Exploring high temperature responsive novel non coding RNAS and functional annotations from niger (Guizotia abyssinica)
Автор: Shafia Hoor F., Nagesh Babu R.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 3 т.20, 2024 года.
Бесплатный доступ
Introduction: MicroRNAs (miRNAs) are small non-coding RNAs which regulate gene expression by cleavage or repression of target genes at post-transcriptional level by translational inhibition/ mRNA degradation. Niger (Guizotia abyssinica) is an important oilseed crop widely grown in India. Identification and expression of non-coding RNAs during abiotic remains unclear till date. Methodology: Small RNA library was constructed by high throughput sequencing from control and stress tissues. Target genes of identified miRNAs were predicted using psRNATarget and their GO terms were annotated. The results were validated using RT-qPCR.
Abiotic stress, growth factors, high throughput sequencing, transcription factors
Короткий адрес: https://sciup.org/143182796
IDR: 143182796
Текст научной статьи Exploring high temperature responsive novel non coding RNAS and functional annotations from niger (Guizotia abyssinica)
Niger (Guizotia abyssinica) is an important but neglected edible oil seed crop widely grown in India. Niger is grown in an area of 2.53 lakh hectares with the production of 0.83 lakh tonnes and the productivity of 326 kg/ha (Suryanarayana et al., 2018). The crop of dry areas grown mostly by tribals and desired attention was not accorded on the biotic and abiotic stress conditions. Being a rain fed crop, Niger is exposed many abiotic stresses like drought, high temperature, salt and low nutrients which adversely affect the plant productivity. To date, the reports pertaining to biochemical effects of high temperature or role of mi NAs under high temperature on Niger cultivar is sparse. Thus, it is necessary to elucidate stress tolerance mechanisms by the involvement of mi NAs during high temperature to develop/improve the tolerance cultivar. Small NAs have emerged as ubiquitous key molecules regulating gene expression at the post-transcriptional level, either by repressing m NA translation or mediating the degradation of the targeted m NAs depending on their degree of complementarity (Dugas and Bartel 2004). Precursor stem-loop secondary structures are characteristic features of mi NAs and are conserved across species (Bartel and Sunker 2005; Carrington & Ambros 2003). In plants, small NAs and more specifically, micro NA (mi NA)s (~21 nt), have been functionally associated with development, biotic and abiotic stress (Khraiwesh et al., 2012). egulation of mi NAs by abiotic stresses was initially reported independently by Jones- hoades et al. (2006) and Sunkar et al (2004). Subsequently, a number of reports have been published which echoed that mi NAs are themselves regulated by abiotic factors and they in turn, control the levels of target genes involved in governing the stress responses. Two of the most featuring examples are mi 398 and mi 395, which have been repeatedly shown by independent groups to regulate cellular responses in many different stresses (Lelandais-Briere et al. 2009; Li et al. 2010; Li et al. 2010). Bharadwaj et al. 2014 had identified 15 conserved mi NAs in heat stressed Brasicca libraries and validated the expression of mi 395 which is indented as another mi NA involved in heat stress response other than mi 398 as established in Arabidopsis (Lu et al 2013) and French bean (Naya et al., 2014). ecently, Kavya et al. (2020) have reported the up-regulation of mi 166a, mi 156, mi 6173, mi 169e-5p, mi 6478 and mi 166U under salinity stress. However, till date no reports of mi NA characterization in Niger drawn us towards elucidating the role of in adaptive strategies employed by the oil seed plant to overcome the climatic cues. In this view, we ensued with heat treating the plants at 48 ºC for 8 h and profiled their small NA expression using high throughput sequencing and validating the results with qPC . We identified 125 conserved mi NAs belonging to 45 families. The cumulative studies of relative quantification using T-qPC . Our results emphasize that differential expression would render stress tolerance and has important implications for gene regulation under abiotic stress conditions.
MATERIALS AND METHODS
Plant material and high temperature stress treatment Niger seeds were surface sterilized and grown under controlled conditions at 28 °C day/25 °C night with 12 h light/12 h dark photo period. After 6 day of germination, seedlings were exposed to high temperature stress (42 °C for 1 h (induction); 45 °C for 1 h and 48 °C for 6 h). Tissues (shoot) were harvested immediately and stored at -80 °C for further analysis.
Small RNA library Construction and sequencing: Following NA extraction, small NA library (control and stress) was prepared according to the True Seq small NA sample prep Kits protocol (Illumina San Deigo USA). The quality and quantity of total NA were analyzed using Agilent 2100 bio-analyzer. Ten to thirty nt s NAs were purified from 15% denaturing polyacrylamide gel and then ligated with the 5’ and 3’ adapters. After being reverse transcribed by Superscript II reverse transcriptase (Invitrogen, USA), s NAs were amplified by PC . High throughput sequencing was performed using Nextseq500 platform (Illumina, USA).
Identification of miRNAs, target predictions and GO analysis: After Illumina sequencing, high quality small NA reads were extracted from raw reads through filtering the adapter dimers and low-quality tags.
Subsequently, unique sequences with 18~25 nucleotides length were mapped with ESTs of Niger precursors in mi Base 21.0 by BLAST search to identify conserved and novel mi NAs. The potential candidate mi NAs were identified by folding the flanking EST sequence of unique small NAs using mfold web server (Zuker et al. 2004). Parameters were set based on the criteria for annotation of plant mi NAs by Meyers et al (2008). To identify novel mi NAs, the mi Deep -P program was used to obtain all candidate precursors with hairpin-like structures that were perfectly mapped by sequencing tags DP/) (Yanget al.2011). Target predictions were performed using the ps NATarget web server with ESTs of Niger NATarget/analysis) using default parameters with a maximum of 3 expectation GO terms of the target genes were annotated according to their biological process, molecular functions, or involvement as cellular components using Blast2GO. The enzyme mapping of the annotated sequences was performed directly using the GO terms and KEGG orthologs.
qRT-PCR analysis of miRNA: T-qPC was used to validate the results obtained from the high throughput sequencing of mi NAs. NA was isolated using Trizol (Invitrogen) as per manufacturer’s instruction. 1 µg Total NA was reverse transcribed using stem-loop primers designed according to Chen 2005 and gene specific primers for target genes using One Step PrimeScript mi NA cDNA Synthesis Kit (Takara, Japan). t-qPC was performed using SYB premix ExTaq (Takara, Japan) and all the primers used were listed in Supplementary file 1. Small nuclear NA U6 and GAPDH were used as internal controls to normalize the mi NA expression and target genes expression, respectively. Subsequently, the quantification was carried out using (CFX-96, Bio ad). Three biological replicates were used per sample in addition to technical replicates along with a no template control and no T-enzyme control. The data were analyzed using 2-ΔΔCT method and reported as means ± standard errors (SE) of three biological replicates. Fold changes were determined by using the ratio of normalized expression
of stress against controlsamples and represented as log 2 values.
RESULTS AND DISCUSSION
High-throughput sequencing of small RNAs: Small NA libraries from stress and control seedlings were screened using Nextseq 500 (Illumina Inc, USA) generated nearly twenty million total raw reads. After removing low-quality sequences, adapters, and small sequences (< 17 nt long), 18,445,935 and 19,445,620 representing high quality sequences were obtained from stress and control libraries respectively. Further to determine the stress specific mi NAs, the sequences were filtered against the control library. Only reads found in stress library were considered for small NA identification. An in-house database comprising of noncoding NAs expect mi NAs from fam (12.0) was created and used to filter small NA fragments corresponding to non-coding NAs such as t NAs, r NA, snc NAs etc., The small NA length distribution (16-30 nt) of each library showed that the most abundant and diverse species were those 21-24 nt in length, a typical size range for Dicer-derived products (Fig 1) and termed as unique sequences which were further considered for identification of conserved mi NAs.In order to identify the conserved mi NAs, the unique sequences were mapped against mature mi NAs in mi Base (v 21). Following Blastn searches and further sequence analysis, a total of 125 mi NAs belonging to 45 families were identified. mi 156, mi 399 and mi 169 (Fig 1) represents the most abundant mi NA family (Fig 2). All the foresaid conserved mi NA families, possessed mi NA* sequences, identified with their stem-loop structures (Supplementary file of structures) prompts as additional evidence in support of the authenticity of the mi NAs. Since, the precursors and mature mi NAs are highly conserved in plants, it is feasible to define the MFEI values of newly identified conserved mi NAs based on their homologs used as reference sequences.
High temperature has drastically affected the distribution and productivity of agriculturally important crops worldwide. To overcome the adverse effect of climatological extremes, plants have developed many strategies at physiological and molecular levels. Gene regulation through post transcriptional modulation is mediated by small non-coding NAs was first evidenced with the advent of let-4 in C.elegans. Till date mi Base is populated with more than 20 thousand entries, of which more than 7000 accounts for plant stress associated mi NAs. In this study, we aimed to gain an insight to characterize mi NAs expressed under high temperature using next generation sequencing. To realize this objective, we constructed NA libraries and high-throughput sequence data revealed the presence of 125 mi NAs belonging to 45 families. Most of the mi NAs that were obtained in our library had preference for the 5′-U and has been reported in other plants, which is in accordance with the defined structures of the mature mi NAs.
Target prediction and GO analysis: To elucidate the biological functions of high temperature stress associated mi NAs were assessed using the ps NATarget software. A total of 750 potential target genes were identified, based on their perfect or nearperfect complementarity to their target m NA sequences. Most of the predicted targets were extensively involved in different biological process involving a large number of gene families. Some of these encoded transcription factors, DNA replication proteins, and those involved in cellular metabolism in addition to wide range of stress response associated proteins. Detailed annotations of these results are presented in (Supplementary File 4).mi 156 and mi 157 targets SPL protein and mi 166 targets Homeobox-leucine zipper family protein respectively.
The GO terms of the targets were annotated according to their biological function and highest percentage of genes falls in to cellular components. The enzyme mapping of the annotated sequences was performed using direct GO for the enzyme mapping and the Kyoto Encyclopedia of Genes and Genomes (KEGG) for the definitions of the KEGG orthologs. The mi NA targeted genes belonged to various biological process, cellular components and molecular functions as depicted in (Fig 3). The maximum numbers of target genes were involved in biological process, including both metabolic and cellular process. However, target genes in binding were the most abundant (80%) within the molecular function category. We have also found many genes representing transporters, receptors and signalling molecules including kinases. The compartmentalization of target genes revealed most of them are membrane proteins and localized in nucleus and cytoplasm.
The putative mi NA targets in Niger were predicted using the ps NATarget program. The target genes (approximately 750 different transcripts) were extensively involved in different biological processes involving a large number of gene families. Some of these genes encoded transcription factors, DNA replication proteins and those that are involved in cellular metabolism in addition to a variety of stress response-associated proteins. mi 156, mi 166 and mi 319 target genes encode Squamosa promoterbinding protein, Homeobox-leucine zipper protein and MYB domain proteins respectively, as previously reported (Ferdous et al., 2015). SPL genes forms one of the most targeted gene and we found 09 SPL genes belonging to Clade-I as major targets from the family. SPLI proteins constitute diverse family of transcription factors which are crucial in plant growth and development. Many studies established the role of SPL proteins in transition of juvenile to adult phase, reproductive transition, trichome development, apical dominance, inflorescence branching, fruit ripening, pollen sac development, and copper homeostasis (Unte et al. , 2003; Manning et al. , 2006; Wu and Poethig, 2006; Schwarz et al. , 2008; Wang et al. , 2009; Yamaguchi et al. , 2009; Yamasaki et al. , 2009; Jiao et al. , 2010; Miura et al. , 2010; Preston and Hileman, 2010; Yu et al. , 2010).
Previous studies established the involvement of mi 156, mi 157 and mi 398 in plant stress response mechanism, specifically in maintaining redox equilibrium under heat stress and copper homeostasis (Lu et al., 2012; Preston and Hileman 2013). They demonstrated that, the repression of mi 398b and mi 398c in Arabidopsis would result in loss of degradation of CSD1 and CSD2, which would support channeling of limited copper to support photosynthetic activity under high temperature induced oxidative stress and copper deficiency. They also showed that the repression of mi 156 and mi 157 would lead to activation of SPLII genes involved in copper independent photosynthetic pathways. The other major transcription factor targeted by conserved mi NAs is found to be MYB domain proteins. MYB is one among the vital proteins in plants which involved in development, metabolism, hormone signal transduction, disease resistance and environmental cues (Pashkovskiy and yazansky 2013). AtMYB44 was demonstrated to act at front line of salt stress induced cellular re-programming and suggested AtMYB44 prevent stress triggered tissue collapse by rapid removal of OS (Persak and Pitzschke 2014). The most widely targeted class of genes is encoding Toll/Interleukin-1 receptor-nucleotide binding site-leucine-rich repeats (TI -NBS-L ). Members of the TI -NBS-L gene family are established as genuine targets for mi 2118. Nucleotide Binding Site -Leucine ich epeats (NBS-L ) gene products confers disease resistant in plants and found to harbor secondary small NAs (tasi NAs). The mi 482 mediated gene cleavage would activate a cascade of defense reactions upon pathogen attack. In cotton, it is demonstrated that, the expression of NBS-L was induced to promote the generation of tasi NAs which mediate small NA mediated gene silencing, a probable mechanism of plant defense against viral and fungal attack. mi 482 and mi 2118 were clustered in Medicago trancatula and targets NBS-L disease resistant gene (Jagadeeswaran et al. 2009).
We also observed 10 ESTs coding G F proteins targeted by mi 396. mi 396, which targets growth regulating factors (G Fs) are an important class of transcriptional regulators involved in the control of cell proliferation in leaves (Khraiwesh et al. 2010). T-qPC profile showed strong up-regulation of mi 396 (1 fold) which appears to play a negative role in cell proliferation due to reduced cell division and it is reasonable to hypothesize that, as in Arabidopsis, mi 396 restricts G F expression, contributing to proliferation arrest in expanding cells ( odruiguez et al. 2010). ecent studies imply an intriguing regulatory network between mi 164, mi 396, and mi 319 in Arabidopsis: mi 319 regulates five members of the TCP transcription factor family (TCP2, 3, 4, 10, 24) that inhibits cell proliferation and directly bind the mi 164 promoter region (Palatnik et al 2007; Martin-Trillo and Cubas 2010).
Expression analysis and validation of high temperature responsive conserved miRNAs:
Total NA from the tissues of control and stressed Niger plants were used to validate the mi NAs. The total NA was converted to cDNA using stem – loop primers. The expression of 10 conserved mi NAs were considered for the study based on their read count/abundancy using q T-PC . The expression levels of the Niger mi NAs under high temperature were significantly altered compared with those of the control seedlings. Based on the sequencing data, we selected 10 mi NAs, which were also reported to as stress responsive mi NAs in other species. Of the 10 mi NA we found only 3 of them were found up-regulated. mi 395 was found to be highly responsive and was induced by 12 folds when compared with the control seedlings. mi 319 and mi 396 were up-regulated by 2folds. Among the down regulated mi NAs, 8-fold depression was observed with mi 156, 4-fold repression was found with mi 169 and mi 398, and an average of 2-fold repression was observed with other mi NAs (Fig 4). To validate the expression of targets the expression analysis was carried out with selected conserved targets of mi NAs. Since mi NAs were conserved across the kingdom, the genes targeted is also conserved with few exceptions. Since Niger lack complete genome data, we selected conserved targets, for the analysis. The expression profile substantiated the previous observations of negative correlation with their respective mi NAs. ATP sulpharylase was highly repressed by 14 folds, followed by DCL1 and G F3. DUO1 and SPL7 were induced by 11 and 9 folds. However, the target genes exhibited marginal changes in their expression. This may be due to the involvement of transcription regulatory factors other than mi NAs whose expression may not alter due to stress induction (Fig 5).
In the present study, T-qPC was carried out to study the expression of randomly selected conserved mi NAs representing the most stress responsive mi NA families. All the mi NAs showed sensitivity towards high temperature and our results evidences the mi NA abundance and their expression trends which is consistent with the previous results. We also observed induction of mi 395, mi 166/167 by 2-fold repression of mi 156, mi 171 which discern the effects of high temperature on Niger. Many mi NAs were temperature sensitive, for instance, mi 160, mi 166, mi 167 and mi 393 were up-regulated in barley and wheat upon heat treatment. Differential expression trends of mi 156, mi 159, mi 396 and mi 398 were also observed in Arabidopsis (Jagadeeshwaran et al., 2009) and Broccoli (Tian et al., 2014). mi 398 was the most extensively studied mi NA with respect to heat stress. It is demonstrated that the repression of the mi 398 under high temperature could render plant tolerant to heat induced oxidative damages (Naya et al., 2014, Lu et al., 2012 and Yu et al., 2012).
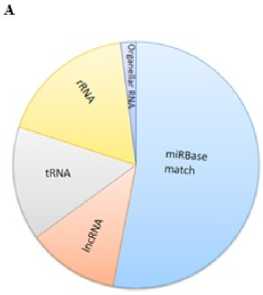
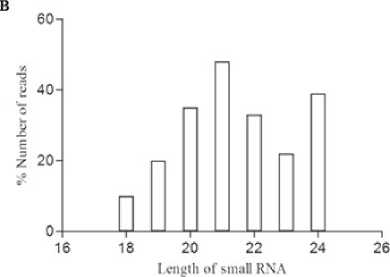
Figure 1. Data filtering and Length distribution. A . Pie plot of data filtering B . Sequence read length distribution of mappable high temperature stress responsive small NAs (s NAs).
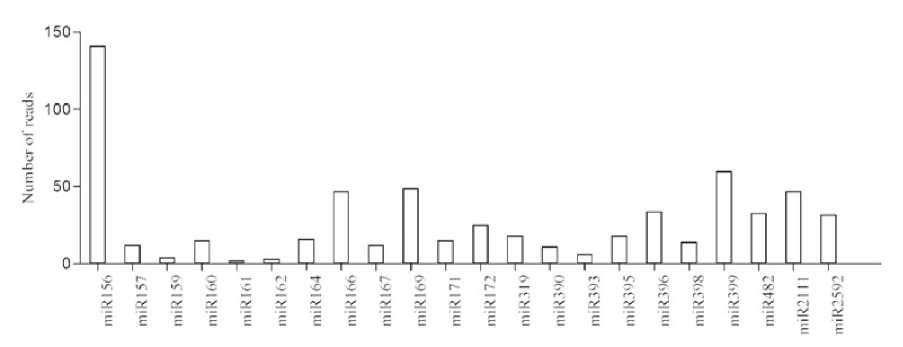
Figure 2. Expression levels of known mi NA families. The expression levels of the mi NA families were normalized by the total number of reads in each of the respective libraries.
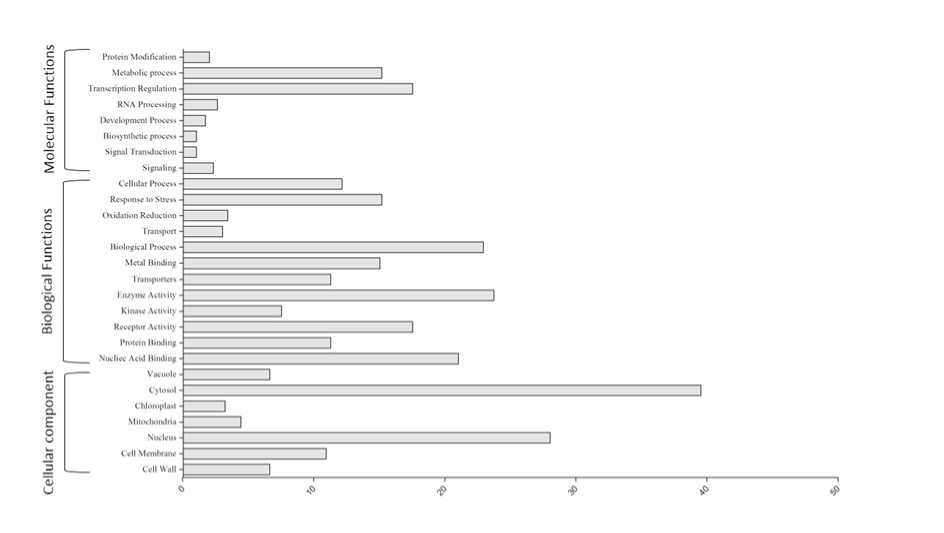
Figure 3. Gene ontology of differentially expressed mi NAtargets
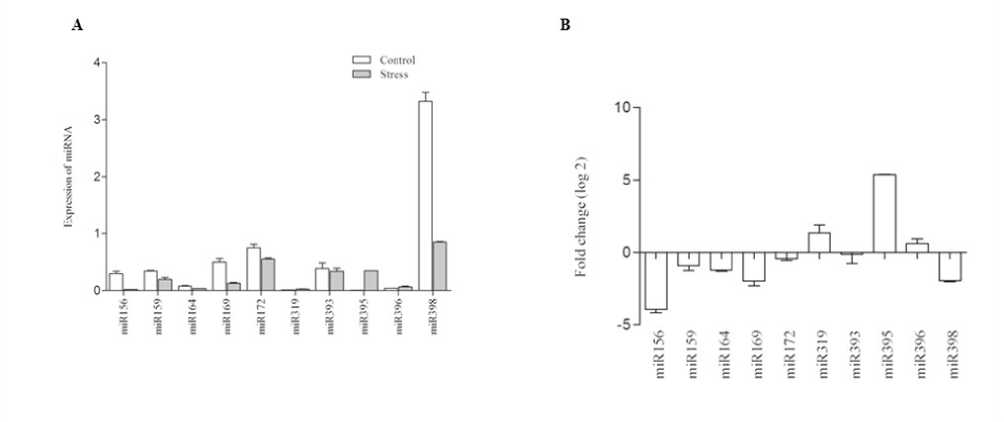
Figure 4. A. Quantification of mi NA abundance via T-qPC Differential expression of 10 mi NAs following high temperature stress. B. Fold changes (log2) in known mi NA levels in response to high temperature stress.
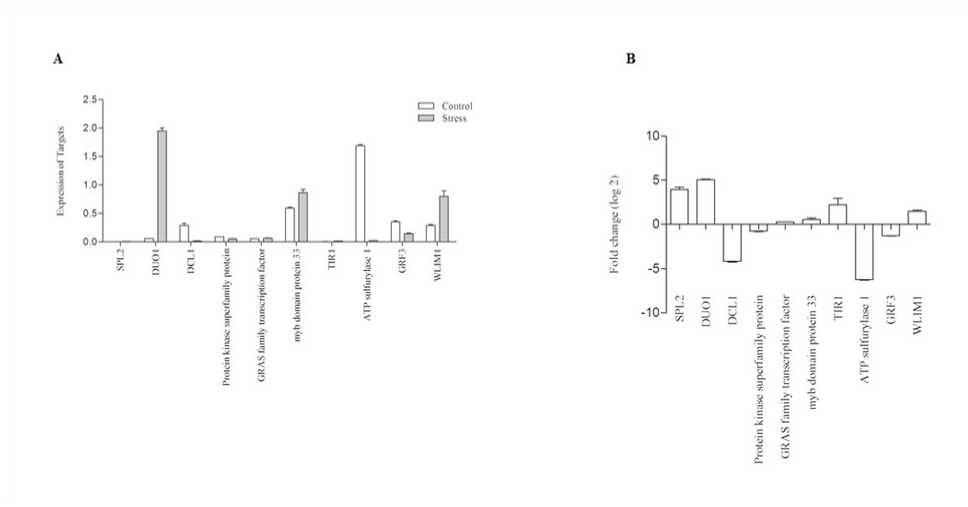
Figure 5. A. Determination of mi NA target gene expression via T-qPC analysis of NigerwholeControl and heat stressed seedlings B. Fold changes (log2) 10 selected mi NAs and their targets determined by T-qPC
CONCLUSION
Collectively, this is an integrated analysis of mi NA-mediated gene expression under high temperature by using the high-throughput sequencing. qPC experiment was used to confirm the expression of these mi NAs. Furthermore, the GO annotation and pathway analysis for predicted targets have implicated the putative roles of the abundant mi NAs. Notably, this investigation insights a useful resource for further characterization of regulatory functions of mi NAs to design genetically modifying plants.
ACKNOWLEDGMENTS
The authors would like to thank DST-FIST, UGC-CPE and DBT Star College Scheme for Instrumentation facility and consumables.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.
Список литературы Exploring high temperature responsive novel non coding RNAS and functional annotations from niger (Guizotia abyssinica)
- Arenas-Huertero, C., Pérez, B., Rabanal, F., Blanco-Melo, D., De la Rosa, C., Estrada-Navarrete, G., and Reyes, J. L. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant molecular biology, 70(4), 385-401.
- Barozai, M. Y. K., Irfan, M., Yousaf, R., Ali, I., Qaisar, U., Maqbool, A., and Riazuddin, S. (2008). Identification of micro-RNAs in cotton. Plant Physiology and Biochemistry, 46(8-9), 739-751.
- Barrera-Figueroa, B. E., Gao, L., Diop, N. N., Wu, Z., Ehlers, J. D., Roberts, P. A., and Liu, R. (2011). Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC plant biology, 11(1), 127.1-17.
- Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell, 136(2), 215-233.
- Bita, C., and Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stresstolerant crops. Frontiers in plant science, 4, 273.
- Ding, D., Zhang, L., Wang, H., Liu, Z., Zhang, Z., and Zheng, Y. (2009). Differential expression of miRNAs in response to salt stress in maize roots. Annals of botany, 103(1), 29-38.
- Dugas, D. V., & Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Current opinion in plant biology, 7(5), 512-520. and Reyes, J. L. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in
- Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., and microRNAs targeting genes involved in fruit ripening. Genome research, 18(10), 1602-1609.
- Carrington, J. C. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PloS one, 2(2), e219.
- Fujii, H., Chiou, T. J., Lin, S. I., Aung, K., and Zhu, J. K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology, 15(22), 2038-2043.
- Griffiths-Jones, S., Grocock, R. J., Van Dongen, S., Bateman, A., and Enright, A. J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research, 34 (suppl_1), D140-D144.
- Jia, X., Wang, W. X., Ren, L., Chen, Q. J., Mendu, V., Willcut, B., and Tang, G. (2009). Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant molecular biology, 71(1-2), 51-59.
- Naik, H. K., & Varadahalli, R. D. (2020). Genomic identification of salt induced microRNAs in niger (Guizotia abyssinica Cass.). Plant gene, 23, 100242.
- Vierling, E., and Scharf, K. D. (2007). Complexity of the heat stress response in plants. Current opinion in plant biology, 10(3), 310-316.
- Microarray analysis of transcriptional responses to abscisic acid and salt stress in Arabidopsis thaliana. International Journal of Molecular Sciences, 14(5), 9979-9998.
- Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., and Griffiths-Jones, S. (2008). Criteria for annotation of plant MicroRNAs. The Plant Cell, 20(12), 3186-3190.
- Moxon, S., Jing, R., Szittya, G., Schwach, F., Pilcher, R. L. R., Moulton, V., and Dalmay, T. (2008). Deep sequencing of tomato short RNAs identifies
- Nageshbabu, R., Jyothi, M. N., and Sharadamma, N. (2013). Expression of miRNAs regulates growth and development of French bean (Phaseolus vulgaris) under salt and drought stress conditions. ISCA Journal of Biological Sciences, 2(1), 52-56.
- Nageshbabu. R., Jyothi, M.N., Usha, Sharadamma.N,Rai D.V and Devaraj V. R. (2014) Identification of miRNAs from French bean (Phaseolus vulgaris) under low nitrate stress. Turk J Biochem, 39 (1),1-8.
- Naya, L., Paul, S., Valdés-López, O., Mendoza-Soto, A. B., Nova-Franco, B., Sosa-Valencia, G., and Hernández, G. (2014). Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS One, 9(1), e84416.
- Pashkovskiy, P. P., and Ryazansky, S. S. (2013). Biogenesis, evolution, and functions of plant microRNAs. Biochemistry (Moscow), 78(6), 627637.
- Shen, J., Xie, K., and Xiong, L. (2010). Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Molecular Genetics and Genomics, 284(6), 477-488.
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. Journal of experimental botany, 58(2), 221-227.
- Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in plant science, 12(7), 301-309.
- Sunkar, R., Kapoor, A., and Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. The Plant Cell, 18(8), 2051-2065.
- Tagami, Y., Inaba, N., Kutsuna, N., Kurihara, Y., and Watanabe, Y. (2007). Specific enrichment of miRNAs in Arabidopsis thaliana infected with Tobacco mosaic virus. DNA research, 14(5), 227233.
- Trindade, I., Capitäo, C., Dalmay, T., Fevereiro, M. P., & Dos Santos, D. M. (2010). miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta, 231(3), 705-716.
- Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., and Schein, J. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313(5793), 1596-1604.
- Valdés L., O. ., Huertero, C. A., ., Ramirez, M., Girard, L., Sanchez, F., Vance, C. P., and Hernandez, G. (2008). Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus deficiency signalling in common bean roots. Plant, Cell & Environment, 31(12), 1834-1843.
- Valdés L, O., Yang, S. S., Aparicio F. R., Graham, P. H., Reyes, J. L., Vance, C. P., and Hernández, G. (2010). MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytologist, 187(3), 805-818.
- Wang, Q. L., and Li, Z. H. (2007). The functions of microRNAs in plants. Front Biosci, 12, 3975-3982.
- Xin, M., Wang, Y., Yao, Y., Xie, C., Peng, H., Ni, Z., and Sun, Q. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC plant biology, 10(1), 1-11.
- Yu, X., Wang, H., Lu, Y., de Ruiter, M., Cariaso, M., Prins, M., and He, Y. (2012). Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. Journal of Experimental Botany, 63(2), 1025-1038.
- Zhang, N., Yang, J., Wang, Z., Wen, Y., Wang, J., He, W., and Wang, D. (2014). Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PloS one, 9(4), e95489.
- Zhang, Y. (2005). miRU: an automated plant miRNA target prediction server. Nucleic acids research, 33(suppl_2), W701-W704.
- Zhou, Z. S., Wang, S. J., and Yang, Z. M. (2008). Biological detection and analysis of mercury toxicity to alfalfa (Medicago sativa) plants. Chemosphere, 70(8), 1500-1509.
- Zhou, Z. S., Zeng, H. Q., Liu, Z. P., and Yang, Z. M. (2012). Genome wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant, cell & environment, 35(1), 86-99.
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research, 31(13), 3406-3415.


