chain, tubulin β-1. |
LEA, ASR, nsTLP, Defensin -like protein, SOD, CYSTEINE PROTEINASE INHIBITOR 8-like, POD, PIP, PAP, SiRLK, SiRLK, SiCAT, SiAOS, PPDK, NADP, ME, QUINOR, GAPH, SiFBA, SiFBA, SiPOD, SiTubu |
CAT, GST, Thioredoxin, MEG, AQUAPORIN, PIP2, HSP-70, EXPANSIN -B3-LIKE, WD40, PAD |
Pan, et al. , 2018 |
||
|
Drought |
Lignin metabolism, metabolism of fatty acids |
Cui, et al. , 2023 |
|
(cutin & wax) |
||||||
|
Salinity |
ROS SCAVENGING |
Upregulation of APX |
Saleem, et al. , 2023 |
|||
|
Low potassium stress |
SiMYB3, AP2, NAC,HOMEOBO X,bHLH,WRKY,B zip,NF,PLATZ,M ADS |
Cao, et al. , 2019 |
||||
|
Setaria italica. L. & S. viridis. L. |
Low potassium stress |
SiSnRK2.6 (ABA SIGNALING) |
Ma, et al. , 2024 |
|||
|
Stress responsiv e |
CaH,PEPC,PPD K,NADP-dependent MDH, NADP-ME |
Muthamilorasa n et al. , 2020 |
||||
|
Pennisetu m glaucum L. (pearl millet) 2n=14 |
DROUGH T |
Root architecture, stay green property, leaf rolling, stomatal closure, decreased photosynthesis, water use efficiency, transpiration efficiency, accumulation of Osmoprotectants proline, betaine, reduced accumulation of ROS, APX, SOD, CAT, NADH, PROLINE, CAROTENOIDS, GSH, KAT2, HAB1, EDT1, RGS1, SRK2C, TPS1, ERD1, GolS2, P5CS1 |
AtBG1, GIG1, GIG2, PYL9, AAO3, CYP707A1, CYP707A3, NCED, PIP2, PIP1, OST2, ADAP, FAR1, HSPA1b, AREB1, SNAC1, MYB6 |
Chakraborty, et al. , 2022 |
||
|
Pennisetu m glaucum L. (pearl millet) 2n=14 |
HEAT |
upregulation of glycerophospholipid metabolism, down regulation of photosynthesis antenna proteins |
HSPs |
Sun, et al. , 2022 |
||
|
Pennisetu m glaucum L. (pearl millet) 2n=14 |
salinity |
C4 photosynthesis Enzymes, efficient enzymatic and non-enzymatic antioxidant system, lower Na+/K+ ratio |
Jha, et al. , 2022 |
|||
|
Pennisetu m glaucum L. (pearl millet) 2n=14 |
Drought |
REC increased, ROS scavenging SOD, CAT, APX |
Upregulation of NADP-ME, Zinc finger, |
bHLH,bZIP,MYB, NAC |
Zhang, et al. , 2021 |
|
Eleusine coracana (finger millet) |
Heat |
Decreased electrolyte leakage, ROS Scavenging, SOD, CAT, GR, POD, APX, MDA |
EcDREB2A |
Singh, et al. , 2024 |
||
|
Panicum miliaceum L. (Proso millet) |
Drought |
POD, SOD, CAT, MDA, JA |
Zhang, et al. , 2019 |
■ I National Library of Medicine
Login
’ National Center for Biotechnology Information
NCBI MSA Viewer 1.26.0 Help Release Notes
Organism
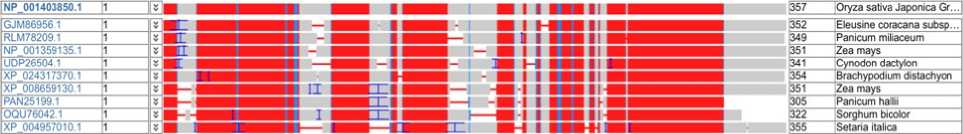
Figure 1: Multiple sequence alignment of bZip transcription factors.

PROTEIN: 1 - 357 (357r shown) - meta NP_001403850.1 / 0 Rows shown: 10/10
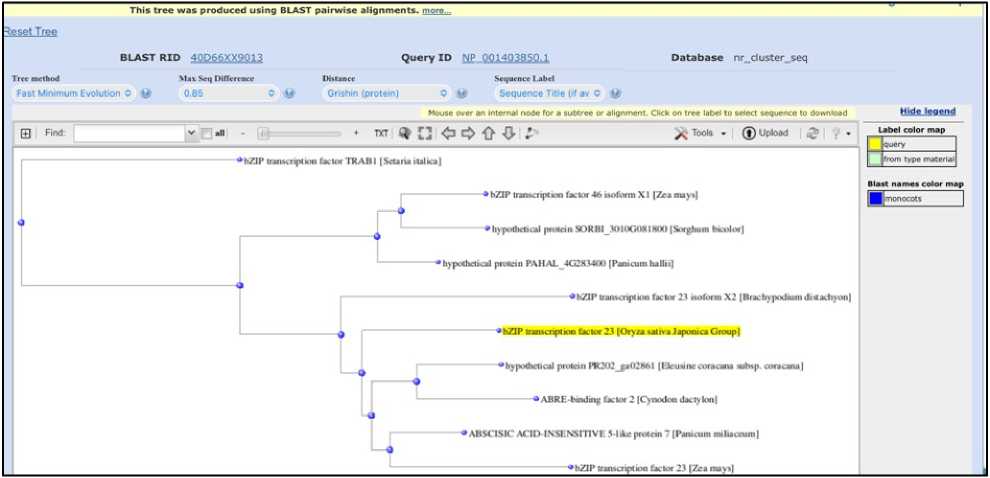
Figure 2: Phylogenetic tree showing the evolutionary relationship of ABA responsive bZIP transcription factors.
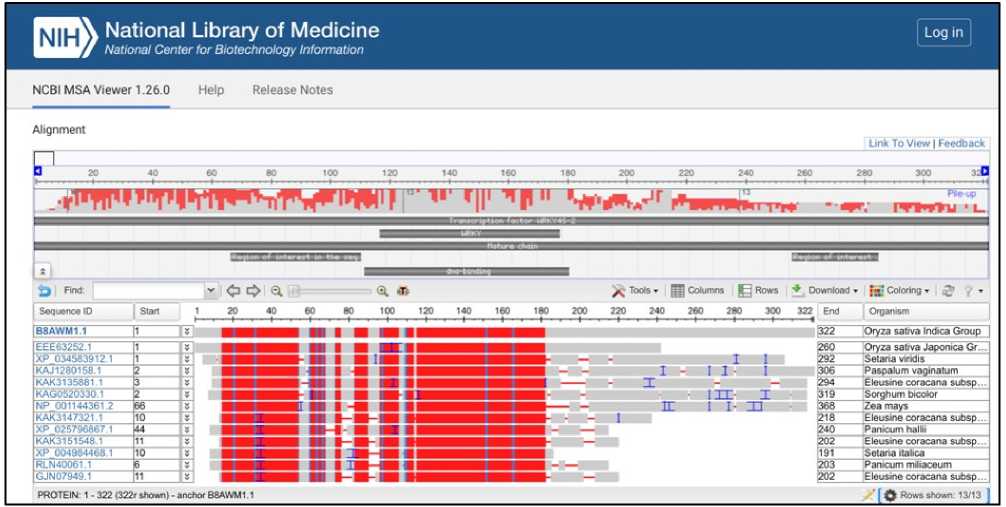
Figure 3: Multiple sequence alignment: WRKY transcription factor proteins are involved in stress responses.
This tree was produced using BLAST pairwise alignment». ■ i
RfiSSlTrsg
BLAST RID 42EQTMJHQ16
Query ID B8AWM1.1
Database nr_c!uster_seq
Tre* method Mn Srg lliflrrtncr l>nt» Fast Minimum Evolution 0 M 0.85 С V Grishin (protein) Sequence Title (if tv 0 Mouse over an internal node for a subtree or alignment Click on tree label to select sequence to download HyiS J«g« ng 0 Find txt Q ФФО о (DUpted Э hypothetical pruaaaQOZM) 1ВСХ12ЯОи2О||-Тешак coracana сиЫр coracana) 'hypothetical proteinQO290 IAGO24711O |Ekuwne coracana nibcp. coracana) 'hypothetical prolan Ж>>2_£а25)12б (Неимпссосасапа ш!мр core canal ♦ WRKY inmcnpUon factor 55 like | Ph галит haUu| 'putative WRKY transcription factor 70 isofcinn X2 IPaniciun miiiaccuml ♦ pruhaNe WRKY tran* r^eiun (actor 70iMiformX2 |Setana italical hypothetical prainn QOZM)_5BG * unchar ас lai/ed proton L ОС 100277272 (Zea mays) iranccnptton factor WRKY45 2 (Setana viridB) hypothetical prolan BS7RO4G210200 (Paspak mil «hypothetical ptuaeia BIMWJMIGOM1001Sukhum bwolcrl ♦ RecName l ulUTrasKT^tun factor URKY4S 2. Shwt= Figure 4: Phylogenetic tree of WRKY proteins. XP_021302434 1 [AHZ30614 1 Elousr»o coracana Eragrostis curvuia TVU188881 RCV18201 1 Setana । fa Itca AAV90624 1 NP_0012928f 3.1 KQK06813 1 AEY70S121 0 Rom PROTEIN 1 -268 (2Wr shown) -anchor AHZ30814 Figure 5: Multiple alignment: DREB transcription factor proteins. Alignment PU266520 1 XP О J1'- .■: : ■ ' •• ^глнси 1 AFI712931 NP_001292874 1 Panicum ha Hu var hallii Setana itai»ca Setana vindis Sorghum bicoior Sorghum bicolor________ Zea may» Cenchrus amencanus Paapaium notatum var $ Z— may» Brachypodium Astachyon Cenchrus amencanus Figure 6: Phylogenetic tree DREB. Cwncbr TVU22064 Brachypodum Astachyon Oryza Mbv* Japonic# Gr Eragrostis curvuia Eragrostis curvuia __ Clawtine coracana aubop. Sorghum bicotor Setana rtalca___________ NCBI MSA Viewer 1.26.0 Help Release Notes Alignment Link To View | Feedback C ФФ A ^ Tools • У Columns E Rows ♦, Download <2^ 4ft £ Coloring 130 140 152 End Organem X National Library of Medicine у National Center for Biotechnology Information Log in Ф Rows shown 10/10 PROTEIN 1-152 (152r shown)-anchor AFR67587 1 NP001307766 1 1 MSLVRRS-NVFDPFSMDLWDPFDTMFR SIVP-SAT-sTNSETAAFASARIDWKETPEAHVFKADLPGVKKEEVKVEV 74 RLN22070.1 1 MSLVRRS-SVFDPFSLDLWDPFDSMFR SWP-SAG DSETAAFASARIDWKETPEAHVFKADLPGVKKEEVKVEV 72 XP 003568220 1 1 MSLVRRG-SVFDPFSQDLWDPIDSIFR SIVP-AAAasSDFDTAAFVNARMDWKETPEAHVFKADLPGVKKEEVKVEV 75 NP 001388405.1 1 MSLVRRS-NVFDPFA-DFWDPFDGVFR SLVP-ATS---DRDTAAFANARVDWKETPESHVFKADLPGVKKEEVKVEV 71 TW»W.l 1 MSLQRVR-----PFLLDLWDPFDNMFR SIVPsASS DSDTAAFANARIDWKETPEAHVFKADLPGVKKEEVKVEV 69 KAK3147709.1 1 MSLIRRS-NIFDPFSLDLWDPFQ-GFP(9]SLFP-RTS—SESETAVFAGARIDWKETPEAHVFKADVPGLKKEEVKVEI 81 XP 021311361 1 1 MSLVRRStNVFDPFA-DFWDPFD-VFR SIVP-AAS—TDRDTAAFANARIDWKETPEAHVFKADVPGVKKEEVKVEV 72 XP 004984666 1 1 MSLIRRS-NVFDPFSLDLWDPFQ-GFP(6 JSLFP-RIP—SDSETAAFAGARIDWKETPEAHVFTADVPGLKKEEVKVEV 78 NP001307766 1 75 EDGNMLVISGQRSREKEDKDDKWHRVERSSGQFVRRFRLPENTKVDQVKAGLENGVLTVTVPKAEEKKPEVKAIEISG 152 RLN22070.1 73 EDGNVLVISGQRSREEEDKNDRWHRVERSSGQFKRRFRLPENAKVDQVKAGLENGVLTVTVPKAEEKKPEVKSIOISG 150 XP003568220 1 76 EDGNVLWSGERSREKEDKNDKWHRVERSSGKFVRRFRLPENAKVEQVKAGLENGVLTVTVPKSEVKKPEVKAIEISG 153 NP 001388405 _L 72 EEGNVLVISGQRSKEKEDKNDKWHRVERSSGQFMRRFRLPENAKVDQVKASMENGVLTVTVPKAEVKKPEVKAIEISG 149 TVU22064.1 70 EDGNVLVISGERSKEKEDKNDKWHRVERSSGQFMRRFRLPENAKVDQVKAGLENGVLTVTVPKAEVKKPEVKAIEISS(17 1] 318 КАК314770Э.1 82 EDGNVLQISGERNKEQEEKNDKWHRVERSSGKFMRRFRLPENAKTEQIKASMENGVLTVTVPKEEVKKPEVKSIQISG 159 XP 021311361 1 73 EDGNVLVISGERRKEKEDKDDKWHRVERSSGRFMRRFRLPENAKTEEVKAGLENGVLTVTVPKAEVKKPEVKSVEIAG 150 XP 004984666 A 79 EDGNVLQISGERSKEQEEKNDKWHRVERSSGKFLRRFRLPENAKTEQIKASMENGVLTVTVPKEEVKKPEVKPVQITG 156 Figure 7: Multiple sequence alignment: Heat Shock Proteins. Accession Description NP_001307766.1 RLN22070.1 XP_003568220.1 NP 001388405.1 TVU22064.1 KAK3147709.1 XP 021311361.1 XP_004984666.1 16.9 kDa class I heat shock protein 1 [Zea mays] 16.9 kDa class I heat shock protein 1 [Panicum miliaceum] 16.9 kDa class I heat shock protein 1 [Brachypodium distachyon] 16.9 kDa class I heat shock protein 3 [Oryza sativa Japonica Group] hypothetical protein EJB05_31740, partial [Eragrostis curvuia] hypothetical protein QOZ80_3BG0285820 [Eleusine coracana subsp. coracana] 16.9 kDa class I heat shock protein 3 [Sorghum bicolor] 17.4 kDa class I heat shock protein [Setaria italica] Figure 8 Phylogenetic tree showing the evolutionary relationship of HSP. DISCUSSION The paper reports the study on evolutionary relationship of ABA responsive bZIP, WRKY, DREB and HSP transcription factors in millets (Eleusine, Setaria, Panicum, Cenchrus, Sorghum) and cereal grasses (Maize and Rice). ABA bZIP TFs are functionally conserved among millets, suggesting shared drought adaptation strategies. WRKY TFs in millets evolved under similar stress response selection pressures, while Sorghum and Maize show divergence. High sequence similarity across millets and cereal grasses show the conserved role of WRKY TFs in drought responses. DREB TFs evolved early in monocots but diversified functionally in stress prone millets and provided adaptive advantage in arid climates. High sequence similarity in HSP TFs shows evolutionary conservation and their role in heat stress resilience. Millets (Eleusine, Setaria, Panicum) share strong evolutionary ties across all major stress related TF families. Cenchrus aligns with Panicum/ Setaria, but shows independent divergence in some TFs. Sorghum and Maize form a distinct cluster. Rice appears as a distant relative but useful as a reference but closely related to stress resilient millets. Stress responsive proteins in millets have future scope to be incorporated into contemporary crops for improving their abiotic stress tolerance. CONCLUSIONS Millets (Eleusine, Setaria, Panicum) share strong evolutionary ties across all major stress related TF families. Cenchrus aligns with Panicum/ Setaria, but shows independent divergence in some TFs. Sorghum and Maize form a distinct cluster. Rice appears as a distant relative but useful as a reference and is closely related to stress resilient millets. Foxtail millet (Setaria italica) has excellent abiotic stress tolerance due to its efficient ROS scavenging mechanism. In Pearl millet (Cenchrus americanus/Pennisetum glaucum) drought responsive genes belong to 8 functional groups – ABA signaling, hormone signaling, ion and osmotic homeostasis, TF mediated regulation, molecular adaptation, signal transduction, physiological adaptations, detoxification. Finger millet (Eleusine coracana) is an important climate smart crop with nutraceutical potential grown for creating sustainability in feed and fodder. CONFLICTS OF INTEREST All authors declare that they have no conflicts of interest.