Expression of miRNAs confers enhanced tolerance to drought and salt stress in finger millet ( Eleusine coracona)
Автор: Nageshbabu R., Usha , Jyothi M.N., Sharadamma N., Rai D.V., Devaraj V.R.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 3 т.9, 2013 года.
Бесплатный доступ
Plants respond to the environmental cues in various ways, recent knowledge of RNA interference in conferring stress tolerance had become a new hope of developing tolerant varieties. Here we attempt to unfold the molecular mechanism of stress tolerance through miRNA profiling and expression analysis in Finger millet (Eleusine coracona) under salt and drought stress conditions. The expression analysis of 12 stress specific conserved miRNAs was studied using semi-quantitative real time PCR and Northern blot assay. Our studies revealed that, although most of the miRNAs responded to the stresses, the expression of particular miRNA differed with the nature of stress and the tissue. The expression analysis was correlated with the existing data of their target genes. Abiotic stress up-regulated miRNAs are expected to target negative regulators of stress responses or positive regulators of processes that are inhibited by stresses. On the other hand, stress down-regulated miRNAs may repress the expression of positive regulators and/or stress up-regulated genes. Thus the current study of miRNAs and their targets under abiotic stress conditions displays miRNAs may be good candidates to attribute the stress tolerance in plants by transgenic technology.
Drought, finger millet, mirna, salt stress
Короткий адрес: https://sciup.org/14323767
IDR: 14323767
Текст научной статьи Expression of miRNAs confers enhanced tolerance to drought and salt stress in finger millet ( Eleusine coracona)
MicroRNAs (miRNAs) are an important class of endogenous small silencing RNAs in regulating the expression of a wide range of target genes that are involved in many important biological processes in both plant and animals. These small (generally 2122 nt) RNA molecules, originated from primary “hairpin” transcripts, can induce translational suppression or direct mRNA cleavage. Similar to regular mRNAs, the expression of miRNAs is highly regulated. Their expression pattern could provide critical clues in understanding miRNA functions. However, a detailed expression pattern for specific miRNA is difficult to obtain because of their short length, presence of similar miRNA families and intermediate precursor sequences. Many miRNAs show unique tissue-specific, development-related, and stress-induced expression. The miRNAs that are found to be regulated in response to a given abiotic stress could be of three types (1) known miRNAs that are known to be regulated by the stress, (2) known miRNAs that are newly discovered and to be regulated by the stress and (3) new miRNAs that are regulated by the stress. MicroRNAs of the first type are good positive controls and also could reveal unknown aspects of the regulation, such as tissue or organ specificity. Many of the miRNAs discovered till date have confirmed their involvement in regulatory networks associated with plant stress response.Of which 12 were found to be involved in salt and drought stress. These key players include miR156, miR157, miR159, miR166, miR167, miR169, miR171, miR172, miR393, miR395, miR396, miR398 families (Xie et al., 2010; Matts et al., 2010). Finger millet (Eleusine coracona) is the primary food source for millions of people in tropical regions. Till date many of the investigators focused on biochemical and physiological responses under environmental constrains. However, no work has been carried out in unwinding the role of miRNAs from the millet in regulating abiotic stress responses. Here we attempted to divulge these stress induced epigenetic regulators involved in restoring the cellular homeostasis.
MATERIALS AND METHODS
Plant material and stress treatments
Finger millet (Eleusine coracona L MR-1 .) seedlings were grown in a green house at 28 ◦C; 13 h light until 8 day old and were then randomly divided into three groups. Two groups were grown in half-strength modified Hoagland nutrient solution containing two salt concentrations: 200mM NaCl and 400mM NaCl for 48 h and the other group was used for drought (with-holding water for 3 weeks) stresses respectively. Separate control lines were maintained for each stress. The shoots and roots were harvested separately from both control and stress samples frozen immediately in liquid nitrogen until RNA isolation.
RNA isolation and real time PCR analysis
Total RNA was isolated from different tissues of control and stressed seedlings using miRNA Isolation kit (Ambion) according to manufacturer’s instructions. The quality of miRNA was analysed by 15% Urea PAGE, using 14, 24, and 39 nt oligos as size standards.Twelve miRNAs were selected for this study, which included miR156, miR157, miR159, miR162, miR167, miR169, miR172, miR395, miR396, miR397, miR398, and miR399. A majority of these miRNAs have been reported to play a role under stress conditions in model plant species. Semi quantitative real time -PCR was used to characterize the expression of 12 miRNAs in both shoot and root under salt and drought stress conditions. First, the RT reaction was carried out using TaqMan micro RNA reverse transcription kit (Applied Biosystems) the obtained cDNA was used for PCR amplification using family specific miRNA primers. The primers used in these amplification reactions were listed in supplementary table 1.
RNA gel blot analysis
For miRNAs quantification, northern blot hybridization was conducted using high sensitive miRNA Northern blot assay kit (Signosis, USA). 30 µg total RNA of each sample was electrophoresed on 15% polyacrylamide gel and transferred to membrane. Biotin-labeled Antisense RNA (Invitrogen) was used as hybridization probes and U6 RNA as a loading control. Supplementary table-2 lists the biotin labeled probes used for hybridization reaction.
RESULTS
Salt stress altered the expression pattern of miRNAs in Finger millet
To get an insight into possible tissue-dependent roles of miRNAs in Finger millet, the expression patterns of miRNAs in different tissues including root and shoot were examined using semi quantitative real time PCR and Northern blot assay. Among the 12 miRNAs which have been considered for the study, only eight miRNAs revealed different expression patterns with respect to the tissue and nature of stress. The miRNAs responded varied between root and shoot. The expression of miR159 was up-regulated by two - fold in shoots, where as it remain unexpressed in roots. Similarly the down regulation of miR399 was identified only in shoots. Our results revealed that miRNAs responded differentially with the concentration of salt. Although the effect of salt concentration over miRNA expression was negligible with 0.1 to 0.5 fold altered expression in roots (Figure 1A), there was two-three fold variation in case of shoots. Most of the miRNAs were down regulated in shoots except miR156 and miR159. Two fold up-regulation of miR156 was observed, which was dose dependent at the same time miR159 showed 1.5
fold down -regulation at 400mM salt concentration compared to 200mM. The repression of miR157, miR167, miR169, miR398 and miR399 was also found to be dose dependent with 0.5- 1.0 fold down-regulation with increasing concentration. But no expression of miR398 was found at 200mM salt (Figure 1B). The Northern blot assay was performed with these differentially expressed miRNAs. The results confirmed the response of miRNAs towards the stress was tissue specific (Figure 2).
Response of known miRNAs to drought stress in Finger millet
Although Finger millet is a drought tolerant species, the prolonged exposure of seedlings to the stress at the early vegetative stage is found to affect the crop yield significantly. The underlying mechanism of drought tolerance may be attributed to the altered expression of miRNAs. Expression studies of these epigenetic regulators with respect to drought stress were performed by semi-quantitative RT-PCR and validated by Northern blot assay. Among the 12 miRNAs considered for study, 8 miRNAs responded to drought stress. miR171 and miR172 which were unnoticed in salt stress were found to express under water deficient conditions and remain to be stress specific. In roots, miR156 was up-regulated by 0.5 fold and miR171 by 0.3 fold. miR482 was expressed only in drought stressed roots as a secondary miRNA expressed in response to up-regulation of miR171. However, miR166, miR167 were down regulated by 10 fold and miR395 by 9 folds respectively (Figure 3A). In shoots, the response of miRNAs was quiet different. There was no expression of miR156, miR395 and miR171, where as the expression was found with miR397 and miR398 which were down regulated by 10 fold. The miRNAs which were up-regulated include miR159, miR169, miR172, and miR396 and their fold change was in the range of 1.5 to 3.0. miR159 had shown 3.0 fold up-regulation while miR396 was up-regulated by 0.5 fold (Figure 3B). miR396 had shown 0.5 fold in both the tissues and was not either tissue specific or stress specific. The Northern blot assay was carried out with differentially expressed miRNAs which established the tissue specific expression of miRNAs (Figure 4).
Majority of the targets shared by the abiotic stress specific miRNAs were transcription factors such as MYB-Protein, AHAP2, NF-Yα 2, CCAAT binding protein and other zinc finger proteins involved in transcription machinery. There were also target genes encoding proteins involved metabolic pathways such as phosphatidate cytidyltransferase, protein kinases, membrane bound channels, protein conjugating systems and membrane transporters. A description of the target genes were listed in supplementary (Table 3).
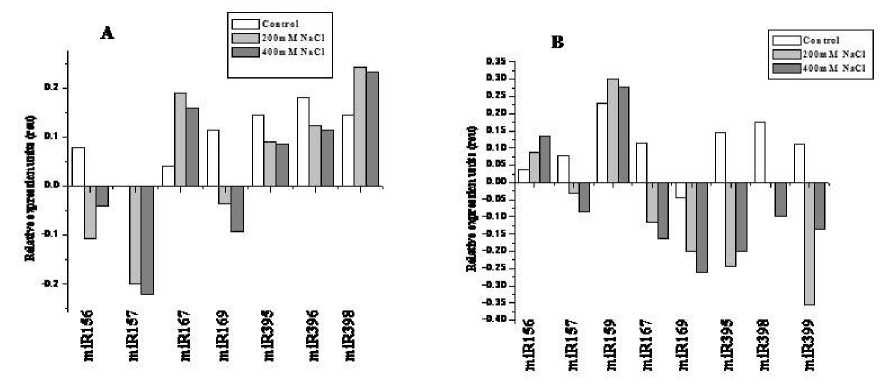
Figure 1. Relative expression levels of conserved miRNAs under salt stressed (root and shoot) Finger millet seedlings.
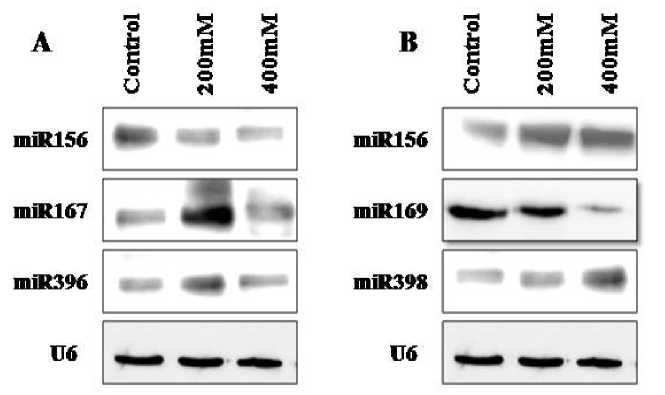
Figure 2. Northern blotting confirming differential expression of miRNAs. Total RNA 30µg from each of three conditions (control and salt) was loaded and hybridized with family specific miRNAs probe. U6 was used as control.
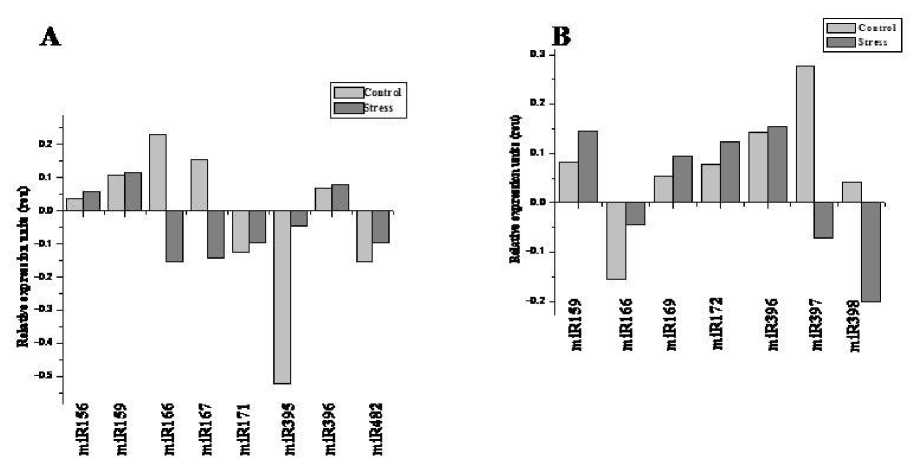
Figure 3. Northern blotting confirming differential expression of miRNAs. Total RNA 30µg from each of three conditions (control and salt) was loaded and hybridized with family specific miRNAs probe. U6 was used as control.
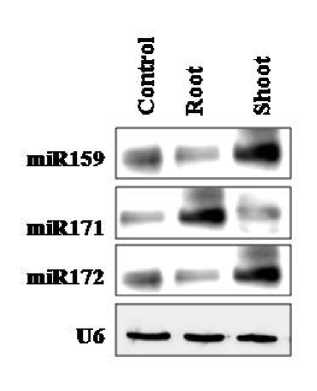
Figure 4. Northern blotting confirming differential expression of miRNAs. Total RNA 30µg from each of three conditions (control and drought) was loaded and hybridized with family specific miRNAs probe. U6 was used as control.
DISCUSSION
Although the miRNAs of several plant species have been studied recently, neither miRNA identification nor an analysis of differential miRNA expression in response to salt and drought stress has been performed in Finger millet. One reason why Finger millet has gained attention as a dedicated food crop is that it can grow on sub-optimal land under salt and drought conditions. Our results indicated that salt stress had a significant effect on the germination rate and growth of Finger millet in almost all tested abiotic stresses. Interestingly, drought stress had no obvious effect on the germination rate; the significant effect of drought stress on Finger millet growth was observed at prolonged water deficient conditions at the early vegetative state. This result suggests that Finger millet has evolved a more effective mechanism to cope with drought stress as opposed to salt stress. Therefore, it is interesting to further investigate the change in gene expression, especially the gene expression regulators, under such stress conditions.
Micro RNAs are an extensive class of newly discovered gene regulators. They have been reported to play important roles under abiotic stress in model plant species. Using qRT-PCR, we studied the expression change of 12 conserved miRNAs in 8 days old Finger millet seedlings exposed to salt and drought stress. Of the 12 miRNAs, 11 have been demonstrated to be involved in salt or drought stress in previous study, eight of these in both the Arabidopsis thaliana and rice . Our results indicate that both salt and drought stresses altered the expression pattern of miRNAs in a dosedependent manner. The expression of an abiotic stress mediated miRNA depends upon the nature of the target gene. MicroRNA targeting the positive regulators of gene encoding proteins that are involved in processes inhibited by stress responses and negative regulators of stress responses were up-regulated and those targeting the positive regulators of stress response and stress up-regulated genes were repressed.
miR156 known to play an important role in multiple biological processes by targeting Squamosal Promoter binding protein-Like (SPL) genes, miR156 has been demonstrated to temporally regulate shoot development (Park et al., 2011), control the development timing from juvenile to adult transition together with miR172 (Xing et al., 2009; Gou et al., 2011). Overexpression of miR156 in Arabidopsis, rice, and maize led to a prolonged vegetative phase together with the production of significantly higher number of total leaves, which resulted in enhanced biomass accumulation (Liu et al., 2008; Zhang et al., 2009), miR156 was demonstrated by microarray-based analysis to response to salt stress but not to drought stress in Arabidopsis; miR156 was induced by 1.6-fold by salinity stress (Zhang et al., 2009). In rice, miR156 was found to respond to drought stress and was down-regulated by 2.1-fold by drought stress (Zhao et al., 2009). Studies have been conducted on maize under salt stress conditions and showed miR156 was not responded (Ding et al., 2011). However, our results demonstrate that expression of miR156 was up-regulated in roots of drought stressed seedlings and shoots of salt stressed samples in contrast to the results found with Arabidopsis, rice and maize. The expression of miR169 differs with species and the nature of stress applied. In Arabidopsis, the miR169 expression is down regulated resulting in the accumulation of NF-YA5 transcription factor which is crucial for the expression of several DRE (dehydration- responsive elements), while in rice, the expression is favored by cold and drought stress. Medicago truncatula showed no expression in miR169 towards drought stress (Wang et al., 2005; Zhao et al., 2010). Reactive oxygen species (ROS) were found to accumulate under drought and salt stress as the oxidative stress occurs concurrently. The accumulated ROS results in membrane damage and lipid peroxidation. The expression of miR398 is down regulated during these induced stresses which lead to increased activities of superoxide dismutase the target genes of miR398. miR171 targets the gene of another family of GRAS transcription factors which is found to be involved in nodule morphogenesis and floral development (Wang et al., 2011). Targets of miR482 were primarily Resistance gene receptor kinases with two out of three targets are defense related kinases and induced bacterial infections. These miRNAs were reported to target NBR-LRR gene their over expression leads to nodulation with increasing lateral root mass (Hue et al., 2010; Eckardt et al., 2012). The expression of different members of the MIR172 family and, consequently, their corresponding mature miRNAs, depends on growth stages and tissue types. In addition, the expression level of miR172 is also affected by day length and temperature (Kwon et al., 2011; Li et al., 2011). In both dicot and monocots, the expression level of miR172 increases as plants grow and after flowering it accumulates in leaves and floral buds (Aukerman and Sakai, 2003; Chuck et al., 2007; Zhu et al., 2009) miR172 sequence that regulates the mRNA abundance and/or translation of the plant specific transcription factor gene APETALA2 (AP2) (Park et al., 2002; Chen, 2004) and a small group of AP2-like genes in Arabidopsis (Aukerman and Sakai, 2003), which contain sequences complementary to miR172 AP2 is a floral organ identity gene while the five AP2-like genes mainly act as flowering repressor (Poethig et al., 2009).
In summary, our results provide insight into the Finger millet miRNAs, highlighting the regulatory network triggered by salt and drought stress. Furthermore, there exists a cross talk between salt and drought stress induced miRNA expression. The plant exhibits varied mechanisms to alter the miRNA expression tissue specifically thereby enhancing the tolerance towards stress.
ACKNOWLEDGEMENT
This work was supported from Department of Science and Technology, Government of India, New Delhi (Ref No; SR/FT/LS-10/2012).
Список литературы Expression of miRNAs confers enhanced tolerance to drought and salt stress in finger millet ( Eleusine coracona)
- Aukerman M.J, Sakai H. (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730-2741.
- Chuck G, Cigan A.M, Saeteurn K, Hake S. (2007) The heterochronic maize mutant Corngrass1 results from over expression of tandem microRNA. Nat Genet 39: 544-549.
- Ding D, Zhang L, Wang H, Liu Z, Zhang Z. (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot 103: 29-38.
- Ding Y, Chen Z, and Zhu C. (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62: 3563-3573.
- Eckardt A. (2012) A MicroRNA Cascade in Plant Defense. Plant cell 24: 840.
- Gou J.Y, Felippes F.F, Liu C.J, Weigel D, Wang J.W. (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512-1522.
- Hui Li, Ying Deng, Tianlong Wu, Senthil Subramanian, Oliver Yu. (2010) Mis-expression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiology 153(4) 1759-1770.
- Kwon H.B, Hwang E.W, Shin S.J, Park S.C, Jeong M.J. (2011) Identification of miR172 family members and their putative targets responding to drought stress in Solanum tuberosum. Genes & Genomics 33: 105-110.
- Li B, Qin Y, Duan H, Yin W, Xia X. (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 62(11): 3765-3779.
- Liu H.H, Tian X, Li Y.J, Wu C.A, Zheng C.C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA-a Publication of the RNA Society 14: 836-843.
- Matts J, Jagadeeswaran G, Roe B.A, Sunkar R. (2010) Identification of microRNAs and their targets in switch grass, a model biofuel plant species. Journal of Plant Physiology 167: 896-904.
- Park C.M, Jung J.H, Seo P.J, Kang S.K. (2011) miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Molecular Biology 76: 35-45.
- Poethig R.S, Wu G, Park M.Y, Conway S.R, Wang J.W. (2009) The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell. 138: 750-759.
- Sunkar R. (2010) MicroRNAs with macro-effects on plant stress responses. Seminars in Cell & Developmental Biology 21: 805-811.
- Tianzuo Wang, Lei Chen, Mingui Zhao, Qiuying Tian and Wen-Hao Zhang. (2011) Identification of drought responsive microRNAs in Medicago truncatula by genome-wide high-throughput Sequencing. BMC Genomics 12: 367-378.
- Wang J.W, Wang L.J, Mao Y.B, Cai W.J, Xue H.W, Chen X.Y. (2005) Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell, 17(8): 2204-2216.
- Wang J.W, Czech B, Weigel D. (2009) miR156 regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 138: 738-749.
- Xie F, Frazier T.P, Zhang B. (2010). Identification and characterization of microRNAs and their targets in the bio-energy plant switch grass (Panicum virgatum). Planta 232: 417-434.
- Xing S, Salinas M, Hohmann S, Berndtgen R, Huijser P. (2010) miR156 targeted and non-targeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell, 22: 3935-3950.
- Zhang B, Pan X. (2009). Expression of microRNAs in cotton. Mol Biotechnol, 42: 269-274.
- Zhao B, Ge L, Liang R, Li W, Ruan K, (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10: 29-38.
- Zhou L, Liu Y, Liu Z, Kong D, Duan M. (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61: 4157-4168.
- Zhu J.K, Li W.X, Oono Y, Zhu J.H, He X.J. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionaly and post-transcriptionaly to promote drought resistance. Plant Cell 20: 2238-2251.


