Estimation of genetic diversity for CLCuV, earliness and fiber quality traits using various statistical procedures in different crosses of Gossypium hirsutum L
Автор: Nazir Amna, Farooq Jehanzeb, Mahmood Abid, Shahid Muhammad, Riaz Muhammad
Журнал: Вестник аграрной науки @vestnikogau
Статья в выпуске: 4 (43), 2013 года.
Бесплатный доступ
In the current studies 70 crosses made among different genotypes were evaluated for CLCuV tolerance, better fiber quality and earliness related traits. For evaluation of these traits cluster, PC and correlation analysis was employed to obtain suitable parents and crosses that can be exploited in future breeding programmes. The earliness related traits had positive and significant association among them and days to 1 st boll opening showed its positive association with CLCuV%. Only negative and significant association was found between GOT% and staple length. Principal component (PC) analysis showed first 3 PCs having Eigen value >1 explaining 64.1of the total variation with days to 1 st square and flower were the most important traits in PC1. Cluster analysis classified 70 crosses into four divergent groups. The genotypes in cluster 1 & 3 comprised of genotypes with less attack of CLCuV%, better GOT% along with better fiber quality. The cluster 2 and 4 comprised of some crosses having fair tolerance against CLCuV% and very good GOT% along with low micronaire values. The crosses in cluster 2 and 3 also exhibited desirable values of earliness related traits. The results concluded that the diversity among the genotypes could be utilized for cultivar breeding and germplasm conservation programs aimed at improving CLCuV% tolerance with better fiber quality.
Cotton, correlation, clcuv, earliness, fiber quality, principle component analysis
Короткий адрес: https://sciup.org/147124092
IDR: 147124092 | УДК: 632.38:575.003.12:311
Текст научной статьи Estimation of genetic diversity for CLCuV, earliness and fiber quality traits using various statistical procedures in different crosses of Gossypium hirsutum L
Cotton is the leading non- food agricultural and industrial fiber crop grown in more than 80 countries (Dutt et al., 2004; Shakeel et al., 2011). Pakistan is the 4th largest cotton producing country in the world after China, USA and India, and 3rd largest consumer of cotton (Akhtar, 2005) yet, Cotton production in Pakistan is stagnant for years due to a number of biotic and abiotic stresses. In Pakistan cotton crop not only face ever alarming threat of cotton leaf curl virus but also fiber quality of the developed cultivars is under question (Farooq et al., 2011). Scientist all over the country made efforts to improve both these factors by exploiting various classic breeding procedures to obtain durable resistance against cotton leaf curl virus and better fiber quality. According to Li et al. 2008 for developing best genotypes efficient utilization of available germplasms in the form of crosses and addition of new germplasms resources are necessary to create sufficient genetic variability. For launching any breeding programme genetic variability is of prime importance. Variability in germplasms not only increase the probability of resistance against various biotic and abiotic stresses but also can provide desirable combinations that can be manifested in future breeding endeavors (Van Esbroeck and Bowman, 1998). Different conventional breeding methods including introduction of exotic germplasm, hybridization, mutation and polploidy can be exploited to obtain the desired variability and thus to find out parental lines that can yield diverse segregation population from where one can select superior progenies (Esmail et al., 2008).
The development of superior cotton genotypes by hybridizing distant parental lines has been reported (Punitha et al., 2004; Akter, 2009). To get precise information on nature and extent of genetic variation depends upon the various techniques used for its estimation, like plant characterization based on agronomical, morphological and physiological traits (Bajracharya et al., 2006). Multivariate analysis based on Mahalonobis’s D2 statistics (MDS), principal component analysis (PCA) and principal coordinate analysis (PCoA) are mostly used to evaluate the magnitude of genetic diversity among the germplasm (Brown-Guedira et al., 2000). Among these biometrical procedures the main edge of principal component analysis (PCA) is that each genotype can be assigned to only one group and it also reflects the significance of largest contributor to the total variability at each axis of differentiation (Sharma, 1998). Genetic variation for morphological traits has been estimated using principal component analysis, which lead to the recognition of phenotypic variability in cotton (Saravanan et al., 2006; Esmail et al., 2008; Li et al., 2008). In the current studies various diverse parents for better fiber quality and CLCuV tolerance were utilized in crossing to obtain desirable combinations that can help to improve cotton productivity. The different statistical procedures utilized in the current studies will be helpful to find out best parental combinations that may result resistance against cotton leaf curl virus with better fiber quality.
MATERIALS AND METHODS
Plant Material & Site Characteristics. A total of 70 crosses were evaluated for this study carried out during the cropping seasons 2012-13 on 25th of May. The experiment was carried out at breeding farm of Cotton Research Institute, Faisalabad, Punjab, Pakistan. The germplasm utilized for crossing have sufficient amount of variability regarding CLCuV%, fiber quality and earliness related traits.
Experimental Design, Plot Size & Cultural Practices. For each entry, plot size measured 4.572 m × 1.524 m, comprising 2 rows set 75 cm apart. Distance between plants within rows was 30 cm. Normal agronomic and cultural practices (irrigation, weeding, hoeing, and fertilizer applications) were adopted as and when required.
Measurement of the studied traits. For measuring the traits 10 representative, undamaged plants were selected in each line and marked for identification. The data regarding appearance of 1st square, 1st flower and 1st boll opening were taken by counting number of days from planting to the appearance of 1st square, 1st flower and 1st boll opened respectively. Cleaned and dry samples of seed cotton were weighed and then ginned separately with single roller electric ginning machine. The lint obtained from each sample was weighed and ginning out turn % was calculated by the following formula:
Ginning outturn (%) = Weight of lint / Weight of seed cotton × 100
Fiber characteristics such as staple length, fiber fineness of each guarded plant were measured by using spin lab HVI-900. This computerized instrument provides us a true profile of raw fiber. It measures the most important characters such as staple length (mm) and fiber fineness (μg/inch) within a quick period of time according to international trading standards.
CLCuV Disease Incidence (%) Methodology. CLCuV disease incidence (%) and reaction of the cultivars was determined using the disease scale (Table 1) described by Akhtar et al. 2010 and Farooq et al. 2011. Then %age of CLCuV disease incidence was calculated by using the following formula:
CLCuV disease incidence (%) = Sum of all disease ratings/total number of plants ×16.16
Statistical Analysis. The average data of both the years were subjected to basic statistics, correlation analysis, cluster analysis and principal component analysis (PCA) using statistical software packages of SPSS version 19 and STATISTICA version 5.0 (Sneath and Sokal, 1973). Cluster analysis was performed using K-means clustering while tree diagram based on elucidation distances was developed by Ward’s method. The D2 statistics was calculated according to Mahalnobis (1936) and Rao (1952). First two principal components were plotted against each other to find out the patterns of variability among genotypes and association between different clusters using SPSS version 19.
RESULTS OF RESEARCH
Correlation studies. The basic statistics of various studied traits demonstrated considerable variability among 70 crosses made among several cotton genotypes Table-2. Simple correlation coefficients revealed some significant associations among 7 studied traits
(Table 3). Days to 1st square exhibited positive and significant association with days to 1st flower and boll opening while with other traits it showed non-significant results. Days to 1st flower had positive correlation with days to 1st boll opening and boll opening days had significant positive association with CLCuV%. The only negative and significant correlation was found between GOT% with staple length.
Principal component analysis. In this study, out of total 7, three principal components (PCs) were extracted having Eigen value >1. These three PCs contributed 64.1% of the total variability amongst the crosses assessed for CLCuV% and other earliness and fiber related traits (Table 4). However, the remaining components contributed only 35.9% towards the total diversity. The PC I contributed maximum towards the variability (25.8%) followed by PC II (22.6%), and PC III (15.7). The traits like days to 1st square and flower showed considerable positive factor loadings on PC I while GOT% had maximum positive loadings on PC II. The negative loadings on PC I and PC II were shown by staple length on both components while the traits like CLCuV% and days to 1st boll opening showed negative loadings only on PC II. The PC III was elucidated by diversity among the genotypes for days to 1st boll opening. A PC biplot Figure-1 showed that variables and crosses are super imposed on the plot as vectors. Distance of each variable with respect to PC-1 and PC-2 showed the contribution of these variables in the variation of genotypes used. The biplot showed that as whole days to 1st square, flower and boll opening contributed maximum towards variability in the crosses.
Cluster analysis. Seventy crosses made among different cotton genotypes were grouped into 4 clusters based on various traits (Table 5). Cluster analysis showed that cluster 1 comprised of 19 crosses, cluster 2 of 7 while cluster 3 had 26 and cluster 4 contained 18 crosses (Table 6). The genotypes in cluster 1 only showed reasonable values of GOT% and CLCuV% but for other traits selection cannot be made. Similarly, the 2nd cluster comprised of genotypes having good values of earliness related traits (Table 5). The members of 3rd cluster were characterized by least values of fiber fineness. The pairwise Mahalanobis distances (D2 statistics) among four clusters of 70 cotton crosses (Table 7) revealed that genotypes of cluster 2 and 3 showed maximum diversity against the members of other studied characters. The first two principal components contributed almost 48.8% towards the total variance. Not a single cluster showed obvious separation. The tree diagram showed more or less similar results comprising of two main groups A and B each of which is further subdivided into two clusters (Figure 2).
Table 1 - Rating scale for cotton leaf curl virus (CLCuV) symptoms
|
Symptoms |
Disease rating |
Disease index (%) |
Disease reaction |
|
Absence of symptoms |
0 |
0 |
Immune |
|
Thickening of a few small veins or the presence of leaf enations on 10 or fewer leaves of a plant |
1 |
0.1-1 |
Highly resistant |
|
Thickening of a small group of veins |
2 |
1.1-5 |
Resistant |
|
Thickening of all veins but no leaf curling |
3 |
5.1-10 |
Moderately resistant |
|
Severe vein thickening and leaf curling on the top third of the plant |
4 |
10.1–15 |
Moderately susceptible |
|
Severe vein thickening and leaf curling on the half of the plant |
5 |
15.1–20 |
Susceptible |
|
Severe vein thickening, leaf curling, and stunting of the plant with reduced fruit production |
6 |
>20 |
Highly susceptible |
(Sum of all disease ratings / total # of plants) × 16.66.
Table 2 - Basic statistics for various traits of 70 crosses between different cotton genotypes
|
Traits |
Minimum |
Maximum |
Mean± S.E. |
Std. Deviation |
Variance |
|
Days to 1st square |
27 |
40 |
34.21±0.27 |
2.26 |
5.13 |
|
Days to 1st flower |
49 |
64 |
55.34±0.37 |
3.06 |
9.36 |
|
Days to 1st boll opening |
79 |
91 |
85.28±0.29 |
2.45 |
6.00 |
|
CLCuV% |
0.0 |
36.76 |
17.18±0.95 |
7.93 |
62.91 |
|
GOT% |
36 |
46 |
41.23±0.28 |
2.34 |
5.48 |
|
SL (mm) |
26.8 |
28.5 |
27.7±0.05 |
0.45 |
0.21 |
|
FF ((µg/inch) |
4.1 |
5.5 |
4.83±0.03 |
0.32 |
0.11 |
Table 3 - Simple correlation coefficients of CLCuV, earliness and quality traits in somecrosses of cotton
|
Traits |
Days to 1st square |
Days to 1st flower |
Days to 1st boll opening |
CLCuV% |
GOT% |
SL (mm) |
|
Days to 1st square |
1.000 |
- |
- |
- |
- |
- |
|
Days to 1st flower |
0.573** |
1.000 |
- |
- |
- |
- |
|
Days to 1st boll opening |
0.268** |
0.228** |
1.000 |
- |
- |
- |
|
CLCuV% |
0.005 |
0.019 |
0.218** |
1.000 |
- |
- |
|
GOT% |
0.130 |
0.155 |
-0.097 |
-0.117 |
1.000 |
- |
|
SL (mm) |
-0.100 |
-0.099 |
0.135 |
0.112 |
-0.382** |
1.000 |
|
FF (µg/inch) |
-0.02 |
-0.153 |
0.087 |
-0.077 |
0.162 |
-0.122 |
Table4 - Principle component analysis of CLCuV, earliness and quality traits in some crosses of cotton
|
n/n |
PC I |
PC II |
PC III |
|
Eigen value |
1.804 |
1.585 |
1.097 |
|
% of total variance |
25.8 |
22.6 |
15.7 |
|
Cumulative variance % |
25.8 |
48.4 |
64.1 |
|
Factor loadings by various traits |
|||
|
Variable |
PC I |
PC II |
PC III |
|
Days to 1st square |
0.621 |
-0.094 |
0.066 |
|
Days to 1st flower |
0.618 |
-0.106 |
0.235 |
|
Days to 1st boll opening |
0.319 |
-0.405 |
-0.489 |
|
CLCuV% |
0.035 |
-0.402 |
-0.290 |
|
GOT% |
0.280 |
0.530 |
-0.093 |
|
SL (mm) |
-0.226 |
-0.541 |
0.023 |
|
Fiber fineness (µg/inch) |
-0.014 |
0.284 |
-0.780 |
Table5 - Cluster analysis of of CLCuV, earliness and quality traits in some crosses of cotton
|
Traits |
Cluster 1 |
Cluster 2 |
Cluster 3 |
Cluster 4 |
|
Days to 1st square |
38.00 |
32.00 |
32.00 |
39.00 |
|
Days to 1st flower |
64.00 |
53.00 |
56.00 |
59.00 |
|
Days to 1st boll opening |
90.00 |
89.00 |
79.00 |
91.00 |
|
CLCuV% |
6.25 |
36.76 |
0.00 |
23.33 |
|
GOT% |
46.00 |
44.00 |
42.00 |
40.00 |
|
SL (mm) |
27.00 |
27.50 |
27.00 |
27.00 |
|
Fiber fineness (µg/inch) |
4.90 |
4.90 |
4.70 |
4.60 |
Table 6 - Cluster membership of various crosses
|
Cluster-1 |
19 |
1004, 1009,1011, 1021,1027,1035,1036,1039, 1051,1058, 1059, 1071, 1078,1079, 1090, 1098, 1103, 1107, 1119 |
|
Cluster-2 |
7 |
1005,1007,1013,1077,1099,1106,1125 |
|
Cluster-3 |
26 |
1006,1010,1012,1014,1019,1024,1034,1038,1040,1041,1042,1045,1047,1050,1053, 1055,1063,1064,1066,1072,1076,1111,1114,1115,1116,1120 |
|
Cluster-4 |
18 |
1008,1017,1033,1037,1044,1054,1056,1067,1074,1081,1085, 1086,1094,1100, 1105,1109,1117,1121 |
Table 7 - D2 statistics among different clusters
|
n/n |
Cluster 1 |
Cluster 2 |
Cluster 3 |
Cluster 4 |
|
Cluster 1 |
0.000 |
- |
- |
- |
|
Cluster 2 |
17.577 |
0.000 |
- |
- |
|
Cluster 3 |
6.530 |
20.534 |
0.000 |
- |
|
Cluster 4 |
9.183 |
8.645 |
12.278 |
0.000 |
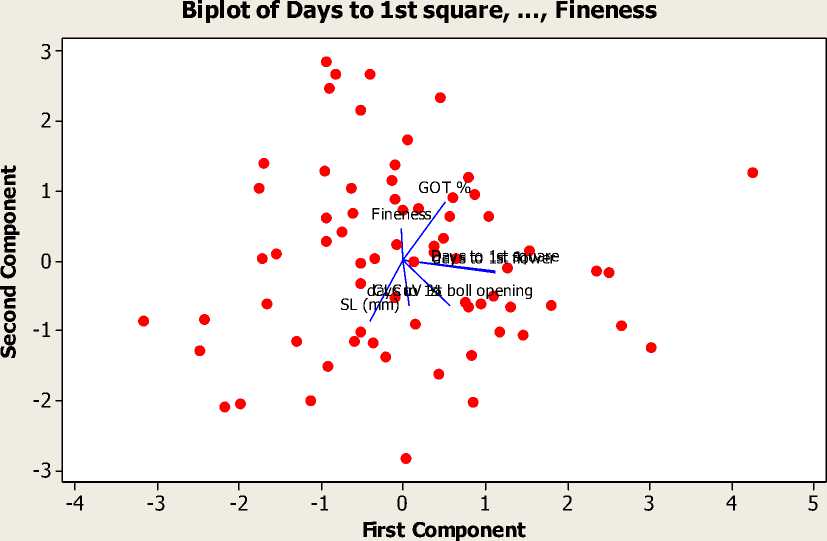
Figure 1 - Biplot between PC-1 and 2 showing contribution of various traits in variability among different crosses
DISCUSSION
Before launching any breeding programme the information regarding association among various traits is a prerequisite as it provides an opportunity for the selection of genotypes having desirable traits simultaneously (Ali et al., 2009c). In the current studies simple correlation analysis revealed some important associations among earliness related traits. The positive correlation among these earliness related traits suggested that these characters are important for direct selection of high yielding genotypes. Farooq et al. 2013 reported positive correlation of days to 1st square and flower under virus intensive conditions. The preservation and utilization of genetic resources could be made by dividing the total variance into its components. It also provides an oppertunity for utilization of appropriate germplasm in crop improvement for particular plant traits (Sneath and Sokal, 1973; Pecetti et al., 1996). The principal component (PC) analysis partition the total variance into different factors. The Principal Components analysis is a powerful tool to obtain parental lines for successful breeding program (Akter et al., 2009). In this experiment, the PC analysis partitioned the total variance into 3 PCs contributing maximum to the total diversity among the crosses due to the study of various traits. Chozin (2007), Mujaju and Chakuya (2008) and Ali et al. 2011 reported important contribution of first PCs in total variability while studying different traits. In our experiment, 1st PC was mainly due to variations in earliness related traits. These results are in agreement with the results of association analysis which showed positive correlation among these traits. PC analysis ultimately confirmed the amount of variation for the traits among the crosses which could be utilized in designing a comprehensive breeding program aimed at improving CLCuD% tolerance, fiber quality and earliness related traits as it is generally assumed that maximum variation yields maximum heterotic effects. Malik et al., 2011 and Ashokkumar and Ravikesavan 2011 reported that the presence of adequate amount of variation in colored cotton genotypes offer sufficient scope for characterization of colored cotton genotypes.
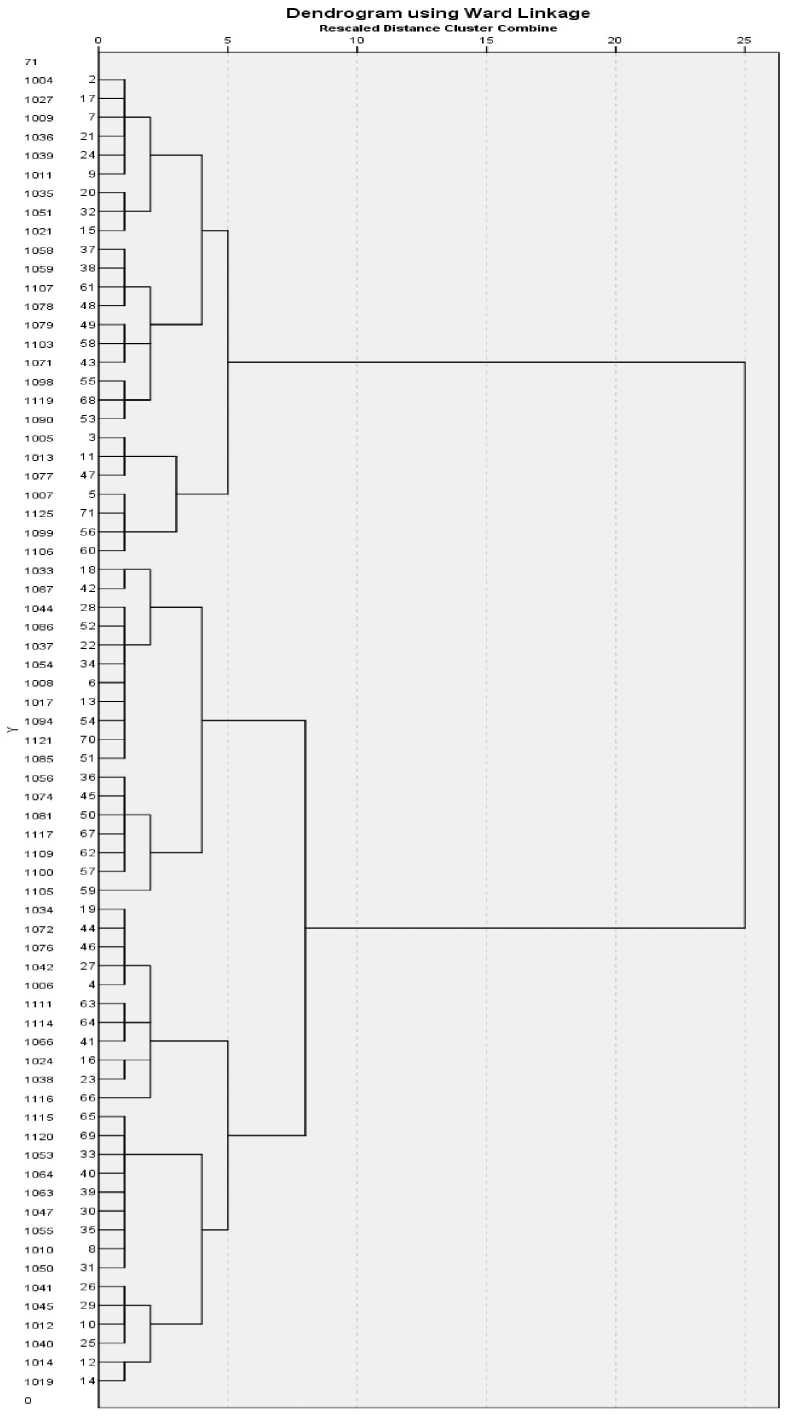
Figure 2 - Tree diagram of 70 cotton crosses based on CLCuV, fiber quality and earliness related traits
The cluster analysis also corroborates sufficient variation for this group of genotypes at various growth stages. The genotypes in cluster 1 & 3 comprised of genotypes with less attack of CLCuD%, better GOT% along with better fiber quality. The cluster 2 and 4 comprised of some crosses having fair tolerance against CLCuV%, very good GOT% along with low micronaire values. The crosses in cluster 2 and 3 also exhibited desirable values of earliness related traits. All the clusters providing some useful information for various studied traits and some of the crosses could be exploited in next generations for better fiber quality, virus tolerance and earliness related traits. The pair wise Mahalanobis distances (D2 statistics) among four clusters revealed that crosses of cluster 2 and 3 showed maximum diversity against the members of other clusters for most of the studied characters so could be exploited mainly to obtain transgressive segregates. The scatter plot showed association between different clusters which could be because a large number of crosses showing variation in adaptation or some sort of similarity between them. However, cluster analysis brings together those crosses showing considerable tolerance against CLCuV% tolerance at one place and on the basis of earliness related traits on another place. Amurrio et al. (1995) and Rabbani et al . (1998) reported lack of relationship between various clusters based on agronomic traits and origins of genotype in peas ( Pisum sativum ) and mustard ( Brassica juncea ) respectively. The tree diagram comprised of group of genotypes showing considerable tolerance against CLCuV% and possessing better yield potential. The occurrence of this wide variation between the clusters is of great genetic value in providing materials aimed at cotton selection for adaptation to CLCuV hit areas. Similar kind of results related to germplasms grouping has been reported by Ayana and Bekele 1998 and Grenier et al. 2001.
CONCLUSION
Correlation, Cluster analysis and PC analysis in the current studies provided facilitation in the identification of genotypes and crosses having tolerance to CLCuD%, better fiber quality and possessing eraliness. Various useful correlations and above mentioned information extracted from cluster and PC analysis will be helpful in designing breeding programmes to obtain high yielding genotypes possessing high degree of CLCuV tolerance and better fiber quality.
Список литературы Estimation of genetic diversity for CLCuV, earliness and fiber quality traits using various statistical procedures in different crosses of Gossypium hirsutum L
- Akhtar KP, Haider S, Khan MKR, Ahmad M, Sarwar N, Murtaza MA, Aslam M (2010) evaluation of Gossypium species for resistance to leaf curl Burewala virus. Ann Appl Biol. 157: 135-147
- Akhtar M (2005) Cotton production in Pakistan and role of trading corporation of Pakistan in price stabilization. ICAC. Research Associate Program. Washington DC, NY. Memphis 10-19 May
- Akter A, Hasan MJ, Paul AK, Mutlib MM, Hossain MK (2009) Selection of parent for improvement of restorer line in rice (Oryza Sativa L.). SAARC J Agri. 7: 43-50
- Ali MA, Jabran K, Awan SI Abbas A, Ehsanullah, Zulkiffal M, Acet T, Farooq J, Rehman A (2011) Morpho -physiological diversity and its implications for improving drought tolerance in grain sorghum at different growth stages. Aust J Crop Sci. 5(3): 311-320
- Ali MA, Nawab NN, Abbas A, Zulkiffal M, Sajjad M (2009c) Evaluation of selection criteria in Cicer arietinum L. using correlation coefficients and path analysis. Aust J Crop Sci. 3: 65-70
- Amurrio JM, de Ron AM, Zeven AC (1995) Numerical taxonomy of Iberian pea landraces based on quantitative and qualitative characters. Euphytica. 82: 195-205
- Ashokkumar K, Ravikesavan R (2011) Morphological diversity and per se performance in Upland Cotton (Gossypium hirsutum L.). J Agric Sci. 3: 107-113
- Ayana A, Bekele E, (1999) Multivariate analysis of sorghum (Sorghum bicolor (L.) Moench) germplasm from Ethiopia and Eritrea. Genet Resour Crop Evol. 46: 273-284
- Bajracharya J, Steele KA, Jarvis DI, Sthapit BR, Witcombe JR (2006) Rice landrace diversity in Nepal: Variability of agro-morphological traits and SSR markers in landraces from a high-altitude site. Field Crops Res. 95: 327-335
- Brown-Guedira, GL (2000) Evaluation of genetic diversity of soybean introductions and North American ancestors using RAPD and SSR markers. Crop Sci. 40: 815-823
- Chozin M (2007) Characterization of sorghum accessions and choice of parents for hybridization. J Akta Agr Edisi Khusus 2: 227-232
- Dutt Y, Wang XD, Zhu YG, Li YY (2004) Breeding for high yield and fibre quality in coloured cotton. Plant Breeding. 123: 145-151
- Esmail RM, Zhang JF, Abdel-Hamid AM (2008) Genetic diversity in elite cotton germplasm lines using field performance and RAPD markers. World J Agric Sci 4: 369-375
- Farooq A, Farooq J, Mahmood A, Batool A, Rehman A, Shakeel A, Riaz M, Shahid MTH, Mehboob S (2011) An overview of cotton leaf curl virus disease (CLCuD) a serious threat to cotton productivity. Aust J Crop Sci. 5(12):1823-1831
- Grenier C, Deu M, Kresovich S, Bramel Cox PJ, Hamon P (2001) Assessment of genetic diversity in three subsets constituated from the ICRISAT sorgham collection using random vs. non-random sampling procedures by using molecular markers. Theor App Genet. 101: 197-202
- Imran M, Shakeel A, Farooq J, Saeed A, Farooq A, Riaz M (2011) Genetic studies of fiber quality parameter and earliness related traits in upland cotton (Gossypium hirsutum L.). AAB Bioflux. 3(3):151-159
- Li Z, Wang X, Yan Z, Guiyin Z, Wu L, Jina C, Ma Z (2008) Assessment of genetic diversity in glandless cotton germplasm resources by using agronomic traits and molecular markers. Front Agric China. 2: 245-252
- Mahalanobis PC (1936). On the generalized distance in statistics. Proc Nat Inst Sci. 12: 301-377
- Malik W, Iqbal MZ, Khan AA, Noor E, Qayyum A, Hanif M (2011) Genetic basis of variation for seedling traits in Gossypium hirsutum L. Afr J Biotechnol. 10: 1099-1105
- Mujaju C, Chakuya E (2008) Morphological variation of sorghum landrace accessions on-farm in Semi-arid areas of Zimbabwe. Int J Bot. 4: 376-382
- Pecetti L, Annicchiario P, Damania AB (1996) Geographic variation in tetraploid wheat (Triticum turgidum spp. Turgidum convar. Durum) landraces from two provinces in Ethiopia. Genet. Resour Crop Evol. 43: 395-407
- Punitha D, Raveendran TS (2004) DNA fingerprinting studies in coloured cotton genotypes. Plant Breed. 123: 101-103
- Rabbani MA, Iwabuchi A, Murakami Y, Suzuki T, Takayanagi K (1998) Phenotypic variation and the relationships among mustard (Brassica juncea L.) germplasm from Pakistan. Euphytica. 101: 357-366
- Rao CR (1952) Advanced Statistical Methods in Biometrical Research. John Wiley and Sons. New York USA
- Saravanan S, Arutchendhil P, Raveendran TS, Koodalimgam K (2006) Assessment of genetic divergence among introgressed culture of Gossypium hirsutum L. through RAPD analysis. J Appl Sci Res. 2: 1212-1216
- Shakeel A, Farooq J, Ali MA, Riaz M, Farooq A, Saeed A, Saleem MF (2011) Inheritance pattern of earliness in cotton (Gossypium hirsutum L.). Aust J Crop Sci. 5(10): 1224-1231
- Sharma JR (1998) Statistical and Biometrical Techniques in Plant Breeding. New Age International (P) Limited Publishers, New Delhi. 432
- Sneath PHA, Sokal RR (1973) Numerical Taxonomy: The Principles and Practice of Numerical Classification. W.F. Free-Man and Co. San Francisco. pp: 573
- Van Esbroeck GA, Bowman DT (1998) Cotton germplasm diversity and its importance in cultivar development. J Cotton Sci. 2: 121-129


