Effect of Stress on Structural Behavior of Periplasmic Membrane In Pathogenic Organism
Автор: Mrunali Patel, Priti Patel
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 3 т.17, 2021 года.
Бесплатный доступ
Microorganisms have an assortment of evolutionary adaptations and physiological advancement mechanism which permit them to survive and stay dynamic in face of environmental stress. The examination propose that all the more proficiently coordinating microbial ecology into biological system nature will require more complete integration of microbial physiological ecology, population biology and process ecology. Microorganisms also have genomic and metabolic plasticity to adapt the numerous stressful conditions they come across during their life. They give a remarkable illustration of adaptation to the most diverse environments. There is still plenty to learn on how pathogens react to host imposed stresses, how environmental microorganisms become acclimated to so viably to the continually changing environments, how metabolic changes at last shape their genome and how all the above can be exploited to our advantage, for example preventing food to spoil, improving food safety, performing industrial synthesis with nominal or no contamination. The revealing insight into fundamental parts of microbial reactions to stress can have commonsense consequences in irrelevant fields, for instance in the fix of human sicknesses. The aim is additionally to provide a strong interdisciplinary climate which will give a gathering to the flow of new various thoughts for better understanding microbial physiology under stress. There are a few stress to organisms include osmotic stress, oxidative stress, pH stress, thermal stress, periplasmic stress, and nutrient and starvation stresses. Environmental stresses are commonly active during the cycle of microbial fermentation and have critical impact on microbial physiology. Microorganisms have built up a progression of systems to oppose ecological anxieties. They keep up the honesty and smoothness of cell films by tweaking their configuration and composition, the penetrability and activities of carriers are changed in accordance with control nutrient transport and ion exchange.
Pathogenesis, Resistance mechanism, Stress, Structural behaviour
Короткий адрес: https://sciup.org/143173902
IDR: 143173902
Текст научной статьи Effect of Stress on Structural Behavior of Periplasmic Membrane In Pathogenic Organism
Life on earth is diverse and interactive: not one biological system harboring a single species has yet been described. Interactions between organisms can be very diverse in nature, ranging from significant through neutral to detrimental. ''stress'' to be something that make physiological difficulties that compromise microbial capacity or survival. Organisms must acclimate to immediate stress by modifying their allocation of resources from growth to survive pathways; a stress too extreme will compel the mint dormancy or kill them.[1] Only a few studies have attempted to build up the complex flow of influence from environmental conditions, through microbial physiological reactions, community composition changes, and on to a definitive biological system scale dynamics.[2],[3] Pathogenic microorganisms must withstand diverse host conditions during infection. Environmental signals, for example pH, temperature, nutrient scarcity and so forth, not just activate adaptive responses within organism to these particular stress conditions yet in addition direct the expression of virulence genes at once and reasonable place. A valuation for stress reactions and their guideline is fundamental for a comprehension of microbial pathogenesis.
A new feature of study of microbial osmoadaptation, including an analysis of the potential functions of a portion of the before referenced osmo stress responsive components in adding to the harmfulness capability of various pathogenic organism. A few parts of clinical host-pathogen interactions involve stress reactions. Adaptive reactions permit pathogenic organism to resist host defences, for example reactive oxygen and nitrogen species, antimicrobial peptides and nutritional limitation during infection.[4]
STRESSES IN THE HOST ENVIRONMENT
Bacteria experience stress from their initial moment of contact with the host. For most pathogen, this involves a variety in temperature. Respiratory pathogen should adapt to a variety of host-determined antimicrobial mediater including bactericidal peptides produced by epithelial cells[5] and may also be essential to adapt to nitrosative stress[6], hyperosmolarity and oxygen limitation.[7] In contrast, enteric microorganisms are ingested and should survive the hostile environment of the stomach, that is notable for an strongly acidic pH and the presence of reactive nitrogen species produced from dietary nitrate. Host inflammatory responses recruit phagocytic cells, exposing microorganisms to oxidative and nitrosative stress.[8] Host sequestration of required metals and different nutrients creates beneficial challenges as intracellular pathogens respond to particular cytoplasmic or phagosomal environment, respectively.[9,10]
To survive these changing environments, microorganism have developed exquisite systems which not only sense these stresses but also trigger reasonable responses that allow survival and propagation under respected conditions.
Acid pH
Pathogenic organism being experience acid pH in the gastrointestinal and genital parcels, skin and endocytic vesicles of the intracellular degradative path. To familarize these acid pH, numerous pathogenic organism express two-component system (TCS), that sense environmental acidic pH and then initiate a signaling cascade permitting adaptation to these conditions. Expressionof a few decarboxylases and genes involved in central metabolism, transport and membrane composition allow the cell to adjust to acidic conditions.
Oxidative and Nitrosative Stress
Flavoproteins and quinones in the electron transport chain generate endogenous oxidative stress by means of the extrinsic univalent decrease of molecular oxygen (O2) to superoxide (O2·-). The low levels of oxidative stress produced in these metabolic processes are predominated by the micromolar amounts of oxyradicals synthesized by the NADPH oxidase during the respiratory explosion of professional phagocytes that helps to damage biomolecules of attacking microbes and it is important to differentiate between microbial defenses designed to respond to low or elevated levels of oxidative stress.[11] Microorganism might be exposed to nitric oxide (NO·) and reactive nitrogen species (RNS, for example, S-nitrosothiols and peroxynitrite (ONOO-)
produced in reaction of NO· with O2, O2-, organic and inorganic radicals, iron or low-molecular weight thiols. NO- is produced enzymatically by microbial nitrate and nitrite reductases in addition to by microbial and host cell nitric oxide synthases. Expression of antioxidant enzymes (catalase, superoxide dismutase) or nitric oxide-detoxifying proteins (NO- reductase, flavohemoglobin) as well as stress-resistant enzymes and cluster repair machinery enhance resistance to oxidative or nitrosative stress.
Envelope Stress
Communal sources of cell envelope stress include antimicrobial cationic peptides, bile, acid pH, misfolded secretions and outer membrane proteins, modification in phospholipids and lipopolysaccharide, and variation in proton motive force (PMF). The conservation of PMF is expected to influence different processes essential for pathogenesis including microbial motility, protection from host-derived antimicrobial motivators, secretion and nutrient acquisition.
Temperature
A shift from ambient environmental temperature to host body temperature can be a key signal for pathogens. The expression of a few virulence genes is thermoregulated. In ram-negative bacteria, the histone-like protein H-NS and nucleoid like protein Hha are included in temperature-dependent regulation. At higher temperatures, the N-terminus of H-NS suffers a conformational change, commanding to the development of dimers as opposed to oligomers and a reduction in cooperative DNA-binding.[12]
MICROBIAL RESPONSES TO ACIDSTRESS
Microorganisms experience acid stress throughout various bioprocesses. Microbial species have built up a variety of resistance mechanisms. The damage caused by acidic environments is diminished through the support of pH homeostasis, cell membrane integrity and fluidity and metabolic regulation. The acid tolerance mechanism can be utilized to shield probiotics against gastric acids during the process of food intake and can improve the biosynthesis of organic acids. The combination of systems and synthetic biology technologies propose new and wide projections for the modern utilizations of microbial acid tolerance mechanism.
Microorganisms have enhanced growth conditions for their cellular functions. Metabolic disorders and even cell death may be caused by changes external environment, for example, pH [13]. Most microorganisms can endure and become acclimated to minor changes in ecological pH, while induced acid tolerance may happen as the environmental pH decline gradually.
Complex mechanism at the physiological and molecular levels have been produced by microorganisms to survive and become accustomed to acid stress[13]-[18] and an assortment of approaches has been deployed to expose acid tolerance mechanisms in different organisms at various levels. [19]-[23]
RESISTANCE MECHANISMS
pH homeostasis pH homeostasis is the regulation of the pH inside and outside the cell and is a significant indicator of the physiological condition of cells in an acidic environment. [24]
A continuous pH gradient is more favorable to most acid-tolerant microbes. This is for the explanation that a huge of energy should be consumed-through to maintain neutral pHi, which severely restrict the growth and metabolism of microbes.[25] The pHi of these acid-tolerant microbes reduction with acidification of the environment, however is maintain at a more elevated level than pHex. When the acid reaches at a definite concentration, the pHi declines sharply and the pH homeostasis is destroyed. This results in protein and DNA damage, with the cells eventually withering.[26] Therefore, sustaining pH homeostasis is necessary for organisms to survive in acidic conditions.
Alteration of cell membranes
The initial target of environmental stress is cell membranes, which help with sustaining cellular activities under acidic conditions in many ways. Low pH ordinarily prompts morphological changes in cells, which is a consequence of the damaged lipoidal cell membrane and decreased fluidity.[27] The viability of cells under stress conditions is constrained by membrane status;
cell membrane confer acid tolerance to cells through protection of their integrity and fluidity because of acid adaptation.[28]
Metabolic regulations
Microorganisms have established complex metabolic regulatory mechanisms to improve their acid tolerance during adaptation to acid environments. They redesign their precursors, cofactors and redox factors for survival, progress and metabolism under acidic conditions by strengthening the glycolytic pathway.[29]
PRODUCTION OF SUPEROXIDE IN BACTERIA IS STRESS AND CELL STATEDEPENDENT
The part of reactive oxygen species (ROS) in microbial metabolism and stress response has arisen as a major theme in microbiology and infectious disease. Receptive fluorescent dyes have the potential to advance the study of ROS in the complex intracellular environment, especially for high-content and high-throughput analyses. Current dye based approaches to measuring intracellular ROS have the potential for significant artifacts.
The recent emergence of fluorescent probes to detects ROS builds on the long term utilization of fluorescent dyes to consider microbial components. In Microorganism, fluorescent probes have been most generally utilized in attempts to define the part of ROS production in response to antibiotic exposures.[30]-[34] An assortment of fluorescent ROS probes are presently accessible, with each probe professing to have diverse reactivity, stability, fluorescence and membrane transport properties.[35],[36] The efficiency and particularity of various dyes for identifying various ROS in vivo have rarely been established with any confidence,[35] substantially less over a range of various organism.
The investigations show that appropriately controlled flow cytometry combined with fluorescent probes delivers precise and accurate quantitative analysis of ROS generation and metabolic changes in stressed organism. The accessibility of fluorescent dyes for the detection and quantification of reactive chemical species in cells has cultivated the development of single-cell, multi-parameter, high throughput analyses in microbiology and antibiotic pharmacology, including both multi-well plate based population assays and flow cytometry assays.
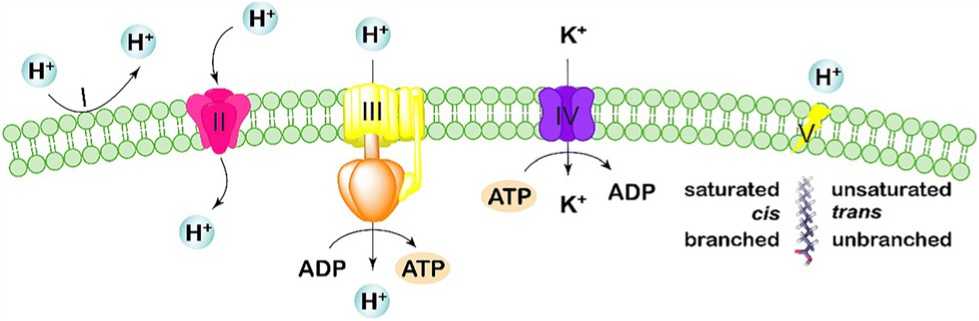
Figure 1: Acid tolerance mechanism related with cell membranes and ion transport systems. Microbial cells maintain pH homeostasis by restricting the inward flow of portions through highly impermeable cell membranes (I) and directing the size of membrane channels (II), deflecting the influx of protons through producing chemiosmotic gradients via potassium ATPases (III), pumping excess protons out from the cytoplasm through proton pump (IV), and maintaining the integrity and fluidity of cell membranes by modulating fatty acid composition (V).
Table 1. Acid-tolerant mechanisms utilized by various microorganisms.
|
Mechanisms |
Microorganisms |
Yeasts |
|
F0F1-ATPase proton pumps |
Escherichia coli, Lactococcus, Streptococcus, Corynebacterium glutamicum, P. acidipropionici, Bacillus |
S. cerevisiae, C. glabrata, Zygosaccharomyces bailii |
|
Decarboxylation and deamination |
Escherichia coli, Lactococcus, Lactobacillus, P. acidipropionici |
|
|
Cell membrane modification |
Escherichia coli, Lactococcus, Lactobacillus |
S. cerevisiae Z. bailii |
|
Metabolic regulation |
P. acidipropionici |
S. cerevisiae, Z. bailii |
|
Macromolecule protection and repair |
Escherichia coli, Lactococcus, Lactobacillus, Streptococcus, C. glutamicum, A. pasteurianus |
S. cerevisiae, Z. bailii |
|
Protection from organelle |
S. cerevisiae |
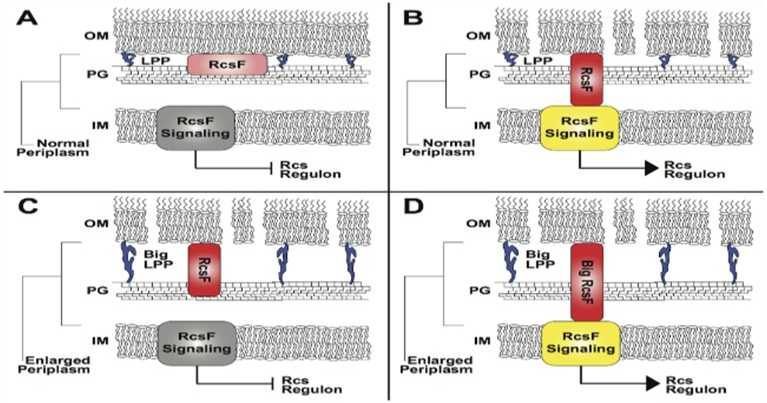
Figure 2: RcsF signalling is altered by a modification in size of the periplasmic space
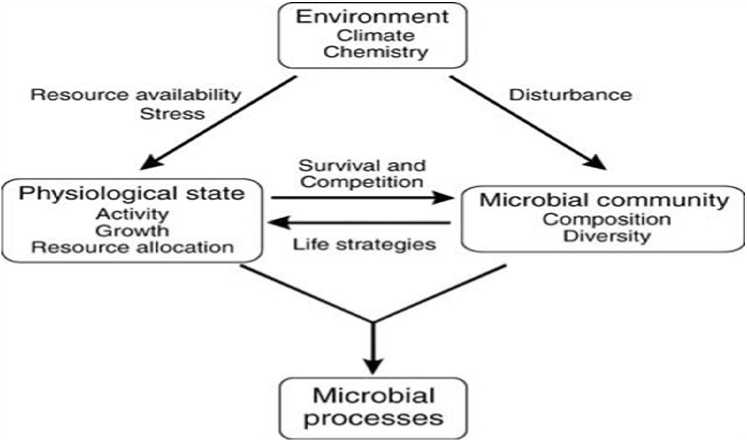
Figure 3: Links among environmental drivers, microbial physiology, community composition, and ecosystem processes.
VIRULENCE AND STRESS-RELATED PERIPLASMIC STRUCTURE PROTEIN (VISP) IN ORGANISM / HOST ASSOCIATIONS ram-negative bacteria have LPS in their external layer. Although LPS is a pathogen-associated molecular pattern known by toll-like receptor 4 (TLR4), many pathogens modify its lipid-A domain to either avoid recognition by the innate immune system or gain resistance to host factors that compromise the integrity of the bacterial cell envelope.[37]
A quality in S. Typhimurium encodes a periplasmic protein, renamed virulence and stress-related periplasmic protein (VisP), that binds to peptidoglycan and interacts with the LpxO-lipid-A-modifying enzyme inhibiting its function. This inhibition of LpxO-mediated lipid-A modifications is fundamental for resistance to intravacuolar stressors to promote intramacrophage replication and systemic disease.
THE GRAM-NEGATIVE BACTERIAL PERIPLASM: SIZE MATTERS ram-negative bacteria have a variety of significant functions that sense membrane damage and poisonous compounds, such as antimicrobial peptides, that damage the outer membrane.[38],[39] These detecting systems include those that allow remodelling of the microbial surface to be more resistant to toxic compounds.[40] Some of these sensing systems are receptors that function as sensor kinases with domains in periplasm to detect specific molecules or damage. One of the exceptional sensor kinase systems, termed the Rcs system which on membrane damage activates synthesis of extracellular polysaccharide to deliver cellular protection and biofilm formation has an outer membrane lipoprotein RcsF, that interacts with signaling proteins with specific periplasmic domains on envelope damage and peptidoglycan stress to stimulate the synthesis of extracellular polysaccharide creation and further pressure related adapting pathways.[41] Thus, envelope damage in some way brings the RcsF lipoprotein in greater proximity to the inner membranesensing system and thus it evolved to detect disorder in the outer membrane or peptidoglycan (Fig 2).
The RcsF outer layer lipoprotein sensor should contact its inner membrane signaling partners to activate sensing. This sensing needs a particular periplasmic distance because lengthening of the Lpp linkages to peptidoglycan increase the distance of the periplasm, and except if RcsF is lengthened, signaling can no longer occur. In pannel A: the state in which RcsF is not activating signaling because no envelope disorder is progressing. In pannel B: envelope diorder leads to RcsF actual interaction with the inner membranesensing system, and the Rcs regulon is activated. In pannel C, in which Lpp has been lengthened and the periplasmic intermembrane distance lengthened, the Rcs regulon can not be activated despite envelope disorder. In pannel D: the defect of the long Lpp is corrected by lengthening RcsF. IM, internal film; Lpp, Braun's lipoprotein; OM, outermembrane; P , peptidoglycan; RcsF, Regulator of capsule synthesis F.
MICROBIAL STRESS RESPONSE PHYSIOLOGY AND ITS IMPLICATIONS FOR ECOSYSTEM FUNCTION
A fluctuating climate makes conditions that can be unpleasant for microorganisms and they are neither unfading, nor impenetrable to stretch. Organisms ought to have physiological acclimation systems to endure and stay dynamic even with pressure or they will bite the dust. Notwithstanding, those transformation and acclimation systems make physiological expenses at the life form level and can change the organization of the dynamic microbial network, making shifts in environment level C, energy and supplement flows.
Moisture stress
Draught
Draught is perhaps the most well-known environmental stress that soil microorganisms experience. As soils dry, substrate diffusion becomes limited and organisms may encounter resource limitation that can slow biogeochemical process rates.
Despite the fact that decreasing water potentials impose direct physiological stress that forces microbes to move resource allocation, potentially altering the nature of C and N flows, as opposed to simply backing them down.
Microbes are small and minor in intimate contact with soil water and have semipermeable membranes. Thus, cellular water potential expediently equilibrates with that of the surrounding water. As soils dry and water possibilities decline, cells should accumulate solutes to reduce their inward water potential to avoid dehydrating and drying. As their underlying osmolytes, microorganisms use simple organics with a decent equilibrium of high solubility and limited direct physiological impacts.
Rewetting
Having accumulated osmolytes, microbes are faced with the challenges of disposing them when the soil rewets. Soil rewetting is fast and if a microorganism does not dispose of its osmolytes, water will flow into the cell, possibly making it split or rupture unless it has strong cell dividers. To prevent this, microbes should dispose of osmolytes quickly on rewetting, either by breathing, polymerizing or transporting them across the cell membrane.[42]
Freezing and freeze-thaw
Freezing
Cold, especially freezing, temperatures, are another basic stress which requires microbial adaptations and acclimations.[43] At low temperature, lipid membrane can harden[44] and ice crystals can rupture cell membranes[45] both potentially lethal. Besides, as bulk soil water freezes and only thin films of water remain on particle surfaces substrate and O 2 diffusion decrease, including substrate limitation and anaerobiosis.
For microbes to survive freezing temperatures and stay active, there are various basic physiological acclimations. They should shift biochemical pathways,[44] change membrane lipids to maintain membrane fluidity, synthesize protective molecules that incorporate proteins and sugars, [46],[47] synthesize antifreeze proteins[48] and perhaps produce compatible solutes to control water potential.[49]
CONCLUSION
There is interaction among stressors; actively growing organisms are more vulnerable against stress, so one stress which reduces growth rates can able to induce tolerance to different stresses. The influences of a wide range of stress operate at both physiological and community composition levels and these interact to generate the general linkage between environmental conditions and bio-geo chemical process. The significance of stress in structuring the composition and function of microbial communities suggest that our current conception of microbial ''functional groups'' based largely on process-based groups to completely represent microbial process response to environmental change. Pathogenic bacteria are able to sense and respond to diverse microenvironmental stresses experienced during contamination. These responses permit pathogens not only to withstand specific stressful conditions but additionally to express virulence- related genes in a spatio-temporally appropriate manner. A total comprehension of bacterial stress responses provides novel insights into the nature of host microenvironments, mechanisms of virulence and stress resistance, and potential focuses for intervention for the counteraction or treatment of infectious diseases.
CONFLICTS OF INTEREST
All authors have declared that they do not have any conflict of interest for publishing this research.
Список литературы Effect of Stress on Structural Behavior of Periplasmic Membrane In Pathogenic Organism
- 1. Suzina, N. E., Mulyukin, A. L., Kozlova, A. N.,Shorokhova, A. P., Dmitriev, V. V., Barinova, E.S., ... & Duda, V. I. (2004). Ultrastructure of restingcells of some non-spore-formingbacteria. Microbiology, 73(4), 435-447.
- Zak, D. R., Holmes, W. E., White, D. C., Peacock,A. D., & Tilman, D. (2003). Plant diversity, soilmicrobial communities, and ecosystemfunction. Ecology, 84(8), 2042-2050.
- Balser, T. C., & Firestone, M. K. (2005). Linkingmicrobial community composition and soilprocesses in a California annual grassland andmixed-conifer forest. Biogeochemistry, 73(2), 395-415.
- Lee, S., Hinz, A., Bauerle, E., Angermeyer, A.,Juhaszova, K., Kaneko, Y., ... & Manoil, C. (2009).Targeting a bacterial stress response to enhanceantibiotic action. Proceedings of the National Academy of Sciences, 106(34), 14570-14575.
- Grubor, B., Meyerholz, D. K., & Ackermann, M. R.(2006). Collectins and cationic antimicrobialpeptides of the respiratory epithelia. Veterinarypathology, 43(5), 595-612.
- Martel, J., Ko, Y. F., Young, J. D., & Ojcius, D. M.(2020). Could nasal nitric oxide help to mitigate theseverity.
- Worlitzsch, D., Tarran, R., Ulrich, M., Schwab, U.,Cekici, A., Meyer, K. C., ... & Döring, G. (2002).Effects of reduced mucus oxygen concentration inairway Pseudomonas infections of cystic fibrosispatients. The Journal of clinicalinvestigation, 109(3), 317-325.
- Fang, F. C., Frawley, E. R., Tapscott, T., &Vázquez-Torres, A. (2016). Discrimination andintegration of stress signals by pathogenicbacteria. Cell host & microbe, 20(2), 144-153.
- Steele, S., Brunton, J., Ziehr, B., Taft-Benz, S.,Moorman, N., & Kawula, T. (2013). Francisellatularensis harvests nutrients derived via ATG5-independent autophagy to support intracellulargrowth. PLoS Pathog, 9(8), e1003562.
- Nairz, M., Ferring-Appel, D., Casarrubea, D.,Sonnweber, T., Viatte, L., Schroll, A., ... & Galy, B.(2015). Iron regulatory proteins mediate hostresistance to Salmonella infection. Cell host µbe, 18(2), 254-261.
- Fang, F. C. (2011). Antimicrobial actions of reactiveoxygen species. MBio, 2(5).
- Ono, S., Goldberg, M. D., Olsson, T., Esposito, D.,Hinton, J. C., & Ladbury, J. E. (2005). H-NS is a partof a thermally controlled mechanism for bacterialgene regulation. Biochemical Journal, 391(2), 203-213.
- Beales, N. (2004). Adaptation of microorganisms tocold temperatures, weak acid preservatives, low pH,and osmotic stress: a review. Comprehensivereviews in food science and food safety, 3(1), 1-20.
- Fernández-Niño, M., Marquina, M., Swinnen, S.,Rodríguez-Porrata, B., Nevoigt, E., & Ariño, J.(2015). The cytosolic pH of individualSaccharomyces cerevisiae cells is a key factor inacetic acid tolerance. Applied and environmentalmicrobiology, 81(22), 7813-7821.
- Hosseini Nezhad, M., Hussain, M. A., & Britz, M. L.(2015). Stress responses in probiotic Lactobacilluscasei. Critical reviews in food science andnutrition, 55(6), 740-749.
- Ju, S. Y., Kim, J. H., & Lee, P. C. (2016). Long-termadaptive evolution of Leuconostoc mesenteroidesfor enhancement of lactic acid tolerance andproduction. Biotechnology for biofuels, 9(1), 1-12.
- Liu, Y., Tang, H., Lin, Z., & Xu, P. (2015).Mechanisms of acid tolerance in bacteria andprospects in biotechnology andbioremediation. Biotechnology advances, 33(7),1484-1492.
- Matsui, R., & Cvitkovitch, D. (2010). Acid tolerancemechanisms utilized by Streptococcusmutans. Future microbiology, 5(3), 403-417.
- He, G., Wu, C., Huang, J., & Zhou, R. (2016). Acidtolerance response of Tetragenococcus halophilus:A combined physiological and proteomicanalysis. Process Biochemistry, 51(2), 213-219.
- Hu, S., Xiao, X., Wu, X., Xia, X., Yu, Y., & Wu, H.(2017). Comparative transcriptomic analysis byRNA-seq of acid tolerance response (ATR) in EHECO157: H7. LWT-Food Science and Technology, 79,300-308.
- Lee, Y., Nasution, O., Choi, E., Choi, I. G., Kim, W.,& Choi, W. (2015). Transcriptome analysis of aceticacid-treated yeast cells identifies a large set ofgenes whose overexpression or deletion enhancesacetic acid tolerance. Applied microbiology andbiotechnology, 99(15), 6391-6403.
- Sandoval, N. R., Mills, T. Y., Zhang, M., & Gill, R. T.(2011). Elucidating acetate tolerance in E. coli usinga genome-wide approach. Metabolicengineering, 13(2), 214-224.
- Zhai Z, Douillard FP, An H, Wang G, Guo X, Luo Y,Hao Y. Proteomic characterization of the acidtolerance response in Lactobacillusdelbrueckii subsp. bulgaricus CAUH1 and functionalidentification of a novel acid stress-related transcriptional regulator Ldb0677. Environ Microbiol.2014;16:1524-1537.
- 24.25. Baker-Austin, C., & Dopson, M. (2007). Life in acid:pH homeostasis in acidophiles. Trends inmicrobiology, 15(4), 165-171.
- Sun, Y. (2016). F 1 F 0-ATPase Functions UnderMarkedly Acidic Conditions in Bacteria.In Regulation of Ca2+-ATPases, V-ATPases and FATPases(pp. 459-468). Springer, Cham.
- Wu, C., Zhang, J., Chen, W., Wang, M., Du, G., &Chen, J. (2012). A combined physiological andproteomic approach to reveal lactic-acid-inducedalterations in Lactobacillus casei Zhang and itsmutant with enhanced lactic acid tolerance. Appliedmicrobiology and biotechnology, 93(2), 707-722.
- Streit, F., Delettre, J., Corrieu, G., & Béal, C. (2008).Acid adaptation of Lactobacillus delbrueckii subsp.bulgaricus induces physiological responses atmembrane and cytosolic levels that improvescryotolerance. Journal of appliedmicrobiology, 105(4), 1071-1080.
- Sohlenkamp, C. (2017). Membrane homeostasis inbacteria upon pH challenge. Biogenesis of fattyacids, lipids and membranes, 1-13.
- Guan, N., Shin, H. D., Chen, R. R., Li, J., Liu, L.,Du, G., & Chen, J. (2014). Understanding of howPropionibacterium acidipropionici respond topropionic acid stress at the level ofproteomics. Scientific reports, 4(1), 1-8.
- Albesa, I., Becerra, M. C., Battán, P. C., & Páez, P.L. (2004). Oxidative stress involved in theantibacterial action of differentantibiotics. Biochemical and biophysical researchcommunications, 317(2), 605-609.
- Goswami, M., Mangoli, S. H., & Jawali, N. (2006).Involvement of reactive oxygen species in the actionof ciprofloxacin against Escherichiacoli. Antimicrobial agents and chemotherapy, 50(3),949-954.
- Kohanski, M. A., Dwyer, D. J., Hayete, B.,Lawrence, C. A., & Collins, J. J. (2007). A commonmechanism of cellular death induced by bactericidalantibiotics. Cell, 130(5), 797-810.
- Wang, X., Zhao, X., Malik, M., & Drlica, K. (2010).Contribution of reactive oxygen species to pathwaysof quinolone-mediated bacterial cell death. Journalof Antimicrobial Chemotherapy, 65(3), 520-524.
- Dwyer, D. J., Camacho, D. M., Kohanski, M. A.,Callura, J. M., & Collins, J. J. (2012). Antibioticinducedbacterial cell death exhibits physiologicaland biochemical hallmarks of apoptosis. Molecularcell, 46(5), 561-572.
- Kalyanaraman, B., Darley-Usmar, V., Davies, K. J.,Dennery, P. A., Forman, H. J., Grisham, M. B., ... &Ischiropoulos, H. (2012). Measuring reactive oxygenand nitrogen species with fluorescent probes:challenges and limitations. Free radical biology andmedicine, 52(1), 1-6.
- Imlay, J. A. (2015). Diagnosing oxidative stress inbacteria: not as easy as you might think. Currentopinion in microbiology, 24, 124-131.
- Raetz, C. R., Reynolds, C. M., Trent, M. S., &Bishop, R. E. (2007). Lipid A modification systemsin gram-negative bacteria. Annu. Rev. Biochem., 76,295-329.
- Grabowicz, M., & Silhavy, T. J. (2017). Envelopestress responses: an interconnected safetynet. Trends in biochemical sciences, 42(3), 232-242.
- Hancock, R. E., & Diamond, G. (2000). The role ofcationic antimicrobial peptides in innate hostdefences. Trends in microbiology, 8(9), 402-410.
- Dalebroux, Z. D., & Miller, S. I. (2014). SalmonellaePhoPQ regulation of the outer membrane to resistinnate immunity. Current opinion inmicrobiology, 17, 106-113.
- Asmar, A. T., Ferreira, J. L., Cohen, E. J., Cho, S.H., Beeby, M., Hughes, K. T., & Collet, J. F. (2017).Communication across the bacterial cell envelopedepends on the size of the periplasm. PLoSBiology, 15(12), e2004303.
- Wood, J. M., Bremer, E., Csonka, L. N., Kraemer,R., Poolman, B., van der Heide, T., & Smith, L. T.(2001). Osmosensing and osmoregulatorycompatible solute accumulation by bacteria. Comparative Biochemistry and PhysiologyPart A: Molecular & Integrative Physiology, 130(3),437-460.
- Walker, V. K., Palmer, G. R., & Voordouw, G.(2006). Freeze-thaw tolerance and clues to thewinter survival of a soil community. Applied andEnvironmental Microbiology, 72(3), 1784-1792.
- Methe, B. A., Nelson, K. E., Deming, J. W., Momen,B., Melamud, E., Zhang, X., ... & Fraser, C. M.(2005). The psychrophilic lifestyle as revealed bythe genome sequence of Colwellia psychrerythraea34H through genomic and proteomicanalyses. Proceedings of the National Academy ofSciences, 102(31), 10913-10918.
- Rivkina, E. M., Friedmann, E. I., McKay, C. P., &Gilichinsky, D. A. (2000). Metabolic activity ofpermafrost bacteria below the freezingpoint. Applied and EnvironmentalMicrobiology, 66(8), 3230-3233.
- Mihoub, F., Mistou, M. Y., Guillot, A., Leveau, J. Y.,Boubetra, A., & Billaux, F. (2003). Cold adaptationof Escherichia coli: microbiological and proteomicapproaches. International journal of foodmicrobiology, 89(2-3), 171-184.
- Kandror, O., Bretschneider, N., Kreydin, E.,Cavalieri, D., & Goldberg, A. L. (2004). Yeast adaptto near-freezing temperatures by STRE/Msn2, 4-dependent induction of trehalose synthesis andcertain molecular chaperones. Molecular cell, 13(6),771-781.
- Muhr, J. (2009). Carbon dynamics under naturaland manipulated meteorological boundaryconditions in a forest and a fen ecosystem (Doctoraldissertation).
- Mindock, C. A., Petrova, M. A., & Hollingsworth, R.I. (2001). Re-evaluation of osmotic effects as ageneral adaptative strategy for bacteria in subfreezingconditions. Biophysical Chemistry, 89(1),13-24.


