Epigenetic effects of enzastaurin - a new aspect in the mechanism of action of an anticancer drug from protein kinase inhibitors
Автор: Maksimova Varvara P., Makus Julia V., Usalka Olga G., Lylova Eugenia S., Bugaeva Polina E., Zhidkova Ekaterina M., Fedorov Dmitry A., Lizogub Olga P., Lesovaya Ekaterina A., Belitsky Gennady A., Yakubovskaya Marianna G., Kirsanov Kirill I.
Журнал: Сибирский онкологический журнал @siboncoj
Рубрика: Лабораторные и экспериментальные исследования
Статья в выпуске: 4 т.19, 2020 года.
Бесплатный доступ
The purpose of the study was to analyze the ability of five antitumor drugs from the pharmaceutical group of protein kinase inhibitors (gefitinib, imatinib, pazopanib, ponatinib and enzastaurin) to reactivate the expression of the epigenetically silenced GFP in HeLa TI cells, and to estimate the effect of epigenetically active drugs on: 1) acetylation and methylation of histones H3 and H4; 2) integral DNA methylation; 3) activity of HAT and HDAC1 enzymes; 4) expression levels of the genes encoding epigenetic regulation enzymes (DNMT1, DNMT3A, DNMT3B; SIRT1, HDAC1; SETD1A, SETD1B, SUV420H1, SUV420H2, SUV39H1, SUV39H2). Material and Methods. The epigenetic activity of antitumor drugs was determined using the HeLa TI test system, a population of HeLa cells with the retroviral vector containing the epigenetically silenced GFP. The level of integral DNA methylation was analyzed using MspI/HpaII methyl-sensitive restriction analysis. Histone modifications were analyzed by Western blotting with antibodies to acetylated and methylated histones H3 and H4. The total activity of HAT enzymes was analyzed using Histone Acetyltransferase Activity Assay Kit. Expression of the epigenetic enzyme genes was analyzed using real-time quantitative RT-PCR. Results. It was shown that only the enzyme inhibitor Cp protein kinase enzastaurin had the ability to reactivate the expression of epigenetically silenced GFP in the HeLa TI cells. We showed that under the action of enzastaurin, the level of integral DNA methylation and expression of DNMT3A and DNMT3B DNA methyltransferase genes decreased. It was also found that enzastaurin reduced the expression levels of histone deacetylases HDAC1 and SIRT1, but did not affect the activity and expression levels of histone acetylases, the level of histone methylation (H3K4me3, H3K9me3, H3K27me3, H4K20me3), and the level of expression of the histone methyltransferases (SUV39H1, SUV39H2, SUV420H1, SUV420H2, SETD1A и SETD1B). Conclusion. The data obtained are important for clarifying the mechanisms of action of 5 protein kinase inhibitors, in particular with respect to enzastaurin, the protein kinase Cp inhibitor, for which the ability to reactivate epigenetically silent genes due to the effect on DNA methylation and histone acetylation was demonstrated.
Protein kinase inhibitors, enzastaurin, epigenetic activity, hela ti, histone modifications, dna methylation, hat, hdac
Короткий адрес: https://sciup.org/140254368
IDR: 140254368 | УДК: 577.21:615.277.3:615.015.4 | DOI: 10.21294/1814-4861-2020-19-4-67-78
Текст научной статьи Epigenetic effects of enzastaurin - a new aspect in the mechanism of action of an anticancer drug from protein kinase inhibitors
Disruption of epigenetic regulation, along with genetic aberrations, plays a significant role in initiation, promotion, and progression of tumor growth. Activation of proliferation, inhibition of apoptosis, intercellular interactions, and cell aging can occur during carcinogenesis as a result of changes in the profile of epigenetic modifications due to impaired histone acetylation and methylation, as well as DNA methylation [1]. These changes can be due to a mutation or dysregulation of the expression of histone-and DNA-modifying enzymes or direct inhibition of the activity of these enzymes and activation of their degradation. Epigenetic regulatory enzymes include histone acetyltransferases (HATs) and deacetylases (HDACs), histone methyltransferases (HMTs) and demethylases (HDMs), as well as DNA methyltransferases (DNMTs). Epigenetic changes are usually reversible: restoration of the activity of enzymes responsible for epigenetic regulation of transcription leads to normalization of the epigenetic profile [2, 3]. Thus, enzymes responsible for epigenetic regulation of transcription represent targets for anticancer therapy. Low-molecular-weight compounds selectively inhibiting the enzymes of epigenetic regulation are already used for the treatment of certain types of cancer.
Currently, the use of azacitidine and decitabine, inhibitors of DNA methyltransferase (DNMT), is one of the most successful epigenetic anticancer strategies. However, these drugs have a number of limitations. Firstly, since these molecules are modified by cytosine analogues, they can be mistakenly incorporated into nascent DNA and RNA, which increases the probability of mutations. Secondly, azacitidine and decitabine are highly toxic and of low chemical stability [4]. Nowadays, the properties of non-nucleoside inhibitors of DNMTs selectively interacting with the catalytic sites of enzymes are actively studied; these inhibitors include RG108, Psammaplin, hydralazine, and procainamide, with non-nucleoside inhibitors being less effective than azacitidine [1]. Histone deacetylase (HDAC) inhibitors are a class of compounds that can modulate the epigenetic regulation of gene expression via histone acetylation. HDAC inhibitors are the most commonly used epigenetically active agents in the treatment of cancer [5]. Over the past few years, a series of clinical trials on the use of HDAC inhibitors in antitumor therapy have begun. By the beginning of 2020, five histone deacetylase inhibitors were approved by the US Food and Drug Administration (FDA) for the treatment of oncological diseases: vorinostat, romidepsin, belinostat, panobinostat, and chidamide [6]. Moreover, a series of perspective agents are currently under the phase II/III clinical trial: valproic acid, mocetinostat, entinostat, trichostatin A, phenylacetate, phenylbutyrate, tacedinaline, etc. [1]. Over the past 15 years, an essential role of impaired activity of histone methyltransferases (HMTs) in tumorigenesis has been demonstrated. For this reason, another class of compounds, namely HMT inhibitors, was introduced among epigenetic modulators [7]. Other important antitumor targets are methyltransferases SUV391H, EZH2, MLL, Nsd1, and RIZ. A series of selective HMT inhibitors are currently under the phase I/II clinical study: pinometostat, GSK2816126, CPI-1205, TCP, and 4SC-202. In 2020, the FDA approved a selective inhibitor of EZH2 methyltransferase, tazemetostat, for the treatment of metastatic or locally advanced epithelioid sarcoma that cannot be completely resected [8]. Thus, the search for new epigenetically active drugs is a promising trend in the field of chemotherapy of oncological diseases.
The search, development, as well as preclinical and clinical studies of a new drug require a lot of funding and last 5–15 years. For this reason, a promising trend in pharmacology in general and, in particular, and oncology is the drug repurposing. The search for agents with inhibitory activity against HDAC, DNMT, and HMT among the approved drugs that are already used in medical practice is undertaken. For this reason, protein kinase inhibitors approved for the treatment of various tumors (gefitinib, imatinib, pazopanib), as well as drugs undergoing clinical trials (ponatinib and enzastaurin), were chosen as the object of the current study. The multi-target action of these small molecules allowed suggesting their potential effect on the system of epigenetic regulation of transcription [9]. Another reason for choosing this group of drugs was the ability of natural polyphenolic compounds known as modulators of epigenetic regulatory enzymes to inhibit protein kinases [10].
Identification of epigenetically active agents among numerous drugs was complicated by the lack of available test systems that allow quick and inexpensive screening of drugs for their ability to exert an epigenetic effect [11]. For this, we decided to use HeLa TI cells, which were obtained for the study of the mechanisms of retroviral epigenetic silencing in the laboratory of prof. A.M. Skalka in 2007. HeLa TI is a population of cells carrying a retroviral vector integrated into various sites of the genome and containing epigenetically repressed GFP reporter gene. Analysis of the mechanisms of the GFP reporter gene suppression revealed the involvement of more than 15 different factors of epigenetic regulation of transcription in this process [12, 13]. HeLa TI cells are sensitive to HDAC and DNMT, as well as HMT inhibitors [14].
The aim of the current work was to study the ability of five antitumor protein kinase inhibitors to reactivate the expression of epigenetically repressed GFP , as well as to analyze the effect of epigenetically active drugs on: 1) acetylation and methylation of histones H3 and H4; 2) integrated DNA methylation; 3) HAT and HDAC1 enzyme activity; 4) expression level of the genes encoding for epigenetic regulatory enzymes.
Material and methods
Cell lines and reagents
The following cell cultures were used in the study: HeLa TI, a polyclonal population of HeLa cells containing a vector integrated into genome (derived from avian sarcoma retrovirus) carrying epigenetically repressed GFP ; CasKi, an epidermoid cervical carcinoma cell line, characterized by higher level of DNA methylation than HeLa cells [15, 16]. The cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (Biosera), L-glutamine (2 mM, PanEco, Russia), and penicillin/streptomycin mixture (50 units, PanEco, Russia) in 5 % CO2 at 37 °C. Upon reaching 80 % confluence, the cells were passaged at a rate of 1:5.
For the study, the following drugs were selected from the pharmacological group of protein kinase inhibitors: gefitinib, imatinib, pazopanib, ponatinib, and enzastaurin (all provided by Sellek). Compounds with epigenetic activity that were used as positive controls, namely trichostatin A and vorinostat (histone deacetylase inhibitors), as well as azacitidine (inhibitor of DNA methyltransferases) were obtained from Sigma Aldrich. All of the above-mentioned compounds were dissolved in DMSO to obtain stock solutions; the solvent concentration did not exceed 0.1% in the culture medium.
Evaluation of drug cytotoxicity
Cytotoxicity of protein kinase inhibitors was measured using the MTT test [17]. The cells were seeded at 5×103 cells per well in 96-well flat-bottom plates and incubated overnight. Next, serial dilutions of the preparations were added in triplicate and incubated for 72 h under standard conditions (37 °C, 5 % CO2). After this, the cells were treated with 3-[4,5-dime-thylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT, Dia-M, Russia). After four hours of exposure to MTT, the medium was removed, and 100 μl of DMSO was added. The optical density of the solution was measured at 540 nm using a Multiskan Sky microplate spectrophotometer (Thermo Scientific). The cytotoxicity index was determined using 0.1 % DMSO as a negative control.
Analysis of the drug ability to reactivate the expression of epigenetically repressed GFP gene in the HeLa TI cell test system
Flow cytometry was used to analyze the ability of the drugs to reactivate the expression of epigenetically repressed GFP. Cells were seeded in 24-well plates at 2.5×104 cells per well and treated with gefitinib, imatinib, pazopanib, ponatinib (25 μM IC20 for all drugs) and enzastaurin (IC20, 30 μM) 24 h later. After 24-hour incubation, the medium in the plates was replaced with fresh one, and the cells were incubated for another 48 h. Next, the cells were detached from the culture plates using 0.25 % trypsin-EDTA solution (PanEco, Russia). The relative number of GFP-positive cells was assessed using BD FACSCanto™ II flow cytometer. Cells treated with trichostatin A (TCA), a histone deacetylase inhibitor, were used as a positive control. An antitumor drug was considered epigeneti-cally active if GFP reactivation was observed in 12 % of the cells or more. The proportion of GFP-positive cells among 0.01 % DMSO-treated cells did not exceed 4.5–5 %.
Histone extraction
Cells were seeded in 60-mm Petri dishes (3×105 cells per dish) and incubated overnight in standard conditions. Next, enzastaurin (30 μM) was added, and the cells were incubated for 4 h, 12 h, 24 h, and 72 h. Abcam acid extraction protocol and trichloroacetic acid precipitation protocol presented by Shechter et al. [18] were used for obtaining a histone fraction.
Analysis of the effect of enzastaurin on the level of histone modifications
Western blotting was used to analyze the effect of enzastaurin on acetylation and methylation of H3 and H4 histones. Histone proteins were separated by 15 % PAGE and transferred to nitrocellulose membranes with a pore size of 0.22 μm (100 mA, 40 min). The membranes were then blocked in TBST buffer containing 5 % skim milk at room temperature for 30 min. Abcam antibodies to the following histone modifications were used in the study: H3acK9 + H3acK14 + H3acK18 + H3acK23 + H3acK27 (ab47915), H3K9me3 (ab8898), H3K4me3 (ab8580), H4K20me3 (ab9053), and Н4 (ab10158). Membranes were incubated with primary antibodies at +4 °C overnight, washed with TBST and incubated with secondary antibodies (ab6721). For protein detection, Clarity™ Western ECL Substrate visualization reagent (Bio-Rad) and ImageQuant LAS 4000 digital imaging system (GE Healthcare) were used. Densitometric analysis of the blots was performed using ImageJ software. The results were calculated as described by Li et. al. [19]. All experiments were performed in triplicate.
Preparation of nuclear extracts
HeLa TI cells were seeded in 60-mm Petri dishes (8×105 cells per dish). After the cells were attached to the substrate, they were incubated with enzastaurin (30 μM) for 24 h. The nuclear fraction of the cells was obtained according to the Abcam protocol (Nuclear extraction and fractionation protocol, https://www. . Cells were washed with PBS, passed through a 26 G needle and centrifuged at 720 g for 5 min for precipitation of a nuclear fraction. Then, the precipitate was resuspended and passed through a 23 G needle, centrifuged in the same conditions, and the resulting precipitate was resuspended in ddH2O. For DNA destruction, the suspension was sonicated for 3 seconds at an amplitude of 20 μm using a Soniprep 150 Plus ultrasonic disintegrator (MSE).
The effect of enzastaurin on HAT activity
The effect of enzastaurin on the overall activity of HAT was analyzed using Histone Acetyltransferase Activity Assay Kit (ab65352, Abcam) according to the manufacturer’s instructions. A total of 50 μg of nuclear extract was incubated with HAT I/II and NADH-generating enzyme in HAT assay buffer for 4 h at 37 °C. The optical density was measured at 450 nm in a Multiskan Sky microplate spectrophotometer (Thermo Scientific), and an active nuclear extract was used as a positive control. HAT activity was expressed in μM/μg as indicated by the manufacturer, after which the activity was calculated in % relative to the negative control. All experiments were performed in triplicate.
The effect of enzastaurinon the level of HDAC1
To analyze the effect of enzastaurin on the level of HDAC1, the nuclear fraction of the proteins was separated by 10 % PAGE, and then standard Western blotting was performed (as previously described). Abcam antibodies to HDAC1 (ab53091) and H3 histone (ab18521), which served as a load control, were used in the study. Protein detection was performed as described above. Densitometric analysis of the blots was conducted using ImageJ software. All experiments were performed in triplicate.
The effect of enzastaurinon the integrated DNA methylation
The effect of enzastaurin on integrated DNA methylation was analyzed using a commercial EpiJet kit (K1441, Thermo Scientific) based on methylationsensitive HpaII/MspI restriction assay. CasKi cells were seeded in 6-well plates (1.5×105 cells per well). After the cells were attached to the substrate, they were incubated with enzastaurin (2 μM) or the demethylating agent azacitidine (1 μM) as a positive control for 72 h. After every 24 h of incubation, half of the culture medium was replaced with fresh medium, and the drugs under study were added to the concentration indicated above. Next, genomic DNA was extracted from the cells using the GeneJET Genomic DNA Purification Kit (K0721, Thermo Scientific), and the enzymatic reaction with HpaII and MspI restriction enzymes was performed according to the manufacturer’s protocol. Restriction products were analyzed by 1 % agarose gel electrophoresis and detected on a Typhoon 9400 scanner (GE Healthcare). Densitometric analysis of the obtained images was carried out using the ImageJ software. All experiments were performed in triplicate.
The effect of enzastaurinon the genes of chromatin-modifying enzymes
Analysis of the expression level of chromatin-modifying enzyme genes was carried out using quantitative real-time RT-PCR. Cells were seeded in 60-mm Petri dishes (8x105 cells per dish) and treated with enzastau- rin at concentrations of 30, 15 and 7.5 μM after 16–20 h. After 24 h, the cells were removed from the substrate, and the total RNA was extracted using the GeneJET RNA Purification Kit (Thermo Scientific). Next, 2 μg of cDNA were obtained using the reverse transcription kit (Synthol). Sequences of the primers for PCR are presented in table 1. Real-time PCR was performed using a reagent kit for RT-PCR (Syntol). Data were analyzed by calculating the threshold cycle value (Ct) with normalization to the expression of the housekeeping gene ACTB in each sample. Next, the samples were normalized to the negative control (0.1 % DMSO).
Таблица 1/Table 1
Nucleotide sequences of the primers used in the study
Нуклеотидные последовательности используемых праймеров
|
Название/ Name |
Последовательность 5'-3'/Sequences 5'-3' |
|
|
HDAC1 |
CACCCATTCTTCCCGTTCTT GGCATTTCAGGAGTTTGTCTTAT |
Forward Reverse |
|
SIRT1 |
GCAAAGAAGAAACAGCATTGAAG ATGAATGCTGAGTTGCTGGAT |
Forward Reverse |
|
DNMT1 |
AGCACAGAAGTCAACCCAAA TGCGTCTCTTCTCCTCCTTT |
Forward Reverse |
|
DNMT3A |
AGCCCAAGGTCAAGGAGATT TACGCACACTCCAGAAAGC |
Forward Reverse |
|
DNMT3B |
CAACAGCATCGGCAGGAA GTCCTCTGTGTCGTCTGTGA |
Forward Reverse |
|
HAT1 |
GCGATAGAGGCACAACAGAA TGTATTGTTCGGCATCACTCA |
Forward Reverse |
|
CREBBP |
CTGGCAGACCTCGGAAAGAA CTGGCGCCGCAAAAACT |
Forward Reverse |
|
EP300 |
CGCTTTGTCTACACCTGCAA TGCTGGTTGTTGCTCTCATC |
Forward Reverse |
|
SETD1A |
GCGGGCTATTCTCTCACTTG CCCTTCATCCGCCTTGGT |
Forward Reverse |
|
SETD1B |
GGATGTTTGTGCGGGTAGAC AGACACACAACGGAAACACT |
Forward Reverse |
|
SUV420H1 |
CCCGTGTAGCATAAAAGCAGC CCAGTTTCACCAAGGAACCAG |
Forward Reverse |
|
SUV420H2 |
CGTGCTTGGAAGAAGAATGA GCAGTCATGGTTGATGAAGG |
Forward Reverse |
|
SUV39H1 |
GCTAGGCCCGAATGTCGTTA TAGAGATACCGAGGGCAGGG |
Forward Reverse |
|
SUV39H2 |
GCAGGACGAACTCAACAGAA CAACCAAAGGTGGCTTCATT |
Forward Reverse |
Relative expression of the genetic locus (Exp) was calculated using the 2-ΔΔCt method.
Statistical methods
Statistical analysis of the data was performed using Microsoft Excel. To assess the significance of differences between the groups, including the expression levels, a paired two-sample Student’s t-test with a level of statistical significance of p<0.05 was used.
Results and Discussion
Impaired regulation of protein kinase activity leads to various pathophysiological disorders, in particular, hyperproliferation of tumor cells, stimulation of inva-
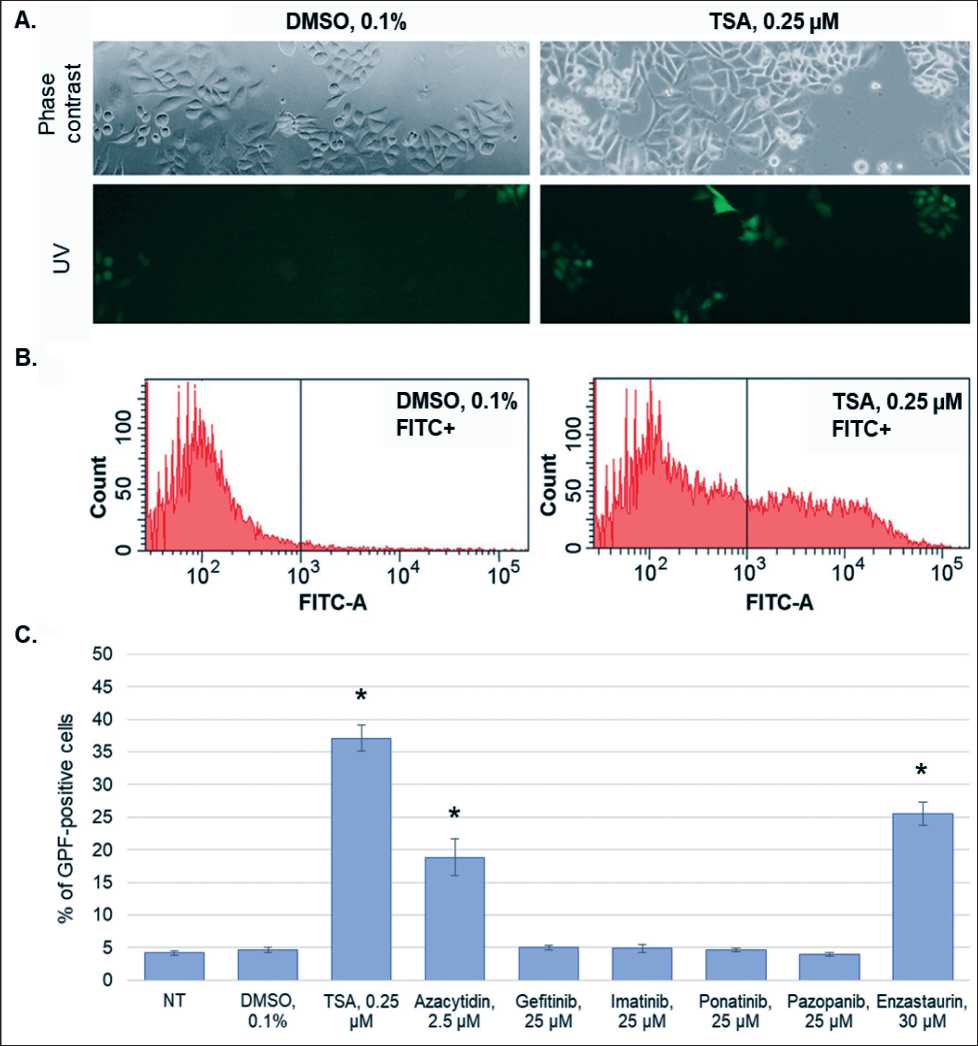
Рис. 1. Анализ эпигенетической активности противоопухолевых препаратов из группы ингибиторов протеинкиназ в тест-системе HeLa TI: А, В. Анализ доли GFP -положительных клеток в популяции HeLa TI при обработке 0,1 % ДМСО и при обработке ингибитором гистоновых деацетилаз трихостатином А с помощью флуоресцентной микроскопии (А) и проточной цитофлуориметрии (В). С. Результаты проточной цитофлуориметрии для исследуемых препаратов. Все результаты представлены в виде M ± SD. Примечание: * – различия статистически значимы по сравнению с контролем (р<0,05)
Fig. 1. Analysis of the epigenetic activity of antitumor drugs of the group of protein kinase inhibitors in the HeLa TI cell test system: A, B. Analysis of the proportion of GFP-positive cells in the HeLa TI cell population treated with 0.1 % DMSO and histone deacetylase inhibitor trichostatin A (TSA) by fluorescence microscopy (A) and flow cytometry (B). C. Flow cytometry results of the drug analysis. All data are presented as M ± SD.
Notes; * – differences are statistically significant as compared to the control (p<0.05)
sion and metastasis [20]. In this regard, inhibition of protein kinases is an effective approach to treat cancer. To date, 37 drugs of this group have been approved for the treatment of various oncological diseases, including non-small cell lung carcinoma, acute lymphoblastic leukemia, chronic myeloid leukemia, breast cancer, etc. [21]. The ability of protein kinase inhibitors to influence gene expression via epigenetic regulation has not been studied yet. The list of the drugs used in the current study includes agents that were approved for antitumor therapy: (1) gefitinib, a selective inhibitor of the epidermal growth factor receptor (EGFR) tyrosine kinase used for the treatment of non-small cell lung cancer; (2) imatinib, a Bcr-Abl tyrosine kinase inhibitor used to treat chronic myeloid leukemia; (3) pazopanib, a non-selective tyrosine kinase inhibitor, which is active against vascular endothelial growth factor receptors (VEGFRs), platelet-derived growth factor receptors (PDGFRs), fibroblast growth factor receptors (FGFRs) and used in the treatment of kidney cancer; as well as two drugs that are currently undergoing clinical trials, namely (4) ponatinib and (5) enzastaurin. Ponatinib is a multi-target inhibitor of tyrosine kinases; it was shown to exert antitumor activity against chronic myeloid leukemia and acute lymphoblastic leukemia [22, 23]. Enzastaurin is a selective inhibitor of Сβ protein kinase and currently undergoing phase III clinical trials in patients with diffuse large B-cell lymphoma [24].
At the first stage, we analyzed the ability of nontoxic doses of gefitinib, imatinib, pazopanib, ponatinib, and enzastaurin to reactivate the expression of epige-netically repressed GFP in HeLa TI test system. As shown by flow cytometry, treatment with enzastaurin results in a 6-fold increase in the proportion of GFP-positive cells. No statistically significant increase in the number of GFP-expressing cells was detected for the other drugs (Fig. 1).
The obtained data indicate that, of the tested drugs, only enzastaurin has an effect on epigenetic regulation of gene expression. For this reason, further work was aimed at analysis of the mechanisms of the epigenetic effect of enzastaurin, in particular, at the study of its effect on the profile of histone modifications and DNA methylation.
Histone acetylation/deacetylation is one of the key mechanisms of chromatin remodeling. Acetylation of histones at lysine residues neutralizes the positive charge of the amino acid and reduces the interaction between the N termini of histones and DNA phosphate groups [3]. This, in its turn, promotes the transformation of silent heterochromatin into transcriptionally active euchromatin. A decrease in the level of lysine acetylation at positions H3K9, H3K14, H3K18, and H4K16 was shown to be associated with promotion and progression of tumor growth, as well as with tumor resistance to chemotherapy [25].
Changes in the global level of H3 histone acetylation caused by enzastaurin were analyzed by Western blotting using antibodies to a number of acetylation sites (H3acK9+H3acK14+H3acK18+H3acK23+H3a cK27). The obtained results indicate that the effect of enzastaurin results in a more than 1.5-fold increase in the level of histone acetylation (Fig. 2A).
Changes in the level of histone acetylation are the result of an altered balance in the activity of histone acetyltransferases (HATs) and deacetylases (HDACs). We demonstrated that treatment of the cells with en-zastaurin did not change the activity of histone acetyltransferases and did not affect the expression level of the corresponding genes (Fig. 2B, 2C).
Among histone deacetylases, enzymes HDAC1 and HDAC2 [26], as well as SIRT1 deacetylase [27], had the greatest contribution to the regulation of histone modifications. We demonstrated that enzastaurin resulted in a decreased HDAC1 expression both at the mRNA and protein levels (1.8-fold decrease). A slight (1.5-fold) decrease in SIRT1 expression was also shown (Fig. 2D, 2E).
In contrast to acetylation, methylation of histones H3 and H4 does not change the total charge of the molecule, and the effect on transcription occurs due to interaction of the effector molecules with modified bases [1]. At the same time, methylation of the amino acid residues of histones can lead to both gene silencing (lysine methylation at H3K9, H3K27, and H4K20 positions) and to induction of gene expression (lysine methylation at H3K4, H3K36, and H3K79 positions), that depends on the modification site and the number of methyl groups (mono-, di-, and trimethylation) [28]. Using Western blotting, we analyzed the effect of enzastaurin on the following sites: H3K4me3, H3K9me3, H4K20me3; no effect of enzastaurin on the methylation level of these sites was observed (Fig. 3A). The data obtained are consistent with the results of real-time PCR, which indicate that the drug has no effect on the expression level of the corresponding histone methyltransferases: (1) SUV39H1 and SUV39H2 (methylation at the H3K9 site), (2) SUV420H1 and SUV420H2 (methylation at H4K20), and (3) SETD1A и SETD1B (methylation of the H3K4 site) (Fig. 3B).
Another major epigenetic mechanism of gene expression regulation is DNA methylation. Hypermethylation of the promoters of tumor suppressor genes inhibits their expression, and, by this way, contributes to the initiation of carcinogenesis and tumor progression [29, 30]. Methylation-sensitive HpaII/MspI restriction assay demonstrated that enzastaurin decreased the level of DNA methylation, which was indicated by a higher degree of DNA cleavage by the methylationsensitive restriction enzyme HpaII (55.8 % compared to the control) (Fig. 4A, 4B). Moreover, the demethylation effect of the drug was comparable with the effect of the positive control 5-azacitidine. These results were consistent with real-time PCR data concerning the drug influence on the expression level of DNA methyltransferases. Enzastaurin was shown to reduce the mRNA level of de novo DNA methyltransferases
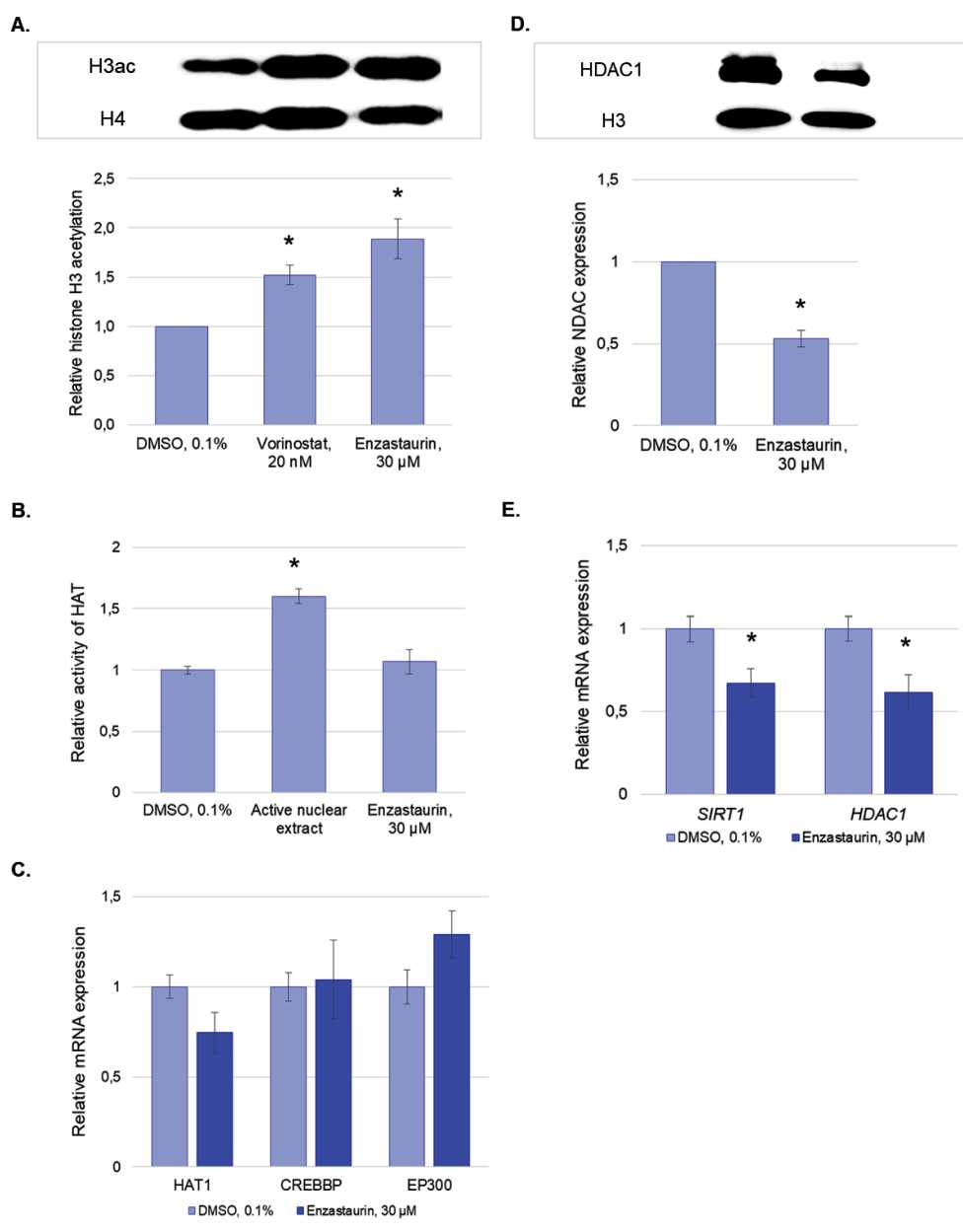
Рис. 2. Анализ эффектов энзастаурина на ацетилирование гистонов:
А. Влияние энзастаурина на ацетилирование гистона Н3, Вестерн-блоттинг и его денситометрический анализ; В. Влияние энзастаурина на тотальную активность ферментов семейства НАТ; С. Анализ влияния энзастаурина на экспрессию генов НАТs;
D. Анализ влияния энзастаурина на экспрессию белка HDAC1, Вестерн-блоттинг и его денситометрический анализ;
E. Анализ влияния энзастаурина на экспрессию генов SIRT1 и HDAC1 методом ОТ-ПЦР в реальном времени. Для анализа данных использовали метод 2-ΔΔCt, в качестве референсного гена использовали АСТВ, далее нормирование проводили по отрицательному контролю. Все результаты представлены в виде M ± SD.
Примечание: * – различия статистически значимы по сравнению с контролем (р<0,05).
Fig. 2. Analysis of the effect of enzastaurin on histone acetylation:
A. Effect of enzastaurin on H3 histone acetylation, Wetsern blotting and densitometric analysis; B. Effect of enzastaurin on the total activity of the HAT family enzymes; C. Analysis of the effect of enzastaurin on the expression of НАТ genes; D. Analysis of the effect of enzastaurin on the expression of HDAC1 protein, Wetsern blotting and densitometric analysis; E. Analysis of the effect of enzastaurin on the expression of SIRT1 and HDAC1 by real-time RT-PCR. The 2-ΔΔCt method was used for data analysis, ÀÑÒÂ was used as a reference gene, and the samples were also normalized to the negative control. All data are presented as M ± SD.
Notees: * – differences are statistically significant as compared to the control (p<0.05)
DNMT3A and DNMT3B (by 2.5-fold and 1.6-fold, respectively), while the expression level of DNMT1 , which was responsible for methylation during replication, remained unchanged.
Thus, screening of the epigenetic activity of five antitumor drugs from the group of protein kinase inhibitors (gefitinib, imatinib, pazopanib, ponatinib, and enzastaurin) allowed us to demonstrate for the first time the ability of enzastaurin to reactivate the expression of epigenetically repressed genes. Our original data concerning enzastaurin effects demonstrate both significant decrease of integrated DNA methylation and inhibition of de novo methyltransferases DNMT3A and DNMT3B expression. Also, an increase in the level of histone acetylation under the action of enzastaurin was shown for the first time, which is consistent with our data on the inhibition of expression of histone deacetylases.
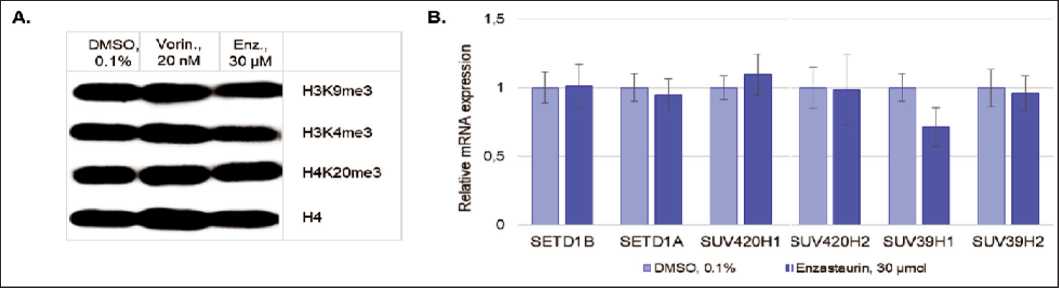
Рис. 3. Анализ эффектов энзастаурина на метилирование гистонов:
А. Влияние энзастаурина на метилирование гистонов Н3 и Н4, Вестерн-блоттинг; B. Анализ влияния энзастаурина на экспрессию генов HMTs методом ОТ-ПЦР в реальном времени. Для анализа данных использовали метод 2-ΔΔCt, в качестве референсного гена использовали АСТВ , далее нормирование проводили по отрицательному контролю. Все результаты представлены в виде M ± SD.
Примечание: * – различия статистически значимы по сравнению с клетками контроля (р<0,05) Fig. 3. Analysis of the effect of enzastaurin on histone methylation:
A. The effect of enzastaurin on the methylation of Н3 and H4 histones, Wetsern blotting; B. Analysis of the effect of enzastaurin on the expression of HMT genes by real-time RT-PCR. The 2-AACt method was used for data analysis, АСТВ was used as a reference gene, and the samples were also normalized to the negative control. All data are presented as M ± SD.
Notes: * – differences are statistically significant as compared to the control (p<0.05)
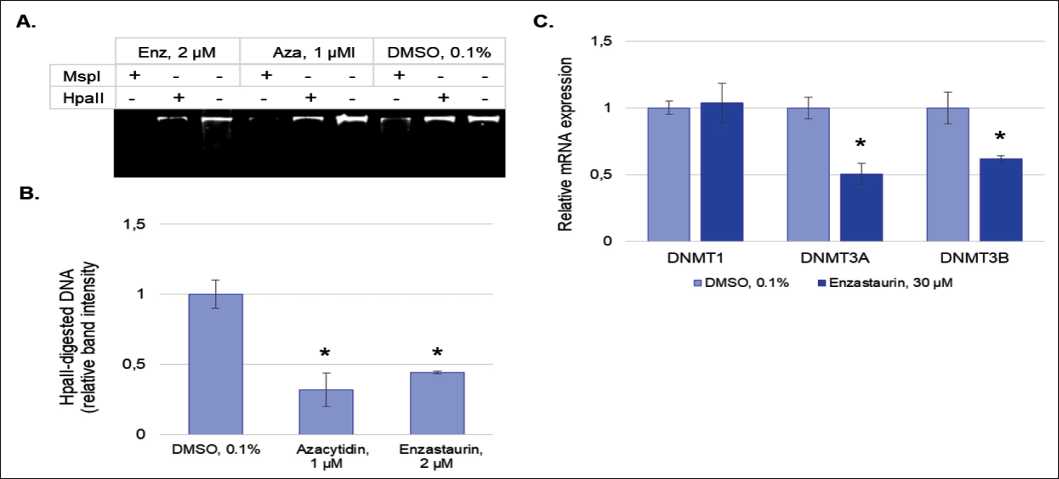
Рис. 4. Влияние энзастаурина на интегральное метилирование ДНК:
А. Электрофореграмма геномной ДНК, обработанной рестриктазами HpaII и MspI; B. Денситометрический анализ полос HpaII-рестрицированной ДНК относительно геномной ДНК; C. Анализ влияния энзастаурина на экспрессию генов DNMTs методом ОТ-ПЦР в реальном времени. Все результаты представлены в виде M ± SD.
Примечание: * – различия статистически значимы по сравнению с контролем (р<0,05)
Fig. 4. The effect of enzastaurin on integrated DNA methylation:
A. Electrophoregram of the genomic DNA treated with restriction enzymes HpaII and MspI; B. Densitometric analysis of the bands of HpaII-restricted DNA relative to the genomic DNA; C. Real-time RT-PCR analysis of the effect of enzastaurin on the expression of DNMT genes. All data are presented as M ± SD.
Notes: * – differences are statistically significant as compared to the control (p<0.05)
Список литературы Epigenetic effects of enzastaurin - a new aspect in the mechanism of action of an anticancer drug from protein kinase inhibitors
- Cheng Y, He C., Wang M, Ma X., Mo F., Yang S., Han J., Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019 Dec 17; 4: 62. doi: 10.1038/s41392-019-0095-0.
- Ravegnini G., Sammarini G., HreliaP., Angelini S. Key Genetic and Epigenetic Mechanisms in Chemical Carcinogenesis. Toxicol Sci. 2015 Nov; 148(1): 2-13. doi: 10.1093/toxsci/kfv165.
- HanM., JiaL., Lv W., WangL., Cui W. Epigenetic Enzyme Mutations: Role in Tumorigenesis and Molecular Inhibitors. Front Oncol. 2019 Mar 29; 9: 194. doi: 10.3389/fonc.2019.00194.
- GnyszkaA., Jastrzebski Z., Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013 Aug; 33(8): 2989-96.
- Li Y., SetoE. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med. 2016 Oct 3; 6(10): a026831. doi: 10.1101/cshperspect.a026831.
- McClure J.J., LiX., Chou C.J. Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Advances in Cancer Research. Academic Press. 2018. 183-211 p.
- Zagni C., Chiacchio U., Rescifina A. Histone methyltransferase inhibitors: novel epigenetic agents for cancer treatment. Curr Med Chem. 2013; 20(2): 167-85. doi: 10.2174/092986713804806667.
- ItalianoA., Soria J.C., ToulmondeM., MichotJ.M.,LucchesiC., Varga A., Coindre J.M., Blakemore S.J., ClawsonA., SuttleB., McDonald A.A., Woodruff M., Ribich S., Hedrick E., Keilhack H., Thomson B., Owa T., CopelandR.A., HoP.T.C., Ribrag V. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018 May; 19(5): 649-659. doi: 10.1016/S1470-2045(18)30145-1.
- VermaN., RaiA.K., Kaushik V., Brünnert D., ChaharK.R., Pandey J., Goyal P. Identification of gefitinib off-targets using a structure-based systems biology approach; their validation with reverse docking and retrospective data mining. Sci Rep. 2016 Sep 22; 6: 33949. doi: 10.1038/ srep33949.
- Ratovitski E.A. Anticancer Natural Compounds as Epigenetic Modulators of Gene Expression. Curr Genomics. 2017 Apr; 18(2): 175-205. doi: 10.2174/1389202917666160803165229.
- Chung F.F., Herceg Z. The Promises and Challenges of Toxico-Epigenomics: Environmental Chemicals and Their Impacts on the Epigenome. Environ Health Perspect. 2020 Jan; 128(1): 15001. doi: 10.1289/ EHP6104.
- Katz R.A., Jack-Scott E., Narezkina A., Palagin I., Boimel P., Kulkosky J., Nicolas E., Greger J.G., Skalka A.M. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J Virol. 2007 Mar; 81(6): 2592604. doi: 10.1128/JVI.01643-06.
- PoleshkoA., Einarson M.B., Shalginskikh N., Zhang R., Adams P.D., Skalka A.M., Katz R.A. Identification of a functional network of human epigenetic silencing factors. J Biol Chem. 2010 Jan 1; 285(1): 422-33. doi: 10.1074/jbc.M109.064667.
- Shalginskikh N., Poleshko A., Skalka A.M., Katz R.A. Retroviral DNA methylation and epigenetic repression are mediated by the antiviral host protein Daxx. J Virol. 2013 Feb; 87(4): 2137-50. doi: 10.1128/ JVI.02026-12.
- Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., Absher D.M., Wold B.J., Myers R.M. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013 Mar; 23(3): 555-67. doi: 10.1101/gr.147942.112.
- Huang Y., Song H., Hu H., Cui L., You C., Huang L. Trichosanthin inhibits DNA methyltransferase and restores methylation-silenced gene expression in human cervical cancer cells. Mol Med Rep. 2012 Oct; 6(4): 872-8. doi: 10.3892/mmr.2012.994.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983 Dec 16; 65(1-2): 55-63. doi: 10.1016/0022-1759(83)90303-4.
- Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat Protoc. 2007; 2(6): 1445-57. doi: 10.1038/nprot.2007.202.
- Li R., Hebert J.D., Lee T.A., Xing H., Boussommier-Calleja A., HynesR.O., LauffenburgerD.A., Kamm R.D. Macrophage-Secreted TNFa and TGFß1 Influence Migration Speed and Persistence of Cancer Cells in 3D Tissue Culture via Independent Pathways. Cancer Res. 2017 Jan 15; 77(2): 279-290. doi: 10.1158/0008-5472.CAN-16-0442.
- Bhullar K.S., Lagaron N.O., McGowan E.M., Parmar I., Jha A., Hubbard B.P., Rupasinghe H.P.V. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018; 17(1): 48. doi: 10.1186/ s12943-018-0804-2.
- KannaiyanR., MahadevanD. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert Rev Anticancer Ther. 2018 Dec; 18(12): 1249-1270. doi: 10.1080/14737140.2018.1527688.
- Huang W.S., Metcalf C.A., Sundaramoorthi R., Wang Y., Zou D., Thomas R.M., Zhu X., Cai L., Wen D., Liu S., Romero J., Qi J., Chen I., Banda G., Lentini S.P., Das S., Xu Q., Keats J., Wang F., Wardwell S., Ning Y., Snodgrass J.T., BroudyM.I., RussianK., Zhou T., CommodoreL., Narasimhan N.I., Mohemmad Q.K., Iuliucci J., Rivera V.M., Dal-garno D.C., Sawyer T.K., Clackson T., Shakespeare W.C. Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methyl-piperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem. 2010 Jun 24; 53(12): 4701-19. doi: 10.1021/jm100395q.
- O'Hare T., Shakespeare W.C., Zhu X., Eide C.A., Rivera V.M., Wang F., Adrian L.T., Zhou T., Huang W.S., Xu Q., MetcalfC.A., Tyner J. W., Loriaux M.M., Corbin A.S., Wardwell S., Ning Y., Keats J.A., Wang Y., Sundaramoorthi R., Thomas M., Zhou D., Snodgrass J., Commodore L., Sawyer T.K., Dalgarno D.C., Deininger M.W., Druker B.J., Clackson T. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009 Nov 6; 16(5): 401-12. doi: 10.1016/j. ccr.2009.09.028.
- Nowakowski G.S., Zhu J., Zhang Q., Brody J., Sun X., Maly J., Song Y., Rizvi S., Song Y., LansiganF., JingH., Cao J., Lue J.K., Luo W., Zhang L., Li L., Han I., Sun J., Jivani M., Liu Y., Heineman T., Smith S.D. ENGINE: a Phase III randomized placebo controlled study of enzastaurin/ R-CHOP as frontline therapy in high-risk diffuse large B-cell lymphoma patients with the genomic biomarker DGM1. Future Oncol. 2020 May; 16(15): 991-9. doi: 10.2217/fon-2020-0176.
- WiltingR.H., Dannenberg J.H. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat. 2012; 15(1-2): 21-38. doi: 10.1016/j.drup.2012.01.008.
- Delcuve G.P., Khan D.H., Davie J.R. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012 Mar 12; 4(1): 5. doi: 10.1186/1868-7083-4-5.
- RifaïK., Judes G., IdrissouM., DauresM., Bignon Y.J., Penault-Llorca F., Bernard-Gallon D. SIRT1-dependent epigenetic regulation of H3 and H4 histone acetylation in human breast cancer. Oncotarget. 2018 Jul 17; 9(55): 30661-30678. doi: 10.18632/oncotarget.25771.
- VolkelP., AngrandP.O. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007; 89(1): 1-20. doi: 10.1016/j. biochi.2006.07.009.
- Koch A., Joosten S.C., Feng Z., de Ruijter T.C., Draht M.X., Melotte V., Smits K.M., Veeck J, Herman J.G., Van Neste L., Van Criekinge W., De Meyer T., van Engeland M. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018; 15(7): 459-466. doi: 10.1038/ s41571-018-0004-4.
- Patnaik S., Anupriya. Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front Pharmacol. 2019; 10: 588. doi: 10.3389/fphar.2019.00588.


