COVID-19 Automatic Detection from CT Images through Transfer Learning
Автор: B. Premamayudu, Chavala Bhuvaneswari
Журнал: International Journal of Image, Graphics and Signal Processing @ijigsp
Статья в выпуске: 5 vol.14, 2022 года.
Бесплатный доступ
Identification of COVID-19 may help the community and patient to prevent the disease containment and plan to attend disease in right time. Deep neural network models widely used to analyze the medical images of COVID-19 for automatic detection and give the decision support for radiologists to summarize the accurate remarks. This paper proposed deep transfer learning for chest CT scan images to detection and diagnosis of COVID-19. VGG19, InceptionRestNetV3, InceptionV3 and DenseNet201 neural network used for automatic detection of COVID-19 disease form CT scan images (SARS-CoV-2 CT scan Dataset). Four deep transfer learning models were developed, tested and compared. The main objective of this paper is to use pre-trained features and converge pre-trained features with targeted features to improve the classification accuracy. It is observed that DenseNet201 noted the best performance and the classification accuracy is 99.98% for 300 epochs. The findings of the experiments show that the deeper networks struggle to train adequately and give less consistency when there is limited data. The DenseNet201 model adopted for COVID-19 identification from lung CT scans has been intensively optimized with optimal hyper parameters and performs at noteworthy levels with precision 99.2%, recall 100%, specificity 99.2%, and F1 score 99.2%.
Deep Transfer Learning, COVID-19, Classification, Pre-trained features.
Короткий адрес: https://sciup.org/15018728
IDR: 15018728 | DOI: 10.5815/ijigsp.2022.05.07
Текст научной статьи COVID-19 Automatic Detection from CT Images through Transfer Learning
The world is impacted by the COVID-19 pandemic and many people were lost their lives since late December 2019. It was identified in Wuhan, China. The United Nations (by means WHO) named it as COVID-19 on February 11, 2020. Later the World Health Organization (WHO) has stated it was a pandemic worldwide. The novel coronavirus was classified as SARS-CoV-2 (severe-acute-respiratory-syndrome coronavirus 2) by the international committee on taxonomy of viruses. It can spread from person to person. The novel coronavirus is infected worldwide with over 42,48,22,078 confirmed cases and over 58,90,312 deaths as of 23rd February 2022 by WHO from national authorities.
The developing and under-developing countries with private health facilities were not even provided the virus diagnostic tests for their citizens. The patients and governments are causing financial problems even for COVID-19 diagnostic tests. While testing the virus detection a small amount of viral RNA is taken from a nasal swab, amplified, and quantified. The gold standard for COVID diagnosing is RT-PCR (reverse transcriptase quantitative polymerase chain reaction). Unfortunately, the RT-PCR diagnostic is time-consuming and manual. The results of the RT-PCR test will take up to 2 to 3 days. Therefore, the patient will not get the prescribed medicine in time. Due to less study on the COVID-19 virus structure and its activities on the human race, sometimes it may damage the infected person in no time. Hence, this situation demands another way of the instant diagnostic test for COVID-19 identification with high accuracy. The high accurate diagnostic test for COVID-19 can help the patient to take therapeutic measures and save the life. Imaging-based diagnostic approaches are adopted to detect the COVID-19 virus infection, which includes X-Ray imaging, CT (computed tomography) imaging, and Ultrasound imaging. There can be software based diagnostic tool to identify COVID-19 patients from radiology images.
Identification of COVID-19 patients in early diagnosis can save lives. The best way to early diagnosis of disease through chest computer tomography (CT) imaging is recommended [1]. The main feature of computer tomography imaging compared to X-ray imaging is the nature of the lesion such as distribution, shape, density, concomitant, and quantity symptoms. Our proposed deep transfer learning neural network architecture can address the issues of CT scan medical image classification. In particular, it is used for automatic detection of COVID-19 disease.
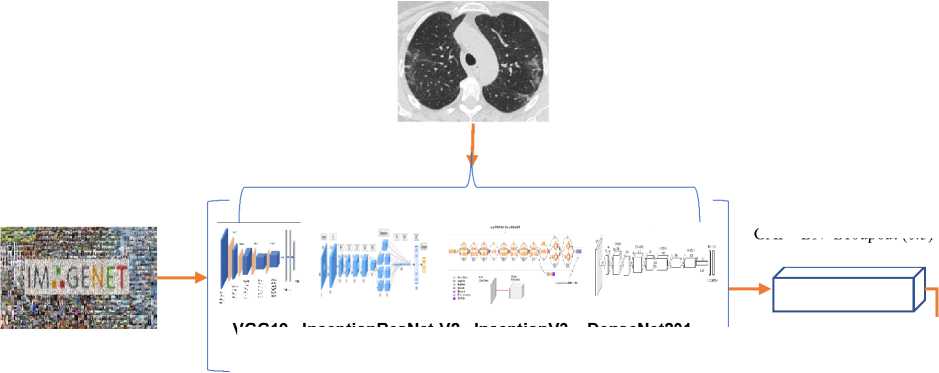
The image processing extracted the lung or chest area and improve the quality of the image. Thereby CT image feature lesion is elevated from the ground truth. The open data set of chest CT scan images of COVID-19 (SARS-CoV-2 data set) is evaluated in our proposed model. The data set was publicly available. The performance matrices of the deep learning model like accuracy, loss, sensitivity, and specificity are outperformed compared to other deep learning classification models. Fig. 1 defines our proposed deep transfer learning neural network generic architecture for image classification.
The primary objective of our study is to provide an automatic COVID-19 detection framework through transfer learning using radiology images. The deep learning model is initialized with pre-weighted from the previous training cycle and later stage weights are fine-tuned with available limited medical image data set. Deep learning and convolutional neural networks are most affective in image classification.
The major key contributions of our study are listed as follows.
1. Introduced deep transfer learning architecture for COVID-19 disease identification through radiology images.
2. The image pre-processing steps to reduce the noise and unwanted non-lung area in CT images. It can improve
3. Implemented the proposed architecture and presented the result analysis compared with existing deep transfer learning models. Further, identified the suitable transfer learning model for selected data set (SARS-CoV-2 CT scan Dataset).
4. Comparative study on several common off the-self CNN models like VGG19, InceptionResNet-V2, InceptionV3, and DenseNet201.
5. Our CNN model with DenseNet201 in combination with image pre-processing pipeline to detect/predict COVID-19 and non-COVID-19 disease with 99.02 accuracy.
2. Related Work
the quality of CT image and minimize the sampling bias.
The COVID-19 infection was identified through numerous attempts. COVID-19 infection can be detected from chest X-ray images or radiology images. COVID-19 can cause bilateral pulmonary parenchymal ground-glass and consolidative pulmonary opacities, sometimes with a rounded morphology and a peripheral lung distribution, according to X-ray findings. The accuracy rate to determine the COVID-19 infection at lung area through X-ray images is acceptable. Software tools like machine learning, deep learning improve the accuracy rate to determine the disease through X-ray images then radiologist. The accuracy of identification cab be improved further by extracting the area of interest in the X-ray images. However, differentiating the COVID-19 infection and pneumonia from X-ray images is very difficult through image processing or computer vision. The alternative and more available radiology image is Computed Tomography (CT). The CT scan medical imaging is also cost effective and accessible to the infected patients and radiologist [1]. The use of a chest CT-scan was strongly advised because imaging findings vary depending on the patient's age, immune state, disease level at the time of scanning, underlying conditions, and pharmacological treatments [2].
Unenhanced chest CT can be taken into consideration for early analysis of viral disease, even from the standard procedure to dragonize COVID-19 infection using RT-PCR (real-time polymerase chain reaction) [3]. Several examples are noted that the standard operating procedure to identify the viral infection results negative in RT-PCR but identified as positive result through CT scan [4]. Compared to chest CT images RT-PCR gives the false-negative outcomes in lots of case. Many studies are noted that chest CT images gave the better accuracy rate for infection identification. Thereby this study was motivated to develop an automatic disease identification using deep learning techniques. In particular, deep transfer learning is selected model for detection. There is a secondary opinion for medical practitioner via deep transfer learning model. Through a preliminary comparison investigation of various common CNN models, we discover an appropriate Convolutional Neural Network (CNN) model. The chosen VGG19 model is then optimized for the picture modalities to demonstrate how the models may be employed for the very limited and difficult COVID-19 datasets [5]. Because of its extensive feature extraction functionality, artificial intelligence (AI) using deep learning technology has recently showed remarkable success in the medical imaging arena. Deep learning was primarily used to detect and distinguish between bacterial and viral pneumonia in pediatric chest radiographs [6] database can dramatically improve identification of COVID-19. This will provide extensive information for training and testing a Deep learning-based system, most likely using some sort of transfer learning.

Transfer learning
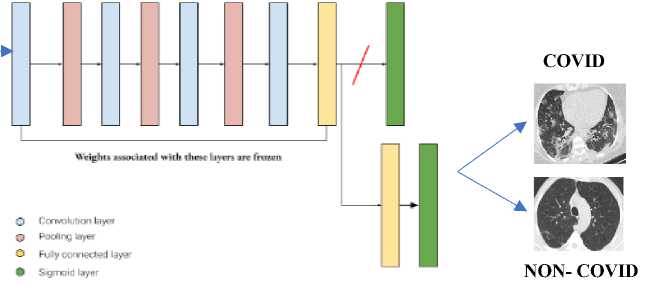
Fig. 1. Automatic COVID-19 disease detection architecture
These approaches will be developed in order to identify COVID-19 characteristics in comparison to various forms of pneumonia or to predict survival [7]. Pleural effusions, lymphadenopathy, pulmonary nodules, and lung cavitation are common chest CT findings, and more than half of patients scanned shortly after symptom start have a normal CT scan [8]. In the assessment of unskilled supply chain management, an Internet-of-things-based architecture for big data evaluation [9] was developed (GSCM) [10, 11]. Medical imaging may be simulated using computational intelligence. Through simulation and visualization, computational intelligence can enable powerful and efficient healthcare research [12]. Meanwhile, because deep recurrent neural networks (DRNNs) have a great capacity to autonomously extract features from time series data, HAR systems based on DRNNs have achieved more successful recognition [13]. Intelligent secure ecosystem based on Metaheuristic and practical link Neural network for edge of factors [14]. The LSTM's convolutional structure enables it to simultaneously record long-term relationships and extract functions from the time–frequency domain [15]. They used CT scans and a three-dimensional convolutional neural network known as VB-net to do automated segmentation to determine the whole lung and infected web locations. Numerous studies have utilized exceptional enhanced learning strategies to identify covid-19[16]. To analyze COVID-19 using X-ray pictures and have worked with VGG16, VGG19, ResNet, DenseNet, and InceptionV3 models. For the study of COVID-19, can be categorized X-ray snap pictures using five distinct feature extraction methods: grey-level co-occurrence matrix (GLCM), local directional patterns (LDP), grey-level run length matrix (GLRLM), grey-level size area Matrix (GLSZM), and Discrete Wavelet transform (DWT) [17]. When analysing COVID-19-infected patients, adopted a transfer of knowledge approach for processing X-ray chest pictures. The remaining one exceeded expectations with a 98 percent accuracy rate in their evaluation. After a thorough assessment of the literature, it's clear that researchers have been increasingly relying on CT imaging to obtain improved results to predict the COVID-19 [18].
VGG19: In 2014, the ImageNet challenge submission used the VGG network topology with 19 layers (VGG19). This model defines several layers for feature extraction and dimensional reduction to minimize the computational cost. It defines 16 Conv layers, 3 fully connected layers, 5 max poll layers and one softmax for final result. For convolutional layers, the filter size 3 x 3 with stride 1 and padding 1 is used. After every convolution, the ReLU activation function is used for non-linearity. The max polling is used for dimension reduction. It uses 2x2 filter, stride 2 and same padding. This deep learning network has 19.6 billion FLOPs [19]. There are other variants of VGG networks for classification like VGG11, VGG16 and other. It is improved the COVID-19 CT image classification performance compared to traditional deep learning models.
InceptionResNet-V2: The network with residual inception block extracts the deep feature space for classification. It uses the cheaper inception blocks for scaling up the dimensionality of filter bank [20]. The InceptionResNet-V2 can retrain to perform new task using transfer learning. Generally, the common approach is to pre-train the network on the ImageNet dataset. However adding it to the ensemble seemed to improve the results marginally [21].
InceptionV3: Another deep learning model trained on image with notable performance is InceptionV3. It is used for image classification deep learning network with accuracy of 78.1% on the ImageNet dataset. The model is constructed using symmetric and asymmetric building elements such as convolutions, average pooling, maximum pooling, concatenations, dropouts, and fully connected layers. The batch normalization is applied to activation inputs and loss of the model is calculated with softmax function [22].
DenseNet201: DenseNet is a relatively recent development in the field of neural networks for visual object detection. DenseNet is relatively similar to ResNet with a few key distinctions. ResNet employs an additive approach that concatenates the output of the previous layer (identity) with the output of the subsequent layer, whereas DenseNet concatenates the output of the previous layer with the output of the subsequent layer. DenseNet was created primarily to address the vanishing gradient effect in high-level neural networks. In simplest terms, because the path between the input and output layers is longer, the information is lost before it reaches its destination [23].
However, the shortcoming of the existing deep learning models for medical image classification using transfer learning on limited data are addressed in our proposed model. A DenseNet201 based deep transfer learning is the best fit for COVID-19 CT image classification and yielded the best results compared with other deep transfer learning models.
3. Network Architecture
The procedure to reuse the previously trained model in the other deep learning networks is known as transfer learning. Fig. 2 illustrates the fundamental concept of transfer learning. The key idea is to start with a somewhat complicated and effectively trained model, trained on a big number of source data, such as ImageNet, and then apply the obtained knowledge to real task (classify covid-19 or non-covid19) with required domain.
Transfer learning provides three important characteristics to make effective deep learning model for classification: (i) To avoid hyper-parameter tuning (ii) extraction of low-level features like textures, contours, tints, edges and shades, and (iii) the last layers of pre-trained model is placed with features of target task (classify covid-19 or non-covid-19) [24].
This paper adopted well known deep learning neural network models for image classification on ImageNet such as VGG19, InceptionResNet-V2, InceptionV3 and DenseNet201 for COVID-19 prediction/detection using Lung CT images. We have shown the comparison results in the section 4 of this article. The results shown that the DesneNet201 surpasses the other transfer learning models. Therefore, we have adopted DenseNet201 based transfer learning model as best for selected dataset. In feature, we will test and validate our model on various modalities of lung CT scan images and report the issues of modalities of same domain feature space data.
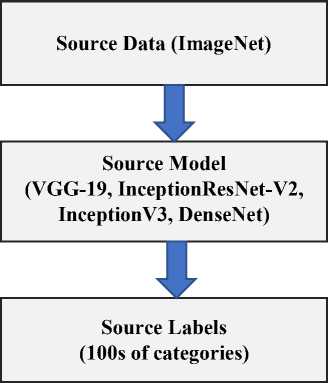
Fig. 2. Semantic view of transfer learning
Convolutional neural networks extract features from the input dataset in the same way that they do in formulation. It can be trained and acquired knowledge with a specified feature space and input that is narrowly constrained. It makes it more difficult for the network to expand deeper and wider. This may result in either an exploding or vanishing gradient. The DenseNet propagates feature maps of preceding layers to the newly created feature maps. Fig. 3 shows the structure of Dense Block in DenseNet (k0 represents the first layer features, k0+k represents the features of second layer and last layer has k0+4k features), in which output of layer l is H ([x ,x ,x ,..., x ]) . The Dense Block is expressed in equations (1), (2), and (3), where, l is the layer index and H means non-linear operation, x represents the feature of the lth layer.
Xt = н (Xt-1)
X = Hi(Xi-1) + Xi-i x, = Hl ([ X0, X1, X 2,...., x,-1])
Input 224x224x3
3x3x512
GAP+ BN+Droupout (0.5)
VGG19 InceptionResNet-V2 InceptionV3 DenseNet201
ReLU
Droupout= 0.2
Fig. 3. Propose Deep Transfer Learning Architecture
DenseNet201 provides concatenations of all feature maps from previous layers, which implies that all feature maps propagate to following layers and are linked to freshly created feature maps. Fig. 4 illustrates the structure of DenseNet201. It contains 4 dense blocks, 3 transition layers and global average pooling(GAP) layer. The dense block performs composite function h, ( . ) . This function performs three consecutive operations such as batch normalization (BN), rectified linear unit(ReLU) and finally a 3 x 3 convolution. The concatenation operation used in dense block is not viable when the size of feature maps changes. But, the essential part of convolutional network is down-sampling layers that change the size of feature-maps. To facilitate down-sampling in our architecture we divide the network into multiple densely connected dense blocks, which are shown in fig. 4. There will a transition layer between dense blocks. These transition layer perform convolution and pooling. The transition layer consists of a batch normalization, 1 x 1
convolutional operation and 2 x 2 pooling with stride of 2. The GAP layer compress the whole slice into a single digit means that compress a feature map of w x w x c to 1 x 1 x c .
Fig. 5 illustrates the structure of DenseNet201. It contains 196 learnable layers in four Dense Blocks (D1, D2, D3, D4), 4 convolutional layers (one from first layer CP, 3 from three transition layers (T1, T2, T3) and 1 fully connected softmax layer. The table 1 shown the details of DenseNet201.
For DenseNet201includes 6 sub blocks in Dense Block 1(D1), 12 sub blocks in Dense Block 2(D2), 48 sub blocks in Dense Block 3 (D2) and 32 sub blocks in Dense Block 4 (D3). Currently, there are four versions of Dense Nets such as DenseNet-121, DenseNet-169, DenseNet-201 and DenseNet-264. Our experiment results noted that DenseNet-201 is most stable for SARS-CoV-2 CT scan Dataset and classification task.
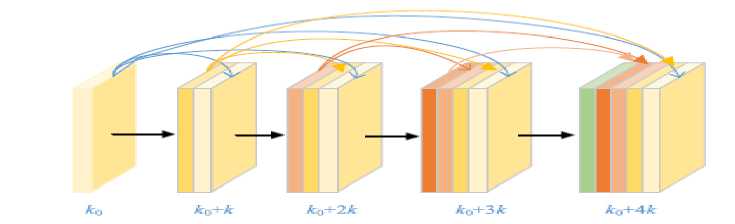
Fig. 4. Dense Block Structure
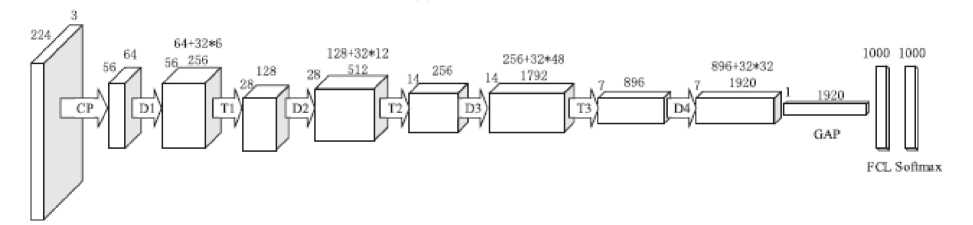
Fig. 5. DenseNet201 Architecture
Table 1. DenseNet201 Layers for ImageNet
|
Layers |
Output Size |
Dimensions of a Layer |
||
|
Input |
224 x 224 x 3 |
|||
|
Convolution (C) |
112 x 112 x 64 |
7 x 7 convolution with stride 2 |
||
|
Pooling (P) |
56 x 56 x 64 |
3 x 3 max pooling with stride 2 |
||
|
Dense Block (D1) (1) |
56 x 56 x 256 |
1 x 1 Convolution 3 x 3 Convolution |
1 , _x 6 |
|
|
Transition Layer(T1) (1) |
56 x 56 x 256 |
1 x 1 convolution |
||
|
28 x 28 x 128 |
2 x 2 average pool with stride 2 |
|||
|
Dense Block (D2) (2) |
28 x 28 x 512 |
1 x 1 Convolution _ 3 x 3 Convolution _ |
x12 |
|
|
Transition Layer (T2) (2) |
28 x 28 x 512 |
1 x 1 convolution |
||
|
14 x 14 x 256 |
2 x 2 average pool with stride 2 |
|||
|
Dense Block (D3) (3) |
14 x 14 x 256 |
1 x 1 Convolution 3 x 3 Convolution |
x 48 |
|
|
Transition Layer (T3) (3) |
14 x 14 x 1792 |
1 x 1 convolution |
||
|
7 x 7 x 896 |
2 x 2 average pool with stride 2 |
|||
|
Dense Block (D4) (4) |
7 x 7 x 1920 |
1 x 1 Convolution 3 x 3 Convolution _ |
x 32 |
|
|
Classification Layer (GAP) (pooling + FCL with softmax) |
1 x 1 x 1920 |
7 x 7 global average pool |
||
|
1000D |
||||
4. Implementation
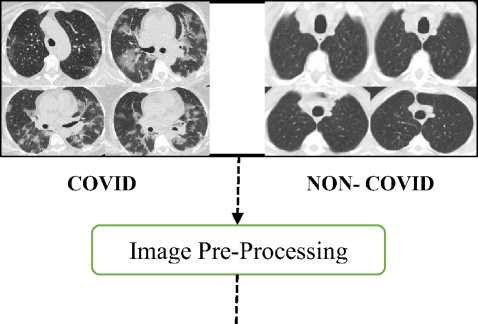
We have used a publicly disseminated CT imaging data set, which is available at https://www. It contains 1252 lung CT images that are positive for COVID-19 and 1229 lung CT images for non-COVID-19 patients. The data set contains a total of 2481 CT images for training, validation and testing. The data set was collected from the real patients from the hospitals of Sao Paulo, Brazil. To train our model, we established a training data set by randomly selecting 80% and leaving the remaining 20% of the data for performance testing. During the training process, 80% of the data samples are randomly selected from training data and remaining 20% of data samples from training data are used to validate the training model. Fig. 6 shows lung CT scan samples images of COVID-19 (a) and Non-COVID-19 (b). The number of samples are increased from the original data set using data augmentation. It is used with rotation up to 15o, horizontal flipping enabled. Further, slant-angle of 0.2o and the mode of filling new pixels as nearest for better robustness. This process enables the diverse data to avoid overfitting.
(b)
Fig. 6. Sample images from dataset (a) COVID-19 (b) Non-COVID-19
For our classification task, we have modified the structure of DenseNet-201. The last fully connected layer was replaced with two neurons, a softmax layer, and a new classification layer with two categories (COVID-19 and Non-COVID-19). Likewise, we have modified the VGG19, InceptionResNet-V2 and InceptionV3 in the last fully connected layer and new classification layer with two classes. The four transfer learning model in deep neural networks are trained with same data set and tested. The models are evaluated on test set with performance metrics of classification. They are (i) Accuracy (ACC) (ii) Precision (PRC) (iii) Sensitivity (SEN) (vi) Specificity (SPC) and (v) F1 score. Table 2 shows the confusion matrices of four models. The comparative analysis among proposed and the existing deep learning based COVID-19 classification models are evaluated on SARS-CoV-2 CT scan Dataset. The selected pretrained model in the paper are VGG19, InceptionResNet-V2 and InceptionV3. The five performance measures are formulated as follows [25]:
Accuracy: In classification problems, accuracy is defined as the number of correct predictions produced by the model over all types of predictions made.
Accuracy ( ACC ) =
TP + TN
TP + TN + FP + FN
Precision: What percentage of positive identification was correct? When the target variable classes in the data are approximately balanced, accuracy is a good metric. If the data is an unbalanced class samples then precision can be considered to measure the level of correct classification.
Pr ecision ( PRC ) =
TP
TP + FP
Sensitivity: It is also called as recall. It is calculated by dividing the number of valid positive findings by the total number of relevant samples.
Sensitivity ( SEN ) =
TP
TP + FN
Specificity: Specificity is diametrically opposed to recall. The specificity refers to the fraction of actual negatives that were projected as negatives.
Specificity ( SPC) =
TN
TN + FP
F1 Score: The F1 Score is defined as the Harmonic Mean of precision and recall(Sensitivity). F1 Score has a range of [0, 1]. It informs you how exact your classifier is (how many times it properly classifies) as well as how resilient it is (it does not miss a significant number of instances).
F 1 Score = 2 *--------1-----—
-------+---- precision recall
I precision 1 + recall 1
F 1 Score = I
I 2
Where TP, TN, FP, and FN stands for true positive, true negative, false positive and false negative respectively.
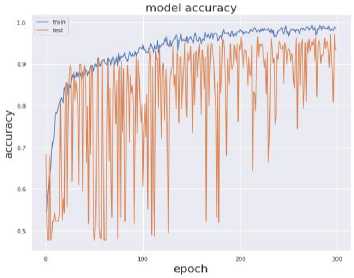
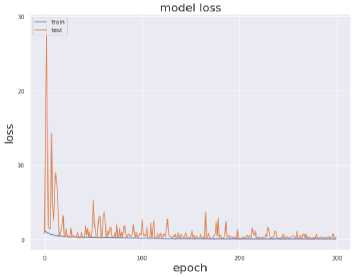
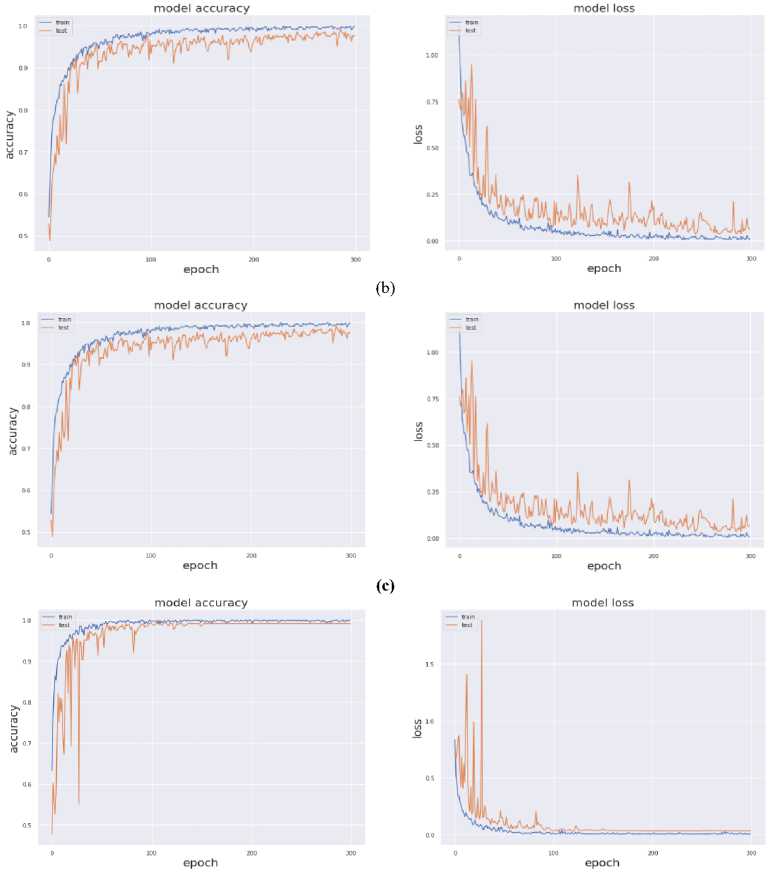
The value of above stated performance metrics should be maximized for best performance. Fig. 8 displays the training and validation accuracy and loss evaluations of the proposed and current deep transfer learning models with regard to number of epochs. The DenseNet201 model achieves much greater accuracy and reduced loss values in 300 epochs. It shows that DenseNet201 convergence on SARS-CoV-2 CT scan Dataset and provided considerably good speed.
5. Performance Analysis
The deep transfer learning classification schemes are trained and tested on SARS-CoV-2 CT scan dataset. The results were compared among VGG19, InceptionResNet-V2, IncepetionV3 and DenseNet-201 image classification models. The google colab is used to run the proposed classification model to determine COVID-19 or Non-COVID-19 .
(a)
(d)
Fig. 7. Training and Testing Accuracy and Loss (a) VGG19 (b) InceptionResNet-V2 (c) InceptionV3 (d) DenseNet-201
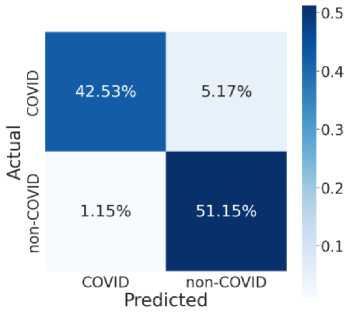
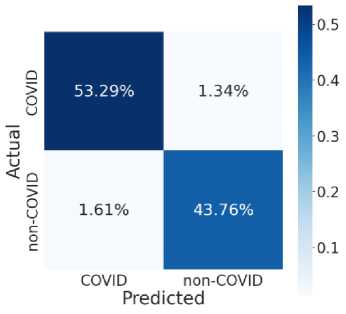
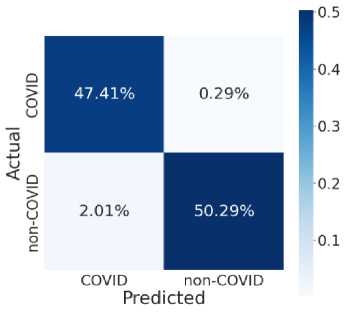
Fig. 7 provides the accuracy and loss analysis of training and testing of proposed deep transfer learning with respect to the number of epochs for four models. The results clearly shown that DenseNet201 model is best performed compared to other models such as VGG19, InceptionResNet-V2, InceptionV3. Hence, the DenseNet201 deep transfer learning model convergence and extracted the required features from lung CT scan images of COVID-19. False positive and false negative outcomes in medical research, especially for severe disorders such as COVID-19, are extremely crucial to minimize. COVID-19 positive patients must not be misclassified as negative because of false negatives for apparent reasons. False negatives have a significant harmful impact on our society. In addition, it is critical to reduce the amount of false positives, i.e., as COVID-19 negative as COVID-19 positive. As a result, individuals and families may suffer from mental distress and financial hardship. For example, DenseNet201 has a 99 percent accuracy rate, and the fig. 8 shows the influence of false positive and negative rates on this accuracy. It is evident that DenseNet201 reduces the occurrence of false positives and negatives. Since the COVID-19 quick test can yield 99 percent accurate results using DenseNet201 deep transfer learning, this model is the best choice for it.
Table 2. COVID-19 classification training analysis
|
Model |
Accuracy |
Specificity |
Recall |
Precision |
F1 Score |
|
VGG19 |
0.988 |
0.972 |
0.984 |
0.983 |
0.972 |
|
InceptionResNet-V2 |
0.987 |
0.981 |
0.974 |
0.981 |
0.961 |
|
InceptionV3 |
0.987 |
0.982 |
0.981 |
0.973 |
0.951 |
|
DenseNet201 |
1.00 |
0.996 |
0.997 |
0.992 |
0.993 |
(a)
(b)
(c)
Fig. 8. Model Testing Confusion Matrices (a) VGG19 (b) InceptionResNet-V2 (c) InceptionV3 (d) DenseNet201
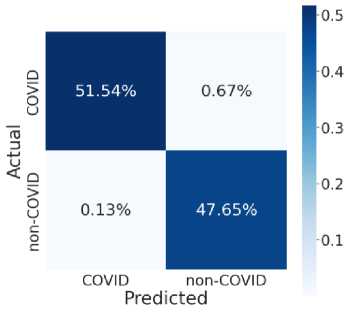
(d)
This model noted the accurate classification of 99.8%. Especially, in medical image analysis application domain demands 100% accuracy to estimate the disease. Fig. 8 illustrate the relationship among the classification and helpful to compute the performance measures of proposed model. Table 2 and table 3 shown the comparison among four models in training and testing of the model. The accuracy of DenseNet201 training and testing are 1.00 and 0.998 respectively. Therefore, DenseNet201 is the best performed and noted the minimum loss and variation between training and validation phases. The deep transfer learning DenseNet201 model can be utilized to classify the COVID-19 or Non-COVID-19 patients using CT scan images.
Table 3. COVID-19 classification testing analysis
|
Model |
Accuracy |
Specificity |
Recall |
Precision |
F1 Score |
|
VGG19 |
0.945 |
0.896 |
0.987 |
0.915 |
0.943 |
|
InceptionResNet-V2 |
0.969 |
0.966 |
0.973 |
0.959 |
0.969 |
|
InceptionV3 |
0.974 |
0.988 |
0.962 |
0.989 |
0.969 |
|
DenseNet201 |
0.998 |
0.992 |
1.00 |
0.992 |
0.992 |
6. Conclusion
This article, presented the COVID-19 disease predication using deep transfer learning using SARS-CoV-2 CT scan images. Four best image classification model such as VGG19, InceptionResNet-V2, InceptionV3 and DenseNet201 are adopted to perform the patient classification. The DenseNet201 based transfer learning model noted the best classification results. It classifies the SARS-CoV-2 chest CT images with the training, validation and testing accuracy as 1.00, 0.999 and 0.998 respectively. The comparison results shown that The DenseNet201 based deep learning model can be utilized for COVID-19 disease predication as compared with other well-known deep transfer learning models. Because CT scans are accessible in the majority of hospitals or diagnostic centers, the suggested approach can enhance the COVID-19 testing procedure. As a result, the suggested model may be used in place of several COVID-19 testing kits.
Further to develop an automatic COVID-19 detection system for medical diagnostics from multi-modality imaging. Instead of applying deep learning techniques directly, it is very much required to extract the deep features of input data like fusion, texture. Vessels, image filtering and etc. when sufficient data available, the current study can be extended to meta-heuristic techniques and multimodal data fusion.
Список литературы COVID-19 Automatic Detection from CT Images through Transfer Learning
- M. Abdel-Basset, V. Chang, and R. Mohamed, “HSMA_WOA: A hybrid novel Slime mould algorithm with whale optimization algorithm for tackling the image segmentation problem of chest X-ray images,” Appl. Soft Comput. J., vol. 95, p. 106642, 2020, doi: 10.1016/j.asoc.2020.106642.
- Y. H. Jin et al., “A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version),” Med. J. Chinese People’s Lib. Army, vol. 45, no. 1, pp. 1–20, 2020, doi: 10.11855/j.issn.0577-7402.2020.01.01.
- Y. Fang and P. Pang, “Senivity of Chest CT for COVID.19: Comparasion to RT.PCR,” Radiology, vol. 296, pp. 15–17, 2020.
- X. Xie, Z. Zhong, W. Zhao, C. Zheng, F. Wang, and J. Liu, “Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing,” Radiology, vol. 296, no. 2, pp. E41–E45, 2020, doi: 10.1148/radiol.2020200343.
- J. Zhang et al., “Viral Pneumonia Screening on Chest X-Rays Using Confidence-Aware Anomaly Detection,” IEEE Trans. Med. Imaging, vol. 40, no. 3, pp. 879–890, 2021, doi: 10.1109/TMI.2020.3040950.
- H. Hou et al., “Pr es s In Pr,” Appl. Intell., vol. 2019, pp. 1–5, 2020, [Online]. Available: http://arxiv.org/abs/2003.13865.
- J. P. Cohen, P. Morrison, and L. Dao, “COVID-19 Image Data Collection,” 2020, [Online]. Available: http://arxiv.org/abs/2003.11597.
- W. Cai, J. Yang, G. Fan, L. Xu, B. Zhang, and R. Liu, “Chest CT findings of coronavirus disease 2019 (COVID-19),” J. Coll. Physicians Surg. Pakistan, vol. 30, no. 1, pp. S53–S55, 2020, doi: 10.29271/jcpsp.2020.Supp1.S53.
- M. V. Moreno et al., “Applicability of Big Data Techniques to Smart Cities Deployments,” IEEE Trans. Ind. Informatics, vol. 13, no. 2, pp. 800–809, 2017, doi: 10.1109/TII.2016.2605581.
- M. Abdel-Baset, V. Chang, and A. Gamal, “Evaluation of the green supply chain management practices: A novel neutrosophic approach,” Comput. Ind., vol. 108, pp. 210–220, 2019, doi: 10.1016/j.compind.2019.02.013.
- B. Huang et al., “Deep Reinforcement Learning for Performance-Aware Adaptive Resource Allocation in Mobile Edge Computing,” Wirel. Commun. Mob. Comput., vol. 2020, 2020, doi: 10.1155/2020/2765491.
- V. Chang, “Computational Intelligence for Medical Imaging Simulations,” J. Med. Syst., vol. 42, no. 1, pp. 1–12, 2018, doi: 10.1007/s10916-017-0861-x.
- X. Li, Y. Wang, B. Zhang, and J. Ma, “PSDRNN: An Efficient and Effective HAR Scheme Based on Feature Extraction and Deep Learning,” IEEE Trans. Ind. Informatics, vol. 16, no. 10, pp. 6703–6713, 2020, doi: 10.1109/TII.2020.2968920.
- B. Naik, M. S. Obaidat, J. Nayak, D. Pelusi, P. Vijayakumar, and S. H. Islam, “Intelligent Secure Ecosystem Based on Metaheuristic and Functional Link Neural Network for Edge of Things,” IEEE Trans. Ind. Informatics, vol. 16, no. 3, pp. 1947–1956, 2020, doi: 10.1109/TII.2019.2920831.
- M. Ma and Z. Mao, “Deep-Convolution-Based LSTM Network for Remaining Useful Life Prediction,” IEEE Trans. Ind. Informatics, vol. 17, no. 3, pp. 1658–1667, 2021, doi: 10.1109/TII.2020.2991796.
- S. Wang et al., “IMAGING INFORMATICS AND ARTIFICIAL INTELLIGENCE A deep learning algorithm using CT images to screen for Corona virus disease ( COVID-19 ),” pp. 6096–6104, 2021.
- A. Narin, C. Kaya, and Z. Pamuk, “Department of Biomedical Engineering, Zonguldak Bulent Ecevit University, 67100, Zonguldak, Turkey.,” arXiv Prepr. arXiv2003.10849., 2020, [Online]. Available: https://arxiv.org/abs/2003.10849.
- E. Soares, P. Angelov, S. Biaso, M. H. Froes, and D. K. Abe, “SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification,” medRxiv, p. 2020.04.24.20078584, 2020, [Online]. Available: https://www.medrxiv.org/content/10.1101/2020.04.24.20078584v3%0Ahttps://www.medrxiv.org/ content/10.1101/2020.04.24.20078584v3.abstract.
- K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” 3rd Int. Conf. Learn. Represent. ICLR 2015 - Conf. Track Proc., pp. 1–14, 2015.
- C. Szegedy, S. Ioffe, V. Vanhoucke, and A. A. Alemi, “Inception-v4, inception-ResNet and the impact of residual connections on learning,” 31st AAAI Conf. Artif. Intell. AAAI 2017, pp. 4278–4284, 2017.
- C. Szegedy et al., “Going deeper with convolutions,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 07-12-June, pp. 1–9, 2015, doi: 10.1109/CVPR.2015.7298594.
- C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, and Z. Wojna, “Rethinking the Inception Architecture for Computer Vision,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2016-Decem, pp. 2818–2826, 2016, doi: 10.1109/CVPR.2016.308.
- G. Huang, Z. Liu, L. Van Der Maaten, and K. Q. Weinberger, “Densely connected convolutional networks,” Proc. - 30th IEEE Conf. Comput. Vis. Pattern Recognition, CVPR 2017, vol. 2017-Janua, pp. 2261–2269, 2017, doi: 10.1109/CVPR.2017.243.
- Raveendra K, Ravi J, "Performance Evaluation of Face Recognition system by Concatenation of Spatial and Transformation Domain Features", International Journal of Computer Network and Information Security(IJCNIS), Vol.13, No.1, pp.47-60, 2021. DOI: 10.5815/ijcnis.2021.01.
- Avijit Kumar Chaudhuri, Arkadip Ray, Dilip K. Banerjee, Anirban Das, "A Multi-Stage Approach Combining Feature Selection with Machine Learning Techniques for Higher Prediction Reliability and Accuracy in Cervical Cancer Diagnosis", International Journal of Intelligent Systems and Applications(IJISA), Vol.13, No.5, pp.46-63, 2021. DOI: 10.5815/ijisa.2021.05.05


