Detection of genetic variations in marine algae Ulva lactuca (Chlorophyta) induced by heavy metal pollutants
Автор: Saleh Basel
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 3 т.11, 2015 года.
Бесплатный доступ
Ulva lactuca (Chlorophyta) green macroalgae has been successfully used as bioindicator for heavy metals pollution in ecosystems. Random amplified microsatellite polymorphism (RAMP) marker was employed to investigate genetic DNA pattern variability in green U. lactuca 5 days after exposure to Cu, Pb, Cd and Zn heavy metals stress. Genomic template stability (GTS%) value was employed as a qualitative DNA changes measurement based on RAMP technique. In this respect, estimated GTS% value was recorded to be 65.215, 64.630, 59.835 and 59.250% for Pb, Cu, Cd and Zn treatment, respectively. Moreover, genetic similarity (GS) induced by the above heavy metals was also evaluated to measure genetic distance between algae treated plants and their respective control. In this respect, estimated GS values generated by RAMP marker ranged between 0.576 (between control and Zn treatment) - 0.969 (for both case; between Pb and Cu and between Cd and Zn treatment) with an average of 0.842. Based upon data presented herein based on variant bands number (VB), GTS% and GS values; the present study could be suggested that Pb and Cu followed similar tendency at genomic DNA changes. Similar finding was also observed with Cd and Zn ions. Thereby, RAMP marker successfully highlighted DNA change patterns induced by heavy metals stress.
Ulva lactuca, algae, ramp marker, dna changes, heavy metal stress
Короткий адрес: https://sciup.org/14323924
IDR: 14323924
Текст научной статьи Detection of genetic variations in marine algae Ulva lactuca (Chlorophyta) induced by heavy metal pollutants
Heavy metals pollution is considered a threat to the environmental ecosystem including seas, rivers and lakes. This field has captured more attention globally and this is due to the fact that, these pollutants have high toxicity and they do not decompose easily in the environment. Therefore, many scientific reports studied the toxicity of heavy metals, their effects on the environment, their ability to enter the food chain and their threat on the health of human being (Rainbow, 1995, 2002; Bing et al., 2013; Khan et al., 2013). Many researches were focused on the response of living organisms when they are exposed to heavy metals, and the possible methods to get rid of their toxicity. The most important advantage of the use of living organisms to monitor the pollution by heavy metals is the ability of heavy metals to be concentrated in the organism's tissues; this allows the monitoring of the moving part of the heavy metals in the environment (Rainbow, 1995). The use of sea creatures as a useful tool in environmental monitoring goes back to the seventies of the last century. The results obtained from those studies form an important base in environmental monitoring. One of the most important problems now is the ability to compare the results obtained from different studies between different creatures, and this is due to the fact that the concentration of a heavy metals in an organism is affected by many physiological and biological factors (Rainbow, 2002). Previous studies have shown that high plants respond differently to heavy metals in their ecosystems. This might be due to the different genetic make-up of the plants (De-Wolf et al., 2004). Aquatic plants have great and noticed ability to concentrate heavy metals in their tissues. Thus, they are known as effective bioindicator to monitor the toxicity caused by heavy metals (Gupta and Sarin, 2009).
Chemical pollutants may cause physical damage to the sequence or the structure of DNA. These damages can be detected using many genetic markers. e.g . Swaileh et al . (2008) applied RAPD marker to investigate genotoxicity induced by wastewater in oat plants ( Avena sativa ). Gupta and Sarin (2009) used RAPD and SCAR markers to investigate Cd, Hg and Cu metals genotoxicity in two aquatic plants. Al-Qurainy et al . (2010) studied genotoxicity of Cd, Pb and Zn ions on Eruca sativa L. using RAPD marker. Cansaran-Duman et al . (2011) employed RAPD marker to detect genotoxicity in Evernia prunastri L. Recently, Raj et al . (2014) investigated tannery effluents impact on mung bean plants using RAPD marker.
Thereby, the current study was carried out with hopeful aim to investigate genotoxicity induced by Cu, Pb, Cd and Zn chemical pollutants in Ulva lactuca macrogreen algae using RAMP marker. Their abundance along the Syrian coast of the Mediterranean Sea was promising factor to use these plants as bioindicator for heavy metals pollution. MATERIALS AND METHODS
Algae sampling and chemical pollution application’s: U. lactuca algae samples were collected along the Syrian coast of the Mediterranean Sea. Samples collection was carried out (35°33ʹ790ʹʹN longitude and 35°43ʹ996ʹʹE latitude) at 4 km North Lattakia - Syria. Sampling was carried out by harvest only individual with the similar size with disposable gloves. Algae were directly washed with seawater where the algae were collected and then transported within a flask with 5 L fresh seawater.
Algae were washed twice upon their arrival to the laboratory, with autoclaved artificial seawater ASW (500 mM NaCl, 10 mM KCl, 30 mM MgSO 4 , 10 mM CaCl 2 and 10 mM Tris-HCl at pH 7.8) medium as previously described by Unal et al . (2010).
Then, they were divided to fresh flask with a fresh ASW previously described solution and kept under controlled laboratory conditions (Temperature of 20˚C, photoperiod of 12/12 h dark/light and illumination of 2950 Lux (~48.7 µmol photons m-2s-1) for 3 days before heavy metals stress application. The mentioned ASW was considered as a control. Chemical stress was applied by adding 18.2 mg/L of Pb2+, 5.8 mg/L of Cu2+, 10.5 mg/L of Cd2+ and 9,9 mg/L of Zn2+ (Standard solution (1000 mg/L) from Fisher Scientific –UK, under their nitrate forms) for each treatment with three replicates/treatment. Experiment was carried out in flask containing 300 mL ASW with or without heavy metals. The same previous described controlled conditions were maintained during the experiment stress application. Five days later, algae were harvested for genetic study.
Total DNA extraction and RAMP marker assay
Genomic DNA was isolated from algal tissues for both of the control and stressed plants by a CTAB (cetyltrimethylammonium bromide) protocol as previously described by Doyle and Doyle (1987) with minor modifications. Tissues (150 mg) were ground in liquid nitrogen, the powder was transferred to a 2 mL Eppendorf tube, homogenized with 900 μL of extraction buffer (100 mM Tris-HCl, pH 8.0, 1.4 M NaCl, 20 mM EDTA, 0.0018 mL β-mercaptoethanol, 2% CTAB), and incubated at 65°C for 20 min. One volume of a chloroform:isoamyl alcohol mix (24:1, v/v) was added, followed by centrifugation at 12,000 g for 10 min at 4°C. The aqueous phase was transferred to a fresh tube, and the DNA was precipitated with an equal volume of cold isopropanol and kept at -20°C for 10 min. Then centrifuged at 12,000 g for 10 min at 4°C, the supernatant was eliminated; DNA was then spooled out and washed with 1 M ammonium acetate and 100% ethanol. The cleaned DNA pellet was air dried and dissolved in 100 μL of 0.1X TE buffer (1 mM Tris-HCl, 0.1 mM EDTA, pH 8.0). After addition of 5 μL of RNase (10 mg/mL), and incubation for 30 min at 37°C. DNA concentration was quantified by DNA Fluorimeter at 260/280 nm and adjusted to final concentration of 10 ng/μL. DNA was stored at -80°C until needed.
RAMP technique consists of the combination of two PCR-DNA based markers (RAPD and ISSR primers). PCR amplification was performed as previously reported by Saleh (2011). RAMP system was applied using 26 (RAPD/ISSR) primer combinations (Table 1) to detect DNA change patterns induced by heavy metals stress compared to their respective control.
RAMP-PCR amplification reactions were performed in 25 µL reaction volume involving 1X PCR buffer, 2 mM MgCl2, 0.25 mM dNTPs, 25 pmoL primer (Operon Technologies Inc. USA), 1.5 U Taq DNA polymerase (Fermentas) and 30 ng template DNA. PCR amplification was carried out in a T-gradient thermal cycler (Bio-Rad, Hercules, USA). It was programmed for 35 cycles after an initial denaturation cycle for 4 min at 94˚C. Where, each cycle consisted of denaturation step 1 min at 94˚C, followed by an annealing step for 2 min at 38˚C, and then an extension step at 72˚C for 2 min, then an extension cycle for 7 min at 72˚C as a final cycle. The PCR products were then separated on a 1.8% ethidium bromide-stained agarose (Bio-Rad, Hercules, USA) in 0.5 X TBE buffer. Separation of PCR products was performed by electrophoresis at 85 V for 2.5 h, and then visualized with a UV transilluminator. A VC 100bp Plus DNA Ladder (Vivantis) ladder standard was used to estimate molecular weight of PCR amplification products.
Amplification patterns yielded by RAMP technique was screened and photographed under UV light. The presence or absence of each size class was scored as 1 or 0, respectively. The estimated percent disagreement values (PDVs) found were used as template to generate a matrix via the Unweighted Pair Group Mean Arithmetic average (UPGMA) using Statistica program (Statsoft 2003). The latter matrix was used to estimate genetic similarity distance (Jaccard, 1908).
Genomic template stability (GTS%) test and genetic similarity distance (GS)
Genomic template stability value was estimated as previously reported by Atienzar et al. (2002) and Cansaran-Duman et al. (2011) according to the following formulate Eq. (1):
GTS% = (1 – a/n) x 100 (1)
Where (a) was RAMP polymorphic profiles detected in each samples treated and (n) the number of total bands in the control. Polymorphism observed in RAMP profiles included disappearance of a normal band and induction of a new band in comparison to the control RAMP profiles.
Jaccard similarity (GS) index based on percent disagreement values (PDV) was used as a comparative analysis to demonstrate the genetic distance among the control and the 4 examined ions. RESULTS
Induced DNA alterations in U. lactuca algae has been evaluated against 4 metals (Cu, Pb, Cd and Zn) toxicity using RAMP marker. Data presented herein mentioned that, one PC (AC)8T/OPR12 combination fail to amplify algal DNA. Moreover, seven PCs (PC2, PC4, PC5, PC9, PC10, PC13 and PC23) combinations gave monomorphic pattern between control and algae treated plants. So, 19 PCs out of 26 PCs combinations were used for monitoring DNA changes induced by heavy metals stress in U. lactuca . As shown in Fig. 1, DNA change patterns induced by applied heavy metals stress as revealed by PC8, PC21 and PC22 RAMP PCs combinations.
The identified variant bands (VB) yielded by 19 RAMP PCs combinations induced in U. lactuca algae by heavy metal (Cu, Pb, Cd and Zn) stress compared to their respective control were presented in Table 2. Where, the total VB generated by the applied four ions were recorded to be 180 bands.
DNA change patterns in U. lactuca induced by chemical pollutants (Cu, Pb, Cd and Zn) has been investigated. From Table 2, it worth noting that, Pb and Cu ions followed the same trend in terms of VB characteristic for these examined ions. Where, VB estimated for the latter mentioned ions were varied between 1 (PC12, PC18, PC24 and PC26) and 5 (PC8 and PC22). As for Cd and Zn, they were also followed the same trend in term of VB. In this respect, VB estimated for these ions were also ranged between 1 (PC12, PC14, PC18, PC24 and PC26) and 5 (PC8 and PC22).
DNA alteration induced by heavy metals stress was expressed by a genomic template stability (GTS %) for each ion treatment (Table 3). In this respect, estimated GTS% values ranged between 28.571 with PC22 combination for all tested ions, and 100% for few PCs combinations according to the tested PCs combinations and examined ions.
Table 1. RAMP primer combinations examined in this study.
|
PC °N |
Primer combination |
Primer Sequence 5' to 3' |
PC °N |
Primer combination |
Primer Sequence 5' to 3' |
|
PC1 |
ISSR1 / OPA18 |
(AC)8T / AGGTGACCGT |
PC14 |
ISSR2 / OPT18 |
(AG)8TC / GATGCCAGAC |
|
PC2 |
ISSR1 / OPB12 |
(AC)8T / CCTTGACGCA |
PC15 |
ISSR2 / OPV03 |
(AG)8TC / CTCCCTGCAA |
|
PC3 |
ISSR1 / OPC07 |
(AC)8T / GTCCCGACGA |
PC16 |
ISSR2 / OPW17 |
(AG)8TC / GTCCTGGGTT |
|
PC4 |
ISSR1 / OPQ18 |
(AC)8T / AGGCTGGGTG |
PC17 |
ISSR2 / UBC159 |
(AG)8TC / GAGCCCGTAG |
|
PC5 |
ISSR1 / OPT18 |
(AC)8T / GATGCCAGAC |
PC18 |
ISSR3 / OPA18 |
(AG)8GTG / AGGTGACCGT |
|
PC6 |
ISSR1 / OPV03 |
(AC)8T / CTCCCTGCAA |
PC19 |
ISSR3 / OPB12 |
(AG)8GTG / CCTTGACGCA |
|
PC7 |
ISSR1 / OPW17 |
(AC)8T / GTCCTGGGTT |
PC20 |
ISSR3 / OPC07 |
(AG)8GTG / GTCCCGACGA |
|
PC8 |
ISSR1 / UBC159 |
(AC)8T / GAGCCCGTAG |
PC21 |
ISSR3 / OPQ18 |
(AG)8GTG / AGGCTGGGTG |
|
PC9 |
ISSR2 / OPA18 |
(AG)8TC / AGGTGACCGT |
PC22 |
ISSR3/ OPR12 |
(AG)8GTG / ACAGGTGCGT |
|
PC10 |
ISSR2 / OPB12 |
(AG)8TC / CCTTGACGCA |
PC23 |
ISSR3 / OPT18 |
(AG)8GTG / GATGCCAGAC |
|
PC11 |
ISSR2 / OPC07 |
(AG)8TC / GTCCCGACGA |
PC24 |
ISSR3 / OPV03 |
(AG)8GTG / CTCCCTGCAA |
|
PC12 |
ISSR2 / OPQ18 |
(AG)8TC / AGGCTGGGTG |
PC25 |
ISSR3 / OPW17 |
(AG)8GTG / GTCCTGGGTT |
|
PC13 |
ISSR2 / OPR12 |
(AG)8TC / ACAGGTGCGT |
PC26 |
ISSR3 / UBC159 |
(AG)8GTG / GAGCCCGTAG |
Notes: PC: Primer combination.
Table 2 . Number of variant bands identified by RAMP marker under heavy metals stress.
|
PCs Name |
Pb |
Cu |
Cd |
Zn |
Total |
|
PC1 |
2 |
2 |
4 |
4 |
12 |
|
PC3 |
2 |
2 |
2 |
2 |
8 |
|
PC6 |
2 |
3 |
4 |
5 |
14 |
|
PC7 |
2 |
2 |
2 |
2 |
8 |
|
PC8 |
5 |
5 |
5 |
5 |
20 |
|
PC11 |
2 |
2 |
2 |
2 |
8 |
|
PC12 |
1 |
1 |
1 |
1 |
4 |
|
PC14 |
0 |
0 |
1 |
1 |
2 |
|
PC15 |
3 |
3 |
3 |
3 |
12 |
|
PC16 |
4 |
4 |
4 |
4 |
16 |
|
PC17 |
3 |
3 |
3 |
3 |
12 |
|
PC18 |
1 |
1 |
1 |
1 |
4 |
|
PC19 |
2 |
2 |
2 |
2 |
8 |
|
PC20 |
2 |
2 |
2 |
2 |
8 |
|
PC21 |
0 |
0 |
2 |
2 |
4 |
|
PC22 |
5 |
5 |
5 |
5 |
20 |
|
PC24 |
1 |
1 |
1 |
1 |
4 |
|
PC25 |
3 |
3 |
3 |
3 |
12 |
|
PC26 |
1 |
1 |
1 |
1 |
4 |
|
Total |
41 |
42 |
48 |
49 |
180 |
Notes: PC: Primer combination.
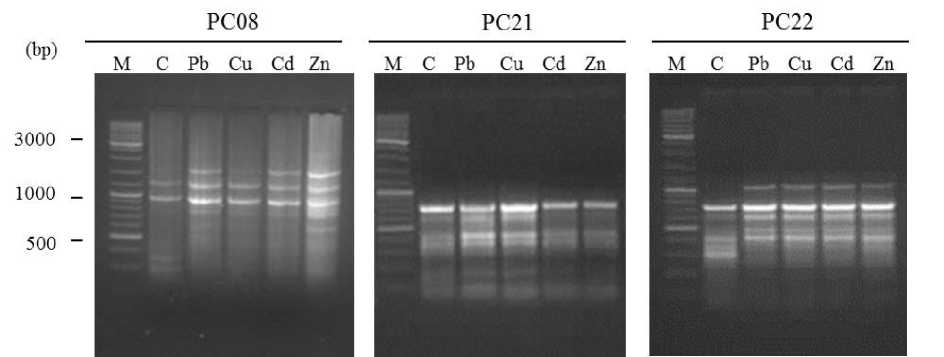
Figure 1. RAMP profile of DNA pattern changes induced after 5 days exposure to Pb, Cu, Cd and Zn heavy metals stress in U. lactuca.
Table 3. Genomic template stability (GTS%) estimated by RAMP marker.
|
PCs Name |
Control |
Pb |
Cu |
Cd |
Zn |
|
PC1 |
100.000 |
80.000 |
80.000 |
60.000 |
60.000 |
|
PC3 |
100.000 |
75.000 |
75.000 |
75.000 |
75.000 |
|
PC6 |
100.000 |
77.778 |
66.667 |
55.556 |
44.444 |
|
PC7 |
100.000 |
71.429 |
71.429 |
71.429 |
71.429 |
|
PC8 |
100.000 |
37.500 |
37.500 |
37.500 |
37.500 |
|
PC11 |
100.000 |
50.000 |
50.000 |
50.000 |
50.000 |
|
PC12 |
100.000 |
75.000 |
75.000 |
75.000 |
75.000 |
|
PC14 |
100.000 |
100.000 |
100.000 |
80.000 |
80.000 |
|
PC15 |
100.000 |
40.000 |
40.000 |
40.000 |
40.000 |
|
PC16 |
100.000 |
50.000 |
50.000 |
50.000 |
50.000 |
|
PC17 |
100.000 |
40.000 |
40.000 |
40.000 |
40.000 |
|
PC18 |
100.000 |
80.000 |
80.000 |
80.000 |
80.000 |
|
PC19 |
100.000 |
71.429 |
71.429 |
71.429 |
71.429 |
|
PC20 |
100.000 |
60.000 |
60.000 |
60.000 |
60.000 |
|
PC21 |
100.000 |
100.000 |
100.000 |
60.000 |
60.000 |
|
PC22 |
100.000 |
28.571 |
28.571 |
28.571 |
28.571 |
|
PC24 |
100.000 |
85.714 |
85.714 |
85.714 |
85.714 |
|
PC25 |
100.000 |
50.000 |
50.000 |
50.000 |
50.000 |
|
PC26 |
100.000 |
66.667 |
66.667 |
66.667 |
66.667 |
|
Mean |
100.000 |
65.215 |
64.630 |
59.835 |
59.250 |
Notes: PC: Primer combination.
Table 4 . Jaccard's similarity matrix among heavy metals treated plants and their control.
|
Sample |
C |
Pb |
Cu |
Cd |
Zn |
|
C |
1.000 |
||||
|
Pb |
0.628 |
1.000 |
|||
|
Cu |
0.626 |
0.969 |
1.000 |
||
|
Cd |
0.578 |
0.928 |
0.919 |
1.000 |
|
|
Zn |
0.576 |
0.900 |
0.930 |
0.969 |
1.000 |
Notes: C: control.
Table 5 . Percent Disagreement Values (PDV) yielded by RAMP marker.
|
Sample |
C |
Pb |
Cu |
Cd |
Zn |
|
C |
0.000 |
||||
|
Pb |
0.360 |
0.000 |
|||
|
Cu |
0.360 |
0.030 |
0.000 |
||
|
Cd |
0.420 |
0.060 |
0.070 |
0.000 |
|
|
Zn |
0.420 |
0.080 |
0.060 |
0.030 |
0.000 |
Notes: C: control.
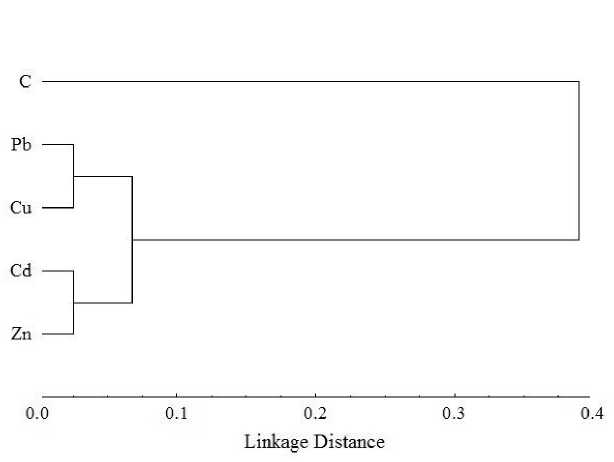
Figure 2. Dendrogram for RAMP marker showing genetic relationship among the U. lactuca control plants and heavy metals treated plants using UPGMA cluster analysis.
DISCUSSION
The unfavorable impact of four heavy metals (Pb, Cu, Cd and Zn) was evaluated in U. lactuca at genetic level. Genotoxicity of the above mentioned ions has been manifested by comparing DNA patterns under treatment compared to their respective control. Where the heavy metals application induced DNA alterations (loss or/and gain bands). According to Atienzar and Jha (2006), in genotoxicity study, the new induced bands could be related to DNA mutation as previously reported by Atienzar et al . (2002), who suggested that DNA mutation is response to a new bands induction, if the mutation occurs at the same locus in sufficient number of cells. While, the disappeared bands could be related to DNA damages.
Nineteen RAMP PCs primer combinations were employed for screening DNA change patterns induced by the mentioned ions. In this respect, PC8 and PC22 combinations yielded the highest VB for the tested four ions compared to the other examined PCs combinations. Overall, the VB induced with the four ions was in the following order of 41, 42, 48 and 49 bands for Pb, Cu, Cd and Zn ion treatment, respectively. Comparative assessment among the 4 examined ions in other investigation in term of total bands (TB) and polymorphic level (P%) (data not shown here) revealed that, TB number was recorded to be 92, 95, 91 and 96 bands with 34.8, 35.4, 40.2 and 40.7% as P% for Pb, Cu, Cd and Zn ion treatment, respectively.
Where, Swaileh et al . (2008) applied 15 RAPD primers to investigate genotoxicity induced by wastewater in oat plants ( Avena sativa ). Al-Qurainy (2009) reported DNA changes induced by toxic Al and Ni ions on Phaseolus vulgaris L. using RAPD marker. The previous study revealed that among 10 RAPD tested primers, 5 produced monomorphic bands and 4 gave unique bands. This investigation allowed identifying genes involved in plant response to Al and Ni toxicity as a starting point. Whereas, Cansaran-
Duman et al . (2011) investigated air pollution using 21 RAPD primers to detect genotoxicity in Evernia prunastri L. collected from different polluted areas in Turkey. The same study mentioned that, 13 out of 21 tested RAPD primers produced reproducible bands. In this respect, the highest polymorphic bands were found in areas closed to the railways and motorways and station near to the iron-steel factory.
Recently, Raj et al . (2014) stated that, the total number of bands were recorded to be 33 as disappeared and 20 as a new produced bands in mung bean plants with untreated effluent compared to their respective control. Indeed, the same investigation indicated the appearance of 12 new bands with disappearance of 18 bands was observed in mung bean plants with untreated effluent compared to their respective control. While, this number was recorded to be 15 and 8 bands for disappeared and new appeared bands respectively in plants treated with effluent.
Previously, Gupta and Sarin (2009) applied 7 RAPD and 4 SCAR primers to investigate DNA changes induced by Cd, Hg and Cu metals in two aquatic plants. The previous study indicated a reduction in GTS was correlated with decrease in chlorophyll and protein content. Similar finding was reported in marine Palnaria palnata (Atienzar et al ., 2000). The latter study stated that GTS was decreased under UV treatment.
RAPD marker successfully applied to detect DNA changes induced in plants by heavy metals (Enan, 2006; Swaileh et al., 2008; Cenkci et al., 2009; Cansaran-Duman et al., 2011; Liu et al., 2012; Raj et al., 2014). In this regards, Al-Qurainy (2010) investigated the genotoxicity of Cd, Pb and Zn ions on Eruca sativa L. after 8 days exposure using 20 ISSR primers. The previous study mentioned that, GS value ranged between 42.8-100%. Otherwise, the same study mentioned that among the 20 ISSR tested primers, only two primers (OPC5 and OPC7) produced the highest bands with 150 mg/L of Cd and Pb treatment. Whereas, Al-Qurainy et al. (2010) reported the genotoxicity of Cd, Pb and Zn ions on E. sativa L. after 8 days exposure using 20 RAPD primers. The previous investigation revealed that, Jaccard similarity value ranged between 47.8395.83%. Moreover, the latter study indicated that among the 20 RAPD tested primers, only three primers (OPC11, OPC12 and OPC13) produced the highest bands with 150 mg/L of Cd and Pb treatment.
Recently, Raj et al . (2014) reported induced DNA changes on mung bean by tannery effluents (CETP) using RAPD marker. The latter study gave a total bands of 87 of which 42 (48%) were polymorphic using 8 RAPD primers. Indeed, Nei’s similarity between control and treated plants was found to be 0.75.
In the current study, cluster analysis test showed the low genetic distance between the 4 tested ions compared to untreated plants. Where, Pb and Cu ions caused similar DNA changes (PDV = 0.03; with GS = 0.969). Similarly, Cd and Zn ions showed the same tendency (PDV = 0.03; with GS = 0.969). Our data were in accordance with previous findings of Enan (2006), who reported a clear distance between control and treated plants of P. vulgaris L. after Cd, Pb, Mn and Cu exposure.
The observed VB detected in the case of Pb and Cu treatment could be suggested that the two previous ions could be caused similar DNA change patterns. Similar tendency could be also observed with Cd and Zn treatment suggesting that these ions exhibited the same impact on DNA alterations. In this regards, Enan (2006) reported that Cu and Cd ions may cause similar DNA modification in P. vulgaris L. treated plants compared to their respective control.
Based upon the estimated VB, GTS% and GS values; the current study could be suggested that Pb and Cu followed similar tendency at genomic DNA changes. Similar finding was also observed with Cd and Zn ions.
In conclusion, genotoxicity of 4 heavy metals (Pb, Cu, Cd and Zn) ions was evaluated on U. lactuca algae using RAMP marker. DNA changes induced by the above mentioned ions were investigated by monitoring loss or gain bands profile yielded under each treatment compared to their respective reference. Number of VB, GTS% and Jaccard’s GS values were estimated to investigate genetic variation induced by the previous heavy metals on U. lactuca algae. Where, Pb and Cu treatment exhibited relatively highest GTS% with lowest number of variant bands reflecting that these ions caused similar DNA changes. Indeed, Cd and Zn ions revealed similar tendency regarding DNA alterations. Thereby, the current study revealed that RAMP marker could be considered as a potential and sensitive tool for investigation induced DNA changes by heavy metals. Based upon our data, it is recommended to use of U.
lactuca algae as useful bioindicator to investigate heavy metals pollution in ecosystems as previously reported in many investigations. Furthermore, heavy metals genotoxicity should be performed by testing different ion concentrations. Otherwise, physiological and biochemical studies performance is needed; that could be considered as a useful support to molecular assay. Thereby, give a clear image about the specific genotoxicity impact of these ions in ecosystems.
Список литературы Detection of genetic variations in marine algae Ulva lactuca (Chlorophyta) induced by heavy metal pollutants
- Al-Qurainy, F. (2009) Toxicity of heavy metals and their molecular detection on Phaseolus vulgaris (L.). Aust. J. Basic. Appl Sci., 3: 3025-3035
- Al-Qurainy, F. (2010) Application of inter simple sequence repeat (ISSR marker) to detect genotoxic effect of heavy metals on Eruca sativa (L.). Afr. J. Biotechnol., 9: 467-474
- Al-Qurainy, F., Alameri, A.A. and Khan, S. (2010) RAPD profile for the assessment of genotoxicity on a medicinal plant; Eruca sativa. J. Med. Plant. Res., 4: 579-586
- Atienzar, F.A., Cordi, B., Donkin, M.B., Evenden, A.J., Jha, A.N. and Depledge, M.H. (2000) Comparison of ultraviolet-induced genotoxicity detected by random amplified polymorphic DNA with chlorophyll fluorescence and growth in a marine macroalgae, Palmaria palmate. Aquat. Toxicol., 50: 1-12
- Atienzar, F.A., Venier, P., Jha, A.N. and Depledge, M.H. (2002) Evaluation of the random amplified polymorphic DNA (RAPD) assay for the detection of DNA damage and mutations. Mutat. Res., 521: 151-163
- Atienzar, F.A. and Jha, A.N. (2006) The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat. Res., 613: 76-102
- Bing, H., Wu, Y., Liu, E. and Yang, X. (2013) Assessment of heavy metal enrichment and its human impact in lacustrine sediments from four lakes in the mid-low reaches of the Yangtze River, China. J. Environ. Sci., 25: 1300-1309
- Cansaran-Duman, D., Atakol, O. and Aras, S. (2011) Assessment of air pollution genotoxicity by RAPD in Evernia prunastri L. Ach. from around an iron-steel factory in Karabük, Turkey. J. Environ. Sci., 23: 1171-1178
- Cenkci, S., Yildiz. M., Ciğerci, I.H., Konuk, M. and Bozdağ, A. (2009) Toxic chemicals-induced genotoxicity detected by random amplified polymorphic DNA (RAPD) in bean (Phaseolus vulgaris L.) seedlings, Chemosphere., 76: 900-906
- De-Wolf, H., Blust, R. and Backeljiau, T. (2004) The use of RAPD in ecotoxicology: A review. Mutat. Res., 566: 249-262
- Doyle, J.J. and Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15
- Enan, M.R. (2006) Application of random amplified polymorphic DNA (RAPD) to detect the genotoxic effect of heavy metals. Biotechnol. Appl. Biochem., 43: 147-154
- Gupta, M. and Sarin, N.B. (2009) Heavy metal induced DNA changes in aquatic macrophytes: Random amplified polymorphic DNA analysis and identification of sequence characterized amplified region marker. J. Environ. Sci., 21: 686-690
- Jaccard, P. (1908) Nouvelles recherches sur la distribution flora. Bull. Sac. Nat., 44: 223-270
- Khan, M.U., Malik, R.N. and Muhammad, S. (2013) Human health risk from heavy metal via food crops consumption with wastewater irrigation practices in Pakistan. Chemosphere., 93: 2230-2238
- Liu, W., Sun, L., Zhong, M., Zhou, Q., Gong, Z., Li, P., Tai, P. and Li, X. 2012. Cadmium-induced DNA damage andmutations in Arabidopsis plantlet shoots identified by DNA fingerprinting. Chemosphere., 89: 1048-1055
- Rainbow, P.S. (1995) Biomonitoring of heavy metal availability in the marine environment. Marine. Poll. Bull., 31: 183-192
- Rainbow, P.S. (2002) Trace metal concentrations in aquatic invertebrates: why and so what? Environ. Poll., 120: 497-507
- Raj, A., Kumar, S., Haq, I. and Kumar, M. (2014) Detection of tannery effluents induced DNA damage in mung bean by use of Random Amplified Polymorphic DNA markers. ISRN Biotechnol., 2014: 1-8
- Saleh, B. (2011) R-ISSR marker as a useful tool for detection of new genomic loci in Arthrocnemum macrostachyum. Biol. Plantarum., 55: 327-330
- Statsoft. Statistica (Data analysis software system), version 6. Statsoft Inc. 2003. www.statsoft.com
- Swaileh, K.M., Hussein, R. and Ezzughayyar, A. (2008) Evaluating wastewater-induced plant genotoxicity using randomly amplified polymorphic DNA. Environ. Toxicol., 23: 117-122
- Unal, D., Isik, N.O. and Sukatar, A. (2010) Effects of chromium VI stress on green alga Ulva lactuca (L.). Turk. J Biol., 34: 119-124


