CRDL-PNet: An Efficient DeepLab-based Model for Segmenting Polyp Colonoscopy Images
Автор: Anita Murmu, Piyush Kumar, Shrikant Malviya
Журнал: International Journal of Image, Graphics and Signal Processing @ijigsp
Статья в выпуске: 4 vol.16, 2024 года.
Бесплатный доступ
Colorectal cancers are the third-largest kind of cancer in the world. However, detecting and removing precursor polyps with adenomatous cells using optical colonoscopy images helps to prevent this type of cancer. Moreover, hyperplastic polyps are benign cancers; adenomatous polyps are more likely to grow into cancerous tumors. Therefore, the detection and segmentation of polyps provide further histological evaluation. However, the main challenge is the extensive range of infected polyp features inside the colon and the lack of contrast between normal and infected areas. To solve these issues, the proposed novel Customized ResNet50 with DeepLabV3Plus Network (CRDL-PNet) model provided a scheme for segmenting polyps from colonoscopy images. The customized ResNet50 extracted features from polyp colonoscopy images. Furthermore, Atrous Spatial Pyramid Pooling (ASPP) is used to handle scale variation during training and improve feature selection maps in an upsampling layer. Additionally, the Gateaux Derivatives (GD) approach is used to segment boundary boxes of polyp regions. The proposed method has been evaluated on four datasets, namely the Kvasir-SEG, ETIS-PolypLaribDB, CVC-ClinicDB, and CVC-ColonDB datasets, for segmenting and detecting polyps. The simulation results have been examined by evaluation metrics, such as accuracy, Intersection-Over-Union (IOU), mean IOU, precision, recall, F1-score, dice, Jaccard, and Mean Process Time per Frame (MPTF) for proper validation. The proposed scheme outperforms the existing State-Of-The-Arts (SOTA) model on the same polyp datasets.
Colonoscopy, deep learning, segmentation, polyp detection, boundary box
Короткий адрес: https://sciup.org/15019460
IDR: 15019460 | DOI: 10.5815/ijigsp.2024.04.02
Текст научной статьи CRDL-PNet: An Efficient DeepLab-based Model for Segmenting Polyp Colonoscopy Images
Colon cancer is a frequent gastrointestinal cancer that typically develops at the intersection of the abdomen and pelvic colon. Although abdomen polyps have a significant chance of developing into colon polyp cancer, the process takes a long time – between five to ten years [1]. The rate of colon polyp cancer early detection can be considerably raised, and the challenge and morbidity of polyp cancer can be minimized if polyps can be found and separated before turning into colon polyp cancer.
There are multiple methods for evaluating the colon, including a barium enema and abdomen plain film. However, colonoscopy is one of the most popular and efficient [2]. A type of fiberoptic endoscope frequently utilized in clinical settings is colonoscopy. Colonoscopy immediately detects intestinal lesions and be inspected the cecum [3]. Even treatment components are combined in certain cutting-edge colonoscopy devices. The technology used for detection and therapy can eliminate lesions as soon as they are located. Therefore, colonoscopy is a reliable method of finding colon polyps [4]. However, standard colonoscopy still has several drawbacks, such as: 1) Patients cannot thoroughly empty their intestines for a colonoscopy examination. The food remnants, fecal deposits, and other interferences always have an impact on the gastrointestinal tract. 2) There are fewer professional medical professionals, which has an impact on the efficacy of colonoscopy. Physicians with less clinical expertise are more likely to overlook some rare colon polyps [5]. Only fewer physicians with substantial clinical expertise are capable of tackling these constraints. As a result, during colonoscopy, up to 25% of polyps are not detected. However, segmentation of infected regions helps to diagnose correctly [6,7].
The Artificial Intelligence (AI) technique is thought to aid in the diagnosis because of the limits of the AI evaluation of the colonoscopy images as stated above schemes. It can long-term overcome the accuracy loss brought on by manual operation. As a result, the AI techniques perform well while processing vast amounts of colonoscopy image data. Deep Learning (DL)-based models are effective for handling image data [8, 46]. It enables a computational scheme made up of many processing phases to learn the data representations at various levels of a abstraction, significantly enhancing the accuracy of voice recognition, a visual object identification, and object detection. It has provided several study findings in the diagnosis and assessment of gastroenterology because of a great ability to analyze medical imaging datasets [9]. Qadir et al. [10] have proposed a precise real-time polyp detection algorithm using singleshot a feed-forward fully convolutional neural networks (F-CNN). Typically, binary masking is used to train F-CNNs for image segmentation. Mangotra et al. [11] have proposed technique in the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) parameters for examining efficiency of polyp segmentation. The model includes a refined, readable ACPD framework (VGG-16) and its real-time software without taking into consideration of bias in selection of the previous open-source colonoscopy image datasets. Shi et al. [12] have proposed a PolypMixer that makes use of MLP-based architectures in both the encoder and the decoder. In order to get around the fixed input scale problem, which can specifically for CycleMLP as the encoder. Additionally, adapting the present CycleMLP as a Multi-head is used multi-head mixer enables the model to investigate rich contextual data from distinct subspaces.
The novel CRDL-PNet is a combination of ResNet50 with DeepLabV3Plus approach for polyp segmentation and performing detection from colonoscopy images that addresses these issues. ResNet50 provides a novel approach to expanding a CNN convolutional layer structure without having any issue of gradients vanishing by using the concept of shortcut connections. The Unet layers have been utilized to extract features from the colonoscopy input images by using the ResNet50 approach to train it faster. Additionally, including the ASPP mechanism which improves the feature section and handles scale variations during training. Additionally, boundary boxes are determined by using the GD mechanism for polyp image segmentation. Specifically, the following contributions are included:
• The lack of contrast between normal and polyp-infected areas detection from colonoscopy images is challenging to segment accurately. A novel customized CRDL-PNet method is proposed to overcome these issues and used polyp segmentation.
• In the proposed system, design a novel CRDL-PNet model for polyp segmentation polyp. Furthermore, ASPP is used during training and improves feature maps in an upsampling layer.
• Additionally, the GD approach is used to segment boundary boxes of polyp regions.
• The experimental evaluation matrices namely, dice, accuracy, recall, jaccard, precision, IOU, F1-score, MPTF, and mIOU show the proposed model outperforms SOTA for polyp colonoscopy images.
2. Literature Survey
3. Methodology
This paper is divided into four sections. In Section 2, the corresponding literature review is described. Section 3 of the proposal approach is covered. The proposed scheme experiments findings and analysis are presented in Section 4. The conclusion and future work are provided in Section 5.
As described in the introduction, DL-based technologies for polyp identification have been reported in several recent studies that analyze novel methods. Some of the published research use polyp detection without localization, i.e., the described algorithms designed to forecast if one (or more) polyps are present in a particular image without providing the precise position of the polyp. Another method of locating polyps in the image frames is by using a bounding box (square or circle) or binary mask to indicate their exact location. Polyp localization is the first task, and polyp segmentation is the second. The positions of multiple polyps in the same frame are frequently predicted using polyp detection or polyp segmentation methods. Moreover, the biomedical imaging technique has used many modalities namely, X-ray, Computer Tomography (CT), and ultrasound, and so on [32, 47]. Chen et al. [13] have proposed an enhanced Faster RCNN network with three processing stages for the feature extraction, area proposal development, and polyp identification. Moreover, include an attention layer to focus on the helpful feature channels and lessen the contribution of the impotent feature channels in order to further enhance the quality. Additionally, feature abstraction capabilities of feature maps are generated by a feature extraction channel. Yue et al. [14] have proposed a new APCNet namely an attention guide pyramid feature network for precise and reliable polyp detection in a colonoscopy scans. Additionally, considering the multiple layers of the network represents the polyp in multiple ways, APCNet is extracts multiple-layer of image features in a pyramidal arrangement, and then uses attention-guide multilayer collection strategy are refine particular features of every layer with the related data of different stages. APCNet is uses a context extraction of feature module, which analyzes the context information of every layer with local data retention and global information compression.
Adjei et al. [15] have proposed modifying and training a pix2pix network to create fictitious colonoscopy images with the polyps to increase the initial collection of data. Then, train the U-Net model and a faster R-CNN algorithm for segmenting and recognizing polyps, respectively. Moreover, build a range of datasets by altering the proportion of traditional augmentation and synthetic samples. Banik et al. [16] have presented a fusion-based adenoma polyp segmentation network (Polyp-Net). CNN has demonstrated tremendous success in field of biomedical image analysis recently because it can make use of in-depth important features with strong discrimination skills. As a result of these realizations, dual-tree wavelet pooled CNN, is an improved form of CNN with a developing pooling mechanism. Apart from the polyp region, the resulting segmented mask also includes several extra high-intensity regions [17]. The gradient weight embedded level-set technique is a new variant of the region-based level-set approach that exhibits a notable decrease in false-positive rate. The pixel-level merging of the two improved techniques demonstrates more potential for segmenting the polyp zones [48]. Mondale et al. [42] have presented an InvoPotNet employs an Involution Neural Network (INN) technique that is 25 times more compact than the Deep CNN approach to automatically recognize potholes in the road. A preprocessed dataset is used to train and test five models: ResNet50, InceptionV3, MobileNetV2, VGG19, and custom Deep CNN. Ye et al. [43] have developed a WDMNet as Wind Dynamics Modeling Network to simulate the various regional wind velocity fluctuations and generate precise multi-step forecasts. WDMNet uses a novel neural stage called INN-based involution gate recurrent unit of partial differential equation as its core component in order to simultaneously capture the geographically diversified variations and distinctions between V-wind and U-wind. These INN-based models have some limitations, this is not able to improve the quality of poly detection in case of unseen data. The detailed related works are discussed in Table 1.
The previous approaches are specific-purpose approaches have traditionally been used to address polyp segmentation challenges in previous attempts. The majority of the provided approaches use two neighboring layers to extract feature information for the polyp representation to solve the issue of shapes and size variations. Actually, deep CNNs contain extensive semantic and spatial data from shallow to the deep stages, and all of these layers are used in performance to represent targets of various sizes and shapes. This proposed scheme is a new segmentation architecture that analyzes and provides information on all layers and feature information of every layer for the polyp segmentation.
Table 1. Literature survey
|
Ref. |
Objective |
Modality |
Datasets |
Techniques |
|
[13] |
Polyp segmentation |
Colonoscopy image |
Colonoscopy image dataset |
Faster R-CNN |
|
[14] |
Polyp segmentation |
Colonoscopy image |
CVC-ColonDB, ETIS, CVC-ClinicDB, Kvasir-SEG, and CVC-T |
APCNet |
|
[15] |
Polyp segmentation and detection |
Colonoscopy image |
ETIS-PolypLaribDB, Kvasir-SEG, CVC-ColonDB, and CVC-ClinicDB, |
U-Net and Faster R-CNN |
|
[16] |
Polyp segmentation |
Colonoscopy image |
CVC-ClinicDB, and CVC-ColonDB |
DT-WpCNN and PolypNet |
|
[17] |
Polyp segmentation |
Colonoscopy image |
Kvasir-SEG, EndoScene, and CVC-ClinicDB |
LFSRNet |
|
[18] |
Polyp detection and segmentation |
Colonoscopy image |
CVC-300, CVC-ColonDB, PICCOLO, CVC-ClinicDB, Kvasir, and ETIS |
Convolutional MLP |
|
[19] |
Polyp segmentation |
Colonoscopy image |
Kvasir-SEG, ClinicDB , ColonDB, EndoScene, and ETIS |
HANet |
|
[20] |
Polyp segmentation |
Colonoscopy image |
Kvasir-SEG |
UPolySeg |
|
[21] |
Polyp segmentation |
Colonoscopy image |
CVC-ClinicDB, CVC-ColonDB, Kvasir-SEG, ETIS-Larib, EndoScene, and PolypDB |
Focus U-Net |
|
[22] |
Polyp detection and segmentation |
Colonoscopy image |
CVC-ClinicDB, ETIS-Larib , and CVC-ColonDB |
Mask R-CNN |
The proposed customized Resnet50 with DeepLabV3Plus (CRDL-PNet) model architecture is shown in Fig. 1. The proposed system is used for polyp detection, segmentation, boundary box of the infected region, and helps to diagnose the poly in early stages. The architecture of the proposed scheme contains convolutional nodes, ResNet50 as a backbone, an activation function node, ASPP, and batch normalization in downsampling. The detail should be described in depth in this section, starting with the network architecture and essential network components. Furthermore, an overview of the proposed loss function and boundary box of the segmented image.
-
3.1. Dataset details
We estimate the proposed approach for polyp segmentation tasks by using four datasets: the Kvasir-SEG [23], CVC-ClinicDB [24], CVC-ColonDB [44], and ETIS-PolypLaribDB [45] datasets.
-
• Kvasir-SEG: This collection includes 1000 images of polyps that were taken using the high-resolution electromagnetic imaging system, as well as bounding box data [23]. The segmentation process uses images and matching ground facts mask images. The collected images have resolutions from 332 × 487 to 1920 × 1072.
-
• CVC-ClinicDB: This dataset is an open-access and has 612 images with 384 × 288 resolution from 31 colonoscopy series [24].
-
• CVC-ColonDB: The 300 colon polyp images and related pixel-level annotation polyp masks in the CVC-ColonDB [44] dataset have been collected from 15 different video sequences. The images are 574 × 500 in resolution.
-
• ETIS-PolypLaribDB: The 196 polyp images in the ETIS-PolypLaribDB [45] collection of 36 distinct polyp types have a resolution of 1225 × 966. The images are taken from colonoscopy videos, and the mask's foundational facts are annotated.
-
3.2. Data augmentation
-
3.3. Polyp segmentation
-
3.3.1. Customized ResNet50 with DeepLabV3Plus model architecture
-
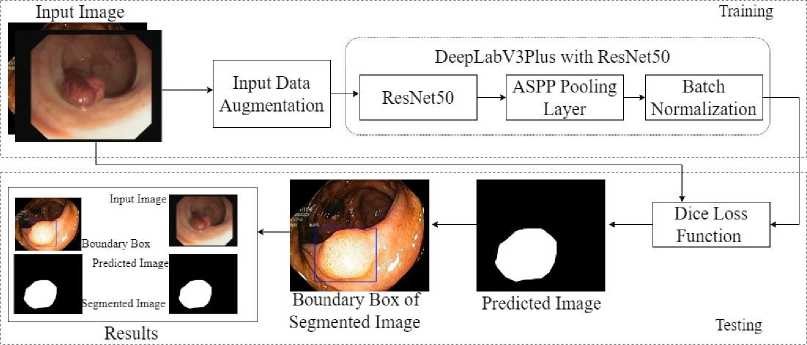
Fig. 1. Proposed methodology
The proposed work involves generating annotated instances of data for biological tasks, and annotating the latest images takes time and is costly. Expensive expertise in medicine is required for high-quality annotations. The earlier research utilized certain datasets that were produced privately. However, sharing those medical records is more
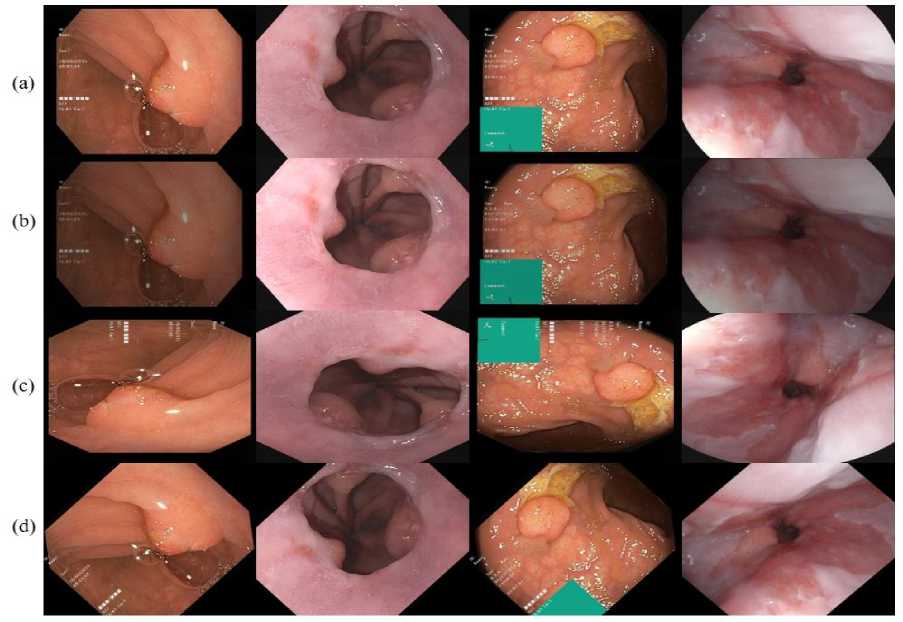
Fig. 2. Data augmentation of polyp image dataset (a) original image, (b) augmented image, (c) rotation with 90o, and (d) rotation with 45o challenging than sharing natural images due to security and ethical concerns. However, there are not many publicly accessible medical imaging databases available. The number of annotated polyp images in each of these datasets is limited to 1000. CNN-based schemes are inherently data, and it is generally acknowledged that larger datasets lead to better performance. Therefore, apply data augmentation we perform handcraft polyp data augmentations on a training dataset. Perform augmentations, namely, random rotation, center crop, vertical flip, horizontal flip, and grayscale conversion, on each image from a training set shown in Fig. 2.
The proposed customized version of ResNet50 with a DeepLabv3Plus Network architecture (CRDL-PNet) is used to increase polyp identification and segmentation while maintaining the model's portability (shown in Fig 2 and Algorithm 1). Resnet50 provides a novel approach to expanding a CNN convolutional layer structure without having the issue of gradients vanishing by using the concept of shortcut connections. This skip connection “skips across” a few layers, changing a standard network into a residual network.
ResNet50 feature map at layer ly is represented as Rt. Then, an updated process between Rt-1 and Rt is considered. In residual block, let r is skip connection and the convolutional kernel is ck, R t-1 can be transformed with R t as follows:
R(,z t ) = relu{r.R(zt -i ) + ck(Z) * R^Z t-i )]
Given that, r. R + ck * R = r (8 + ^7 )
* R , Eq. (1) can rewrite in Eq. (2):
R(Z t ) = X zt-1 К or (8 (Z t — Z t-J + 0. (Z t — Z t- 1))R(Z t- 1)
As such, the ResNet50 can be written in the form of ResNet50 with DeepLabV3Plus as follows in Eq. (3)
r(zn) = YZpathe^ T-v(z) R(j0) = X z pathe —^t M^R(Z0) = X zpath e -p p^^^ R(Z0) (3)
Where, z = zt – zt-1 with V kinetic energy and T potential energy. So, the above Eq. (3) is shown the ResNet50 feature extraction, feature map, and the connection with DeepLabV3Plus.
The Unet layers have been utilized to extract the features from the input colonoscopy images and choose the ROI using the ResNet50 approach to train it faster. Moreover, the DeepLabv3Plus network is an approach for semantic image segmentation using Atrous Spatial Pyramid Pooling (ASPP) and space pyramid modules [25] shown in Eq. (4).
d[i\ = X M=1 Z[i + r.m\M[m\
In order to recover spatial information and enhance boundary segmentation, the DeepLabv3Plus network incorporates an encoder-and-decoder structure. The decoder layers are used to extract specifics and recover geographic
Algorithm 1: Training process of polyp segmentation
|
Inputs: |
Input image Ip, feature map Fp, ResNet50 model RN, loss function Lf, and boundary box bb . |
Output: Probability of segmented polyp is Sp.
Determine the polyp location (PL)
PL ← arg p,q maxF p (p,q)
Obtain feature for Ip
Compute the polyp representative feature
Ψ = median(ψ c )
for each RN from 0 to F p do
Pt ← find point on ψ at RN wrt PL
-
d™ ^ || PL - Pt ||
Apply the model CRDL-PNet [tRN, 5] = CRDL-PNet(dR™)
From line lRN given tRN, δ using loss in Eq. (1)
Determine the boundary box according to Eq. (6)
end
Compute the probability Sp = I^^^F p V RN + Lf,bb information, while the encoder layers are designed to minimize feature loss and collect higher-level semantic data. The DeeplabV3Plus structure extracts features of input image using the residual neural network (ResNet50). The DeepLabv3 design has an enhanced ASPP built in to avoid information loss, making it computationally efficient shown in Fig. 3. Before convolution, ASPP offers a semantic segmentation layer for resampling a particular feature layer at different rates. It also uses numerous filters with enhancing effective fields of view to analyze the original image, collecting objects and meaningful visual context at various scales. The mapping is done by employing several parallel atrous layers of convolution with various sampling rates, as opposed to resampling the features. A bottleneck network ResNet50 and a pyramid pooling mechanism for void space combine the Deeplabv3Plus encoding architecture. To retrieve the semantic data of multiscale images, the empty space pyramid pooling module is linked to the ResNet50 networks terminate. The feature map produced by the network of encoders receives 4-fold bilinear upsampling during decoding by combining the channel-level low-level feature information. The downsampling procedure then recovers the missing spatial information.
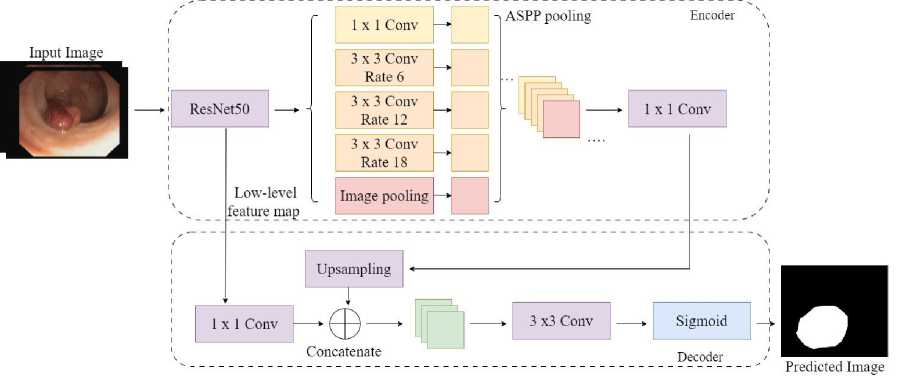
Fig. 3. Customized CRDL-PNet model
The performance of CNN architecture depends on the depth, breadth, and cardinality of the frameworks. Additionally, network architecture design must be looked at to increase a model's expressiveness in addition to these elements. A low-level feature map focusing on relevant traits and suppressing unneeded ones boosts expressiveness. We use channel and low-level feature maps modules to choose the parameters, highlighting significant aspects in the spatial and medium factors of polyp image. Moreover, the dice loss and sigmoid function is used for improve the performance, the Eq. (5) and Eq. (6) shows the dice loss and sigmoid function:
LSdice = 1 - DSC(truey,pred y ) (5)
S(x) = ■ (6)
-
3.3.2. Segmented boundary box
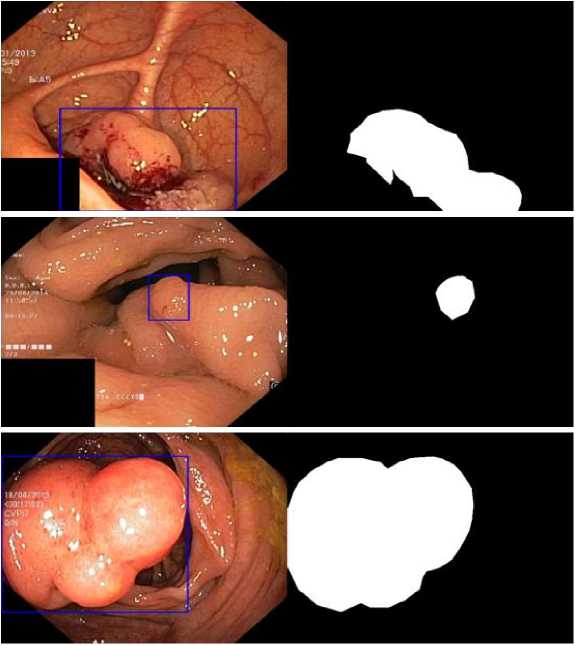
The polyp has been segmented using the proposed CRDL-PNet model. The CRDL-PNet uses performance and colonoscopy images database as a input to globally enhance spatial and optical coherence of the polyps discoveries while maintaining the uniformity of each position's information and pixel's intensity. The proposed method estimates the chances each pixel is to contain a polyp. Additionally, for the infected poly images, the polyp segmentation region boundary box detection by applying GD for recognizing a proper tumor border [6]. Determining the polyps boundaries accurately improves diagnosis shown in Fig. 4.
The changes from region to border integrals. Let's take along the polyp region integral shows in Eq. (7)
P^^) = f^ ptq^dq where q is a function of scaler p and a ω is an open, bounded, and regular subset of Rm with segmented boundary δω of polyp. moreover, region integrals are transformed into line integrals to provide the Euler's for energy in Eq. (8)
ECq,^) = - 1 Kq^a i - a2 )2 + у ф5ш dt
Now, compute the energy of GD by Eq. (9):
‘
D ( t ) = - fn () GDtKM dt
0 ’^
where ϗ shows every pixel mean q curvature of GD. Now, consider a polyp regions dependent in Eq. (8). The abrupt descent system produces an evolutionary form from the minima of Eq. (8) is starting with a polyp initial curve GD(q,0) = GD 0 .
GD t = 9(^ 2 - .J ( ■ + ^ ) M + ^M
Using the edge of polyp information assembled from colon polyp images for boundary box detection of polyp, Eq. (10) provides segmented boundary boxes of polyp shown in Fig 3.
4. Results and Discussion
The proposed system evaluates results by using CVC-ColonDB, ETIS-PolypLaribDB, CVC-ClinicDB, and Kvasir-SEG datasets. The proposed model is tested and trained on an Intel Core i5 processor, 2.8 GHz, 8 GB of RAM, and the graphics processor Unit (GPU) [26]. In addition to TensorFlow and Python 3.8.1, used Keras library to implement the proposed approach. The publicly available dataset has been resized to 224 × 224 pixels. All the collected samples are used in training and only 30% of the data is used in the testing process. The models are trained by using an Adam optimizer 0.001 learning rate, a 0.1 dropout, and a 16-batch size.
-
4.1. Evaluation matrices
The confusion matrix is divided into two sections, each area displaying the categorization results. Each result is shown in a row to emphasize the quantity of information included inside a label's class. The True Positive (TP) and True Negative (TN) phrases, which denote when the estimated result is positive and real outcome is positive or negative, respectively, are used in this study to evaluates the performance of the CRDL-PNet model. Additionally, outcomes projected to be positive but have negative values are referred to as false positive (FP) and false negative (FN). False Positive (FP) are results predicted to be positive but have negative values—the study that serves as the basis for Eqs. (11) to (18).
A.) Accuracy: Accuracy determines the ratio of expected to actual values, independent of whether the samples are positive or negative. The formula is shown below in Eq. (11):
Accuracy =
TP+TN
TP+TN+FP+ FN
B.) Jaccard: The Jaccard used to assess the polyp segmentation (Shown in Eq. (12)):
Jaccard =
TP
FP+TP+FN
C.) Dice: The dice are also used to check the effectiveness of polyp segmentation (determined by using Eq. (13)):
Dice =
2 TP
FP+2TP+FN
D.) Precision: The percentage of all positive samples tested that are absolutely positive is known as the precision and is represented by the formula (shown in Eq. (14)):
TP
Precision = ----
TP+FP
E.) Recall: The recall is the ratio of correct predictions to all positive, as indicated in Formula (shown in Eq. (15)):
Recall =
TP
TP+FN
F.) F1-score: The classification system performance is described by the F1 score. The formula shows compute it using the recall and precision rate (shown in Eq. (16)):
F1 — score =
2xRecall ^Precision
Recall+Precision
G.) IOU and mIOU: An IOU and mIOU is a popular evaluation metric in image segmentation tasks and image detection. An IOU and mIOU provide a score between 0 and 1 depending on the extent to which predicted bounding boxes overlap actual ground truth boxes shown in Eq. (17) and Eq. (18):
IOU =
TP
FP+TP+FN
miou = ^^
M
(a) (b)
Fig. 4. GD-based boundary boxes of segmented polyp regions
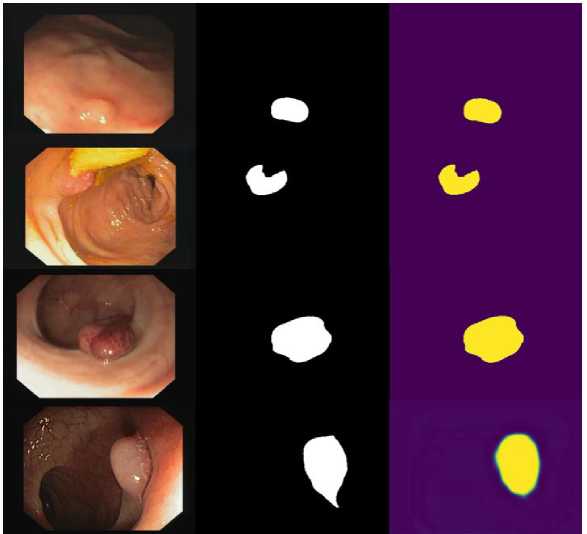
(a) (b) (c)
Fig. 5. Performance results of proposed scheme (a) original image, (b) ground truth, and (c) predicted polyp segmentation
-
4.2. Experimental results
The proposed experiment uses a total of 60 images containing 63 polyps. The training set (70%) and testing set (30%) have been selected according to prior information regarding the boundary detection GD approach. The overall test outcomes of the proposed approach using the training-testing samples are shown in Table 2 and Table 3 with mean and Standard Deviation (SD). The overall test results of accuracy, F1-score, jaccard, dice, recall, precision, mean IOU, IOU, MPTF for the proposed model is presented in Table 2, Table 3, Fig. 7, and Fig. 8, respectively. Moreover, the outcomes are validated with 5-fold cross-validation are shown in Table 4 and Table 5, respectively. There is a minimum of one polyp apparent in every test image. The 55 boundary box images created from segmented images for test-accurate polyp cancer boundary boxes are true positive samples, 17 boundary boxes predicting the intestine wall are false positive samples, and 25 are the missed polyps from false negative samples are provided by the proposed approach, according to assessment findings shown in Fig. 5 and Fig. 6, respectively. The accuracy is 96.30% and 96.98%, the precision is 84.85% and 85.33%, the recall is 92.72% and 92.46%, the F1-score is 80.79% and 81.25%, the Jaccard is 92.59% and 93.12%, and the dice is 92.95% and 94.06% for both false positive and false negative outcomes. The IOU becomes ineffective if the denominator and numerator are of the same sequence and are hence infinitesimal. Therefore, only true positives are affected by every statistical IOU. The overall outcome mean IOU is only 89.12% and 90.82% and mIOU is 90.75% and 92.23% are possibly achieved altogether. The system offers a total 64 boundary boxes for testing-accurate polyp cancer boundary boxes are true positive samples, 5 boxes for predicting gastrointestinal wall, or stool interference as polyps cancer are false positive samples, and 4 are missed polyps false negative samples, according to the evaluation results of the proposed model. Moreover, the proposed scheme is also validated in the unseen dataset. Fig. 9 shows a performance graph of epoch verse loss and accuracy.
Table 2. The performance outcomes of proposed systems for polyp segmentation (Kvasir-SEG and CVC-ClinicDB datasets)
|
Datasets |
Backbone |
Accuracy |
Jaccard |
Dice |
Precision |
Recall |
F1-score |
IOU |
mIOU |
MPTF (ms) |
|
Kvasir-SEG |
ResNet50 |
96.30 |
92.59 |
92.95 |
84.85 |
92.72 |
80.79 |
89.12 |
90.75 |
430 |
|
Mean |
0.963 |
0.925 |
0.929 |
0.848 |
0.927 |
0.807 |
0.891 |
0.907 |
0.430 |
|
|
SD (±) |
0.073 |
0.065 |
0.021 |
0.049 |
0.026 |
0.018 |
0.048 |
0.036 |
0.008 |
|
|
CVC-ClinicDB |
ResNet50 |
96.98 |
93.12 |
94.06 |
85.33 |
92.46 |
81.25 |
90.82 |
92.23 |
317 |
|
Mean |
0.969 |
0.931 |
0.940 |
0.853 |
0.924 |
0.812 |
0.908 |
0.922 |
0.317 |
|
|
SD (±) |
0.062 |
0.084 |
0.056 |
0.048 |
0.021 |
0.039 |
0.076 |
0.043 |
0.006 |
Table 3. The performance outcomes of proposed systems for polyp segmentation (CVC-ColonDB and ETIS-PolypLaribDB datasets)
|
Datasets |
Backbone |
Accuracy |
Jaccard |
Dice |
Precision |
Recall |
F1-score |
IOU |
mIOU |
MPTF (ms) |
|
CVC-ColonDB |
ResNet50 |
97.73 |
93.92 |
94.21 |
84.03 |
90.71 |
81.52 |
89.97 |
90.63 |
390 |
|
Mean |
0.977 |
0.939 |
0.942 |
0.840 |
0.907 |
0.815 |
0.899 |
0.906 |
0.390 |
|
|
SD (±) |
0.062 |
0.061 |
0.031 |
0.029 |
0.037 |
0.028 |
0.061 |
0.041 |
0.009 |
|
|
ETIS-PolypLaribDB |
ResNet50 |
97.09 |
92.37 |
92.77 |
84.58 |
90.16 |
80.73 |
88.73 |
90.52 |
730 |
|
Mean |
0.970 |
0.923 |
0.9277 |
0.8458 |
0.901 |
0.807 |
0.887 |
0.905 |
0.730 |
|
|
SD (±) |
0.071 |
0.026 |
0.043 |
0.085 |
0.096 |
0.047 |
0.059 |
0.077 |
0.006 |
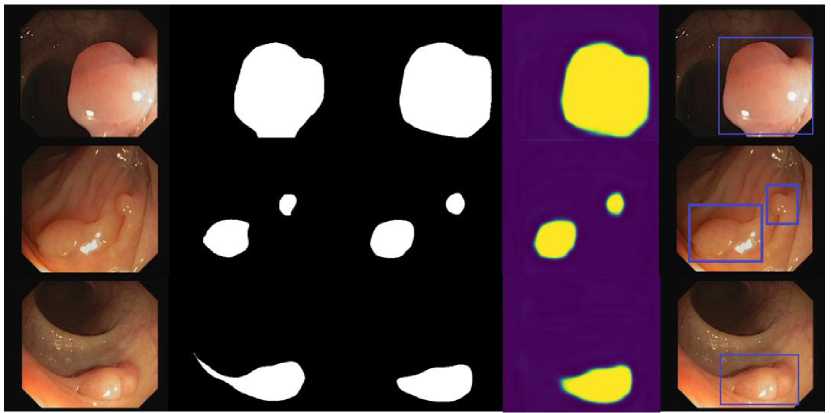
(a) (b) (c) (d) (e)
Fig. 6. Performance results of proposed scheme (a) original image, (b) ground truth, (c) predicted mask, (d) predicted polyp segmentation, and (e) segmented boundary box of polyp
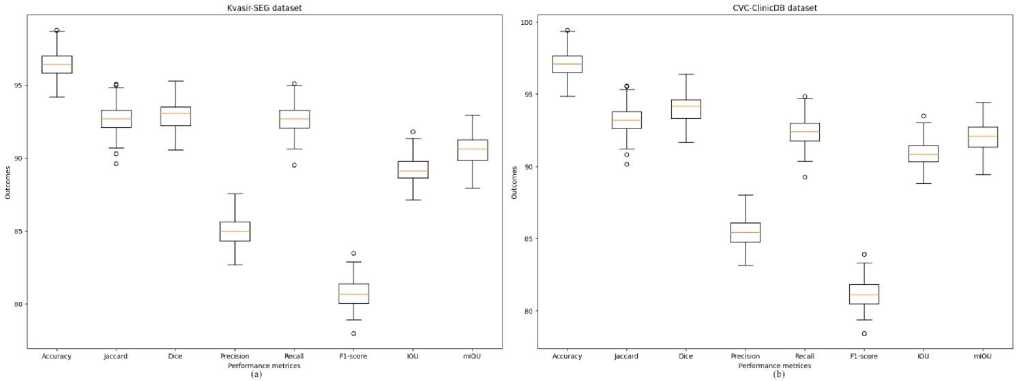
Fig. 7. The performance graph of the CRDL-PNet scheme (a) Kvasir-SEG and (b) CVC-ClinicDB datasets
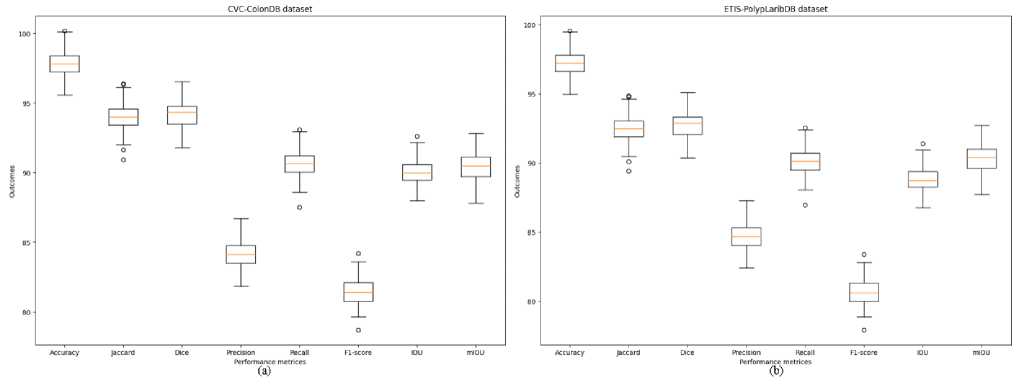
Fig. 8. The performance graph of the CRDL-PNet scheme (a) CVC-ColonDB and (b) ETIS-PolypLaribDB datasets
Table 4. The performance outcomes of CRDL-PNet model for 5-fold cross-validation (a) Kvasir-SEG and (b) CVC-ClinicDB datasets
|
Performance matrices |
Kvasir-SEG dataset |
CVC-ClinicDB dataset |
||||||||||
|
Fold1 |
Fold2 |
Fold3 |
Fold4 |
Fold5 |
Mean Fold |
Fold1 |
Fold2 |
Fold3 |
Fold4 |
Fold5 |
Mean Fold |
|
|
Accuracy |
90.20 |
90.04 |
93.04 |
93.05 |
95.90 |
96.30 |
90.38 |
90.83 |
93.23 |
94.38 |
95.23 |
96.98 |
|
Jaccard |
88.90 |
88.20 |
90.16 |
90.23 |
92.22 |
92.59 |
86.02 |
86.23 |
87.23 |
90.81 |
91.90 |
93.12 |
|
Dice |
87.10 |
88.14 |
89.26 |
91.15 |
91.15 |
92.95 |
88.92 |
89.09 |
90.09 |
92.18 |
93.13 |
94.06 |
|
Precision |
78.91 |
79.10 |
80.13 |
83.05 |
83.08 |
84.85 |
79.63 |
79.16 |
80.16 |
82.18 |
83.75 |
85.33 |
|
Recall |
86.09 |
87.15 |
87.13 |
89.15 |
90.13 |
92.72 |
87.72 |
88.14 |
89.14 |
90.13 |
90.66 |
92.46 |
|
F1-score |
76.91 |
77.15 |
77.90 |
78.90 |
80.14 |
80.79 |
76.08 |
76.14 |
78.14 |
78.89 |
80.33 |
81.25 |
|
IOU |
85.15 |
86.15 |
86.90 |
87.05 |
87.90 |
89.12 |
84.73 |
85.95 |
86.95 |
87.94 |
89.74 |
90.82 |
|
mIOU |
85.92 |
86.19 |
87.15 |
88.13 |
89.22 |
90.75 |
87.10 |
87.95 |
88.89 |
89.94 |
90.28 |
92.23 |
4.3. Results comparison
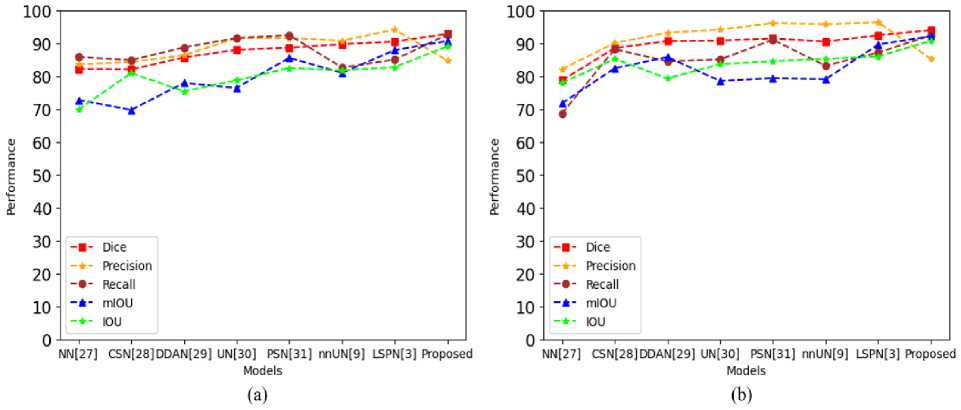
This indicates that the proposed approach has a great deal of promise for helping endoscopists identify and segment polyps during colonoscopy. The approach of the proposed framework does enhance CRDL-PNets performance in comparison to the other model. The results of comparing the proposed models in this paper with the existing SOTA techniques via precision, accuracy, recall, mIOU, IOU, MPTF, and dice for polyp segmentation. The proposed CRDL-PNet outperforms the existing models, as shown in Table 6, Table 7, Fig. 10, and Fig. 11, respectively. Furthermore, CRDL-PNet improves IOU on unseen datasets by 6.29% in Kvasir-SEG, 4.75% in CVC-ClinicDB, 1.77% in CVC-ColonDB, and 10.43% in ETIS-PolypLaribDB datasets in total due to ResNet50 and ASPP in an upsampling layer. Moreover, the proposed scheme outperforms better in comparison to NanoNet [27], ColonSegNet [28], DDANet [29], UNETR [30], PolypSegNet [31], nnU-Net [9], Li-SegPNet [3], FCN [33], Unet [34], Unet++ [35], MultiResUnet [36], ResUnet++ [37], LinkNet [38], Double-Unet [39], BA-Net [40], and Dil.ResFCN [41] in terms of dice, recall, precision, mIOU, and IOU due to the ResNet50, ASPP in an upsampling layer, and dropout being added in the channels. However, in terms of precision Li-SegNet is better than the proposed model. However, the proposed system limitation is the system needs hardware that supports more GPU-accelerated use of parallelization when working with exceeding core sizes datasets and this approach only performs effectively on datasets of medical images.
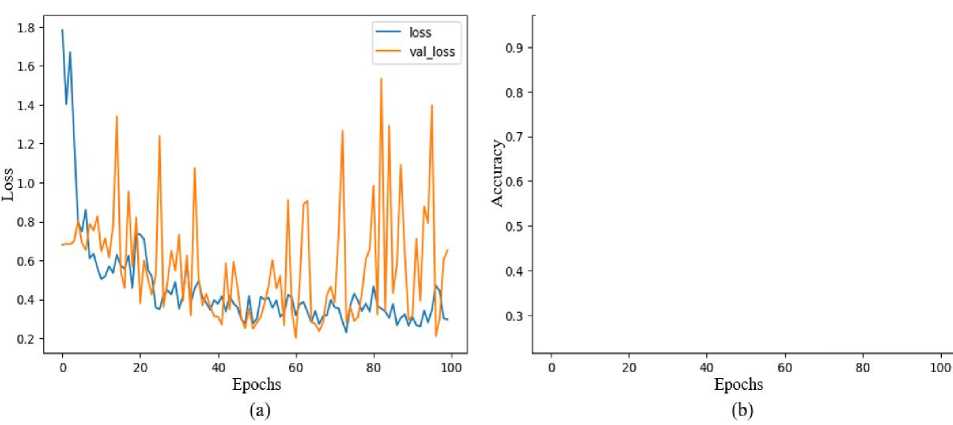
Fig. 9. The performance model graph (a) epoch verse loss and (b) epoch verse accuracy
Table 5. The performance outcomes of CRDL-PNet model for 5-fold cross-validation (a) CVC-ColonDB and (b) ETIS-PolypLaribDB datasets
|
Performance matrices |
CVC-ColonDB dataset |
ETIS-PolypLaribDB dataset |
||||||||||
|
Fold1 |
Fold2 |
Fold3 |
Fold4 |
Fold5 |
Mean Fold |
Fold1 |
Fold2 |
Fold3 |
Fold4 |
Fold5 |
Mean Fold |
|
|
Accuracy |
92.17 |
92.38 |
94.33 |
95.71 |
97.61 |
97.73 |
89.95 |
90.26 |
92.72 |
93.06 |
96.07 |
97.09 |
|
Jaccard |
88.09 |
88.51 |
89.17 |
90.33 |
92.82 |
93.92 |
87.14 |
88.94 |
89.74 |
90.05 |
92.11 |
92.37 |
|
Dice |
88.32 |
88.79 |
89.13 |
91.78 |
93.97 |
94.21 |
85.20 |
86.16 |
87.74 |
87.04 |
90.05 |
92.77 |
|
Precision |
81.63 |
82.08 |
82.73 |
83.13 |
84.03 |
84.03 |
80.65 |
81.31 |
82.98 |
82.13 |
84.13 |
84.58 |
|
Recall |
87.11 |
87.13 |
87.71 |
88.11 |
89.13 |
90.71 |
86.10 |
86.20 |
87.68 |
87.87 |
90.07 |
90.16 |
|
F1-score |
79.36 |
80.01 |
80.08 |
80.10 |
80.97 |
81.52 |
76.22 |
77.68 |
78.68 |
79.74 |
79.18 |
80.73 |
|
IOU |
85.33 |
85.97 |
86.17 |
88.77 |
89.10 |
89.97 |
85.14 |
85.12 |
86.68 |
87.89 |
88.13 |
88.73 |
|
mIOU |
87.79 |
88.14 |
88.71 |
89.33 |
90.07 |
90.63 |
87.08 |
87.91 |
88.13 |
88.94 |
90.33 |
90.52 |
Table 6. The performance of CRDL-PNet comparison with other existing state-of-the-arts models for polyp segmentation (Kvasir-SEG and CVC-ClinicDB datasets)
|
Models |
Kvasir-SEG dataset |
CVC-ClinicDB dataset |
||||||||
|
Dice |
Precisio n |
Recall |
mIOU |
IOU |
Dice |
Precision |
Recall |
mIOU |
IOU |
|
|
NanoNet [27] |
82.27 |
83.67 |
85.88 |
72.82 |
69.88 |
78.80 |
82.20 |
68.70 |
71.80 |
78.00 |
|
ColonSegNet [28] |
82.09 |
84.35 |
84.96 |
69.80 |
81.00 |
88.62 |
90.17 |
88.28 |
82.48 |
85.33 |
|
DDANet [29] |
85.76 |
86.43 |
88.80 |
78.00 |
75.48 |
90.72 |
93.25 |
84.56 |
85.87 |
79.36 |
|
UNETR [30] |
88.10 |
91.61 |
91.66 |
76.50 |
78.80 |
90.90 |
94.24 |
85.15 |
78.60 |
83.70 |
|
PolypSegNet [31] |
88.72 |
91.68 |
92.54 |
85.64 |
82.56 |
91.52 |
96.21 |
91.13 |
79.46 |
84.62 |
|
nnU-Net [9] |
89.77 |
90.85 |
82.71 |
81.06 |
81.78 |
90.60 |
95.87 |
83.25 |
79.12 |
85.32 |
|
Li-SegPNet [3] |
90.58 |
94.24 |
85.09 |
88.00 |
82.83 |
92.50 |
96.42 |
87.26 |
89.69 |
86.07 |
|
CRDL-PNet (Proposed) |
92.95 |
84.85 |
92.72 |
90.75 |
89.12 |
94.06 |
85.33 |
92.46 |
92.23 |
90.82 |
Щ
Е
£
90 ■
80 ■
70 ■
60 ■
50-
40 -
30-
20-
10-
■- Dice
*- Precision
■•■ Recall *- mIOU *- IOU
NN|27] CSN|23]DDAN|29) UN[3O] PSN[31] nnUN(9] LSPN[3] Proposed
Models
Ф
E
£
90 ■
SO- ,
70- Г
60 ■
50 -
40-
30-
20-
Dice Precision Recall mIOU IOU
Fig. 10. The performance model comparison with existing SOTA model (a) Kvasir-SEG and (b) CVC-ClinicDB
NN[27] CSN[28]DDAN[29] UN[3O] PSN[31] nnUN[9] LSPN(3] Proposed
Models (b)
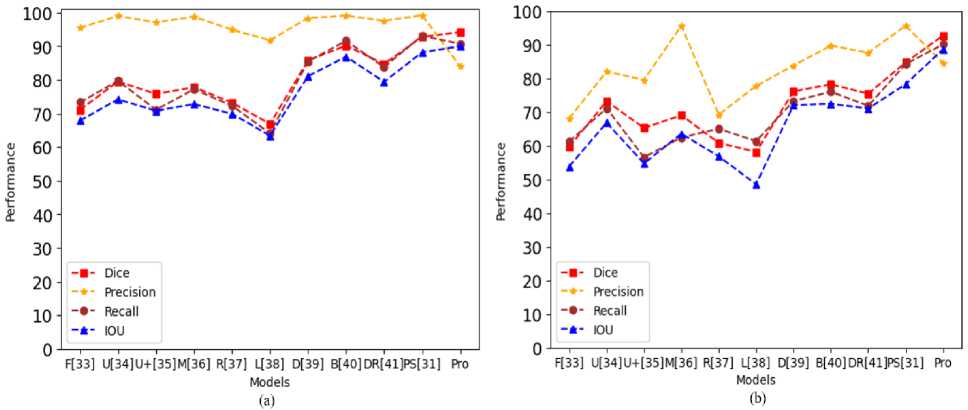
Fig. 11. The performance model comparison with existing SOTA model (a) CVC-ColonDB and (b) ETIS-PolypLaribDB
Table 7. The performance of CRDL-PNet comparison with other existing state-of-the-arts models for polyp segmentation (CVC-ColonDB and ETIS-PolypLaribDB datasets)
|
Models |
CVC-ColonDB dataset |
ETIS-PolypLaribDB dataset |
||||||
|
Dice |
Precision |
Recall |
IOU |
Dice |
Precision |
Recall |
IOU |
|
|
FCN [33] |
71.10 |
95.50 |
73.30 |
67.90 |
59.70 |
68.20 |
61.30 |
53.90 |
|
Unet [34] |
79.50 |
99.00 |
79.70 |
74.20 |
73.30 |
82.10 |
71.20 |
66.90 |
|
Unet++ [35] |
75.80 |
97.10 |
71.10 |
70.80 |
65.30 |
79.60 |
56.60 |
54.80 |
|
MultiResUnet [36] |
77.80 |
98.80 |
77.20 |
72.80 |
69.10 |
95.70 |
62.50 |
63.60 |
|
ResUnet++ [37] |
73.20 |
94.90 |
72.30 |
69.90 |
60.90 |
69.30 |
65.10 |
56.90 |
|
LinkNet [38] |
66.90 |
91.80 |
64.10 |
63.40 |
58.20 |
77.80 |
61.40 |
48.60 |
|
Double-Unet [39] |
85.80 |
98.40 |
85.50 |
81.10 |
76.20 |
83.90 |
73.30 |
72.10 |
|
BA-Net [40] |
90.20 |
99.10 |
91.60 |
86.90 |
78.30 |
89.80 |
76.10 |
72.50 |
|
Dil. ResFCN [41] |
84.70 |
97.60 |
83.80 |
79.40 |
75.50 |
87.70 |
71.90 |
71.10 |
|
PolySegNet [31] |
92.80 |
99.20 |
93.10 |
88.20 |
84.80 |
95.70 |
84.30 |
78.30 |
|
CRDL-PNet (Proposed) |
94.21 |
84.03 |
90.71 |
89.97 |
92.77 |
84.58 |
90.16 |
88.73 |
-
4.4. Discussion
In Table 2 and Table 3, the experimental results from the CVC-ClinicDB, Kvasir-SEG, ETIS-PolypLaribDB, and CVC-ColonDB datasets, respectively, are presented with a state-of-the-art comparison. NanoNet [27], ColonSegNet [28], DDANet [29], UNETR [30], PolypSegNet [31], nnU-Net [9], Li-SegPNet [3], FCN [33], Unet [34], Unet++ [35], MultiResUnet [36], ResUnet++ [37], LinkNet [38], Double-Unet [39], BA-Net [40], and Dil.ResFCN [41] are sixteen of the techniques that are compared with the proposed scheme (CVC-ClinicDB, Kvasir-SEG, ETIS-PolypLaribDB, and CVC-ColonDB datasets) shown in Table 6, Table 7, Fig. 9 and Fig. 10. Since all seven of these techniques are utilized for polyp colonoscopy images. As demonstrated, the proposed system performs better than all existing models in terms of polyp classification.
• The qualitative analysis for performance with Dice values of 92.95%, 94.06%, 94.21%, and 92.77%, Recall of 92.72%, 92.46%, 90.71%, and 90.16%, mIOU of 90.75%, 92.23%, 90.63%, and 90.52%, and IOU of 89.12%, 90.82%, 89.97%, and 88.73% on all four datasets, respectively demonstrate the greatest improvement. The proposed novel CRDL-PNet performs significantly better than NanoNet [27], ColonSegNet [28], DDANet [29], UNETR [30], PolypSegNet [31], nnU-Net [9], Li-SegPNet [3], FCN [33], Unet [34], Unet++ [35], MultiResUnet [36], ResUnet++ [37], LinkNet [38], Double-Unet [39], BA-Net [40], and Dil.ResFCN [41] on polyp image segmentation in real-world scenarios.
• The proposed scheme has the potential to decrease the significance of training time and enhance performance due to its use of the CRDL-PNet approach, which is often used to improve the system performance of polyp classification.
5. Conclusion and Future Work
Overall, the proposed scheme outperforms SOTA techniques and has comparatively minimal learning requirements.
The proposed CRDL-PNet scheme provided a scheme for segmenting polyp from colonoscopy images. The proposed ResNet50 feature extraction extracted from polyp colonoscopy images. Furthermore, an ASPP is used to handle scale variation during training and improve feature selection. Moreover, the GD approach is used to segmented boundary boxes of polyp. As a result, the CRDL-PNet learns additional specifics about the target area of polyp. In the future, CRDL-PNet will be tested on more medical-related tasks including secured feature detection in order to increase the effectiveness of medical image security.
Список литературы CRDL-PNet: An Efficient DeepLab-based Model for Segmenting Polyp Colonoscopy Images
- Bray F, Soerjomataram I, “The changing global burden of cancer: transitions in human development and implications for cancer prevention and control,” Cancer: disease control priorities, vol. 3, pp. 23-44, 2015.
- Yang X, Wei Q, Zhang C, Zhou K, Kong L, Jiang W, “Colon polyp detection and segmentation based on improved MRCNN,” IEEE Transactions on Instrumentation and Measurement, vol. 70, pp. 1-10, 2020.
- Sharma P, Gautam A, Maji P, Pachori R B, Balabantaray B K, “Li-SegPNet: Encoder-decoder Mode Lightweight Segmentation Network for Colorectal Polyps Analysis,” IEEE Transactions on Biomedical Engineering, vol. 70, no. 4, pp. 1330-1339, 2023.
- Gessl I, Waldmann E, Penz D, Majcher B, Dokladanska A, Hinterberger A, Ferlitsch M, “Evaluation of adenomas per colonoscopy and adenomas per positive participant as new quality parameters in screening colonoscopy,” Gastrointestinal endoscopy, vol. 89, no. 3, pp. 496-502, 2019.
- Bae S H, Yoon K J, “Polyp detection via imbalanced learning and discriminative feature learning,” IEEE Transactions on medical imaging, vol. 34, no. 11, pp. 2379-2393, 2015.
- Murmu A, Kumar P, “A novel Gateaux Derivatives with Efficient DCNN-ResUNet Method for Segmenting Multi-class Brain Tumor” Medical and Biological Engineering and Computing, 2023. 10.1007/s11517-023-02824-z.
- Murmu A, Kumar P, “Deep learning model-based segmentation of medical diseases from MRI and CT images,” In TENCON 2021 IEEE Region 10 Conference (TENCON), pp. 608-613, 2021.
- Wu H, Zhao Z, Zhong J, Wang W, Wen Z, Qin J, “Polypseg+: A lightweight context-aware network for real-time polyp segmentation,” IEEE Transactions on Cybernetics, vol. 53, no. 4, pp. 2610-2621, 2023.
- Isensee F, Petersen J, Klein A, Zimmerer D, Jaeger P F, Kohl S, Maier-Hein K H, “nnU-Net: Self-adapting framework for U-Net-based medical image segmentation,” Nature Methods, vol. 18, no. 2, pp. 203–211, 2021.
- Qadir H A, Shin Y, Solhusvik J, Bergsland J, Aabakken L, Balasingham I, “Toward real-time polyp detection using fully CNNs for 2D Gaussian shapes prediction,” Medical Image Analysis, vol. 68, pp. 101897, 2021.
- Mangotra H, Goel N, “Effect of selection bias on automatic colonoscopy polyp detection” Biomedical Signal Processing and Control, vol. 85, pp. 104915, 2023.
- Shi J H, Zhang Q, Tang Y H, Zhang Z Q, “Polyp-Mixer: An Efficient Context-Aware MLP-Based Paradigm for Polyp Segmentation,” IEEE Transactions on Circuits and Systems for Video Technology, vol. 33, no. 1, pp. 30-42, 2023.
- Chen B L, Wan J J, Chen T Y, Yu Y T, Ji M, “A self-attention based faster R-CNN for polyp detection from colonoscopy images,” Biomedical Signal Processing and Control, vol. 70, pp. 103019, 2021.
- Yue G, Li S, Cong R, Zhou T, Lei B, Wang T, “Attention-guided pyramid context network for polyp segmentation in colonoscopy images,” IEEE Transactions on Instrumentation and Measurement, vol. 72, pp. 1-13, 2023.
- Adjei P E, Lonseko Z M, Du W, Zhang H, Rao N, “Examining the effect of synthetic data augmentation in polyp detection and segmentation,” International Journal of Computer Assisted Radiology and Surgery, vol. 17, no. 7, pp. 1289-1302, 2022.
- Banik D, Roy K, Bhattacharjee D, Nasipuri M, Krejcar O, “Polyp-Net: A multimodel fusion network for polyp segmentation,” IEEE Transactions on Instrumentation and Measurement, vol. 70, pp. 1-12, 2020.
- Yue G, Han W, Li S, Zhou T, Lv J, Wang T, “Automated polyp segmentation in colonoscopy images via deep network with lesion-aware feature selection and refinement,” Biomedical Signal Processing and Control, vol. 78, pp. 103846, 2022.
- Jin, Y., Hu, Y., Jiang, Z., & Zheng, Q. (2022). Polyp segmentation with convolutional MLP. The Visual Computer, 1-19.
- Zhang W, Fu C, Zheng Y, Zhang F, Zhao Y, Sham C W, “HSNet: A hybrid semantic network for polyp segmentation,” Computers in biology and medicine, vol. 150, pp. 106173, 2022.
- Mohapatra S, Pati G K, Mishra M, Swarnkar T, “UPolySeg: A U-Net-Based Polyp Segmentation Network Using Colonoscopy Images,” Gastroenterology Insights, vol. 13, no. 3, pp. 264-274, 2022.
- Yeung M, Sala E, Schönlieb C B, Rundo L, “Focus U-Net: A novel dual attention-gated CNN for polyp segmentation during colonoscopy,” Computers in biology and medicine, vol. 137, pp. 104815, 2021.
- Kang J, Gwak J, “Ensemble of instance segmentation models for polyp segmentation in colonoscopy images,” IEEE Access, vol. 7, pp. 26440-26447, 2019.
- Jha Debesh and Smedsrud, Pia H and Riegler, Michael A and Halvorsen, Pl and de Lange, Thomas and Johansen, Dag and Johansen, Hvard D, “Kvasir-seg: A segmented polyp dataset” International Conference on Multimedia Modeling, pp. 451-462, 2020. Available online: https://datasets.simula.no/kvasir-seg/ (accessed on 28 June 2023).
- CVC-ClinicDB, Dataset of Endoscopic Colonoscopy Frames for Polyp Detection, 2021. Available online: https://www.kaggle.com/datasets/balraj98/cvcclinicdb (accessed on 28 June 2023).
- Liu C, Chen L C, Schroff F, Adam H, Hua W, Yuille A L , Li F F, “Auto-DeepLab: Hierarchical Neural Architecture Search for Semantic Image Segmentation,” In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, pp. 15–20, June 2019.
- Prasad S, Kumar P, Sinha K P, “Grayscale to color map transformation for efficient image analysis on low processing devices. In Advances in Intelligent Informatics, pp. 9-18, 2015.
- Jha D, Tomar N K, Ali S, Riegler M A, Johansen H D, Johansen D, Halvorsen P, “Nanonet: Real-time polyp segmentation in video capsule endoscopy and colonoscopy,” In 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), pp. 37-43, 2021.
- Jha D, Ali S, Tomar N K, Johansen H D, Johansen D, Rittscher J, Halvorsen P, “Real-time polyp detection, localization and segmentation in colonoscopy using deep learning,” IEEE Access, vol. 9, pp. 40496-40510, 2021.
- Tomar N K, Jha D, Ali S, Johansen H D, Johansen D, Riegler M A, Halvorsen P, “DDANet: Dual decoder attention network for automatic polyp segmentation,” In Pattern Recognition ICPR International Workshops and Challenges: Virtual Event, pp. 307-314, 2021.
- Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, Xu D, “Unetr: Transformers for 3d medical image segmentation,” In Proceedings of the IEEE/CVF winter conference on applications of computer vision, pp. 574-584, 2022.
- Mahmud T, Paul B, Fattah S A, “PolypSegNet: A modified encoder-decoder architecture for automated polyp segmentation from colonoscopy images,” Computers in Biology and Medicine, vol. 128, pp. 104119, 2021.
- Rahman M, Murmu A, Kumar P, Moparthi N R, Namasudra S, “A novel compression-based 2D-chaotic sine map for enhancing privacy and security of biometric identification systems,” Journal of Information Security and Applications, vol. 80, pp. 103677, 2024.
- Long J, Shelhamer E, Darrell T, “Fully convolutional networks for semantic segmentation,” In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3431-3440, 2015.
- Ronneberger O, Fischer P, Brox T, “U-net: Convolutional networks for biomedical image segmentation,” In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, pp. 234-241, 2015.
- Zhou Z, Siddiquee M M R, Tajbakhsh N, Liang J, “Unet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE transactions on medical imaging, vol. 39, no. 6, pp. 1856-1867, 2019.
- Ibtehaz N, Rahman M S, “MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation,” Neural networks, vol. 121, pp. 74-87, 2020.
- Jha D, Smedsrud P H, Riegler M A, Johansen D, De Lange T, Halvorsen P, Johansen H D, “Resunet++: An advanced architecture for medical image segmentation,” In 2019 IEEE international symposium on multimedia (ISM), pp. 225-2255, 2019.
- Chaurasia A, Culurciello E, “Linknet: Exploiting encoder representations for efficient semantic segmentation,” In 2017 IEEE visual communications and image processing (VCIP), pp. 1-4, 2017.
- Jha D, Riegler M A, Johansen D, Halvorsen P, Johansen H D, “Doubleu-net: A deep convolutional neural network for medical image segmentation,” In 2020 IEEE 33rd International symposium on computer-based medical systems (CBMS), pp. 558-56, 2020.
- Wang R, Chen S, Ji C, Fan J, Li Y, “Boundary-aware context neural network for medical image segmentation,” Medical Image Analysis, vol. 78, pp. 102395, 2022.
- Guo Y, Bernal J, J Matuszewski B, “Polyp segmentation with fully convolutional deep neural networks—extended evaluation study,” Journal of Imaging, vol. 6, no. 7, pp. 69, 2020.
- Mondal J J, Islam M F, Zabeen S, Manab M A, “InvoPotNet: Detecting Pothole from Images through Leveraging Lightweight Involutional Neural Network,” In 2022 25th IEEE International Conference on Computer and Information Technology (ICCIT), pp. 599-604, 2022.
- Ye R, Feng S, Li X, Ye Y, Zhang B, Zhu Y, Wang Y, “WDMNet: Modeling diverse variations of regional wind speed for multi-step predictions,” Neural Networks, vol. 162, pp. 147-161, 2023.
- Figshare, CVC-ColonDB Polyp colonoscopy image dataset [Available online: https://figshare.com/articles/figure/Polyp_DataSet_zip/21221579]
- Silva J, Histace A, Romain O, Dray X, Granado B, “Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer,” International Journal of Computer Assisted Radiology and Surgery, vol. 9, no. 2, pp. 283-293, 2013. DOI:10.1007/s11548-013-0926-3.
- Chen X, Wang X, Zhang K, Fung K M, Thai T C, Moore K, Qiu Y, “Recent advances and clinical applications of deep learning in medical image analysis,” Medical Image Analysis, vol. 79, pp. 102444, 2022.
- Minaee S, Boykov Y, Porikli F, Plaza A, Kehtarnavaz N, Terzopoulos D, “Image segmentation using deep learning: A survey,” IEEE Transactions on pattern analysis and machine intelligence, vol. 44, no. 7, pp. 3523-3542, 2021.
- Gu Z, Cheng J, Fu H, Zhou K, Hao H, Zhao Y, Liu J, “Ce-net: Context encoder network for 2d medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 38, no. 10, pp. 2281-2292, 2019.


