Finding Longest Common Substrings in Documents
Автор: M.I.Khalil, M.A.Hadi
Журнал: International Journal of Image, Graphics and Signal Processing(IJIGSP) @ijigsp
Статья в выпуске: 9 vol.7, 2015 года.
Бесплатный доступ
This paper introduces an algorithm to address the problem of finding the longest common substring between two documents. This problem is known as the longest common substring (LCS) problem. The proposed algorithm is based on the convolution between the two sequences (named major sequence (X) which is represented as array and the minor one (Y) which is represented as circular linked list. An array of linked lists is established and a new node is created for each match between two substrings. If two or more matches in different locations in string Y share the same location in string X, the corresponding nodes will construct a unique linked-list. Accordingly, by the end of processing, we obtain a group of linked-lists containing nodes arranged in certain manner representing all possible matches between sequences X and Y. The algorithm has been implemented and tested in C# language under Windows platform. The obtained results presented a very good speedups and indicated that impressive improvements had been achieved.
Longest Common Substring, Data structures and Algorithms, Documents, Document Similarity
Короткий адрес: https://sciup.org/15013906
IDR: 15013906
Текст научной статьи Finding Longest Common Substrings in Documents
Published Online August 2015 in MECS
There are many methods and algorithms to compare documents or simply to judge similarity between documents. Such algorithms are time and space consuming, for instant the brute force algorithm compares two documents in word by word manner. The classic textbook solution to the longest common substring (LCS) problem is to build the (generalized) suffix tree of the documents and find the node that corresponds to LCS [18]. While this can be achieved in linear time, it comes at the cost of using huge space to store the suffix tree. In applications with large amounts of data or strict space constraints, this renders the classic solution impractical.
Another algorithms compare the subject document with investigated documents by constructing metrics related to the documents such as punctuation, the number of statements, paragraphs [9].
LCS approaches aid the problem of searching for coding regions in DNA sequences yielding the common substring between DNA sequences [10, 15].
The problem of LCS can be defined mathematically as:
Given two documents (strings), S of length m and T of length n , find the longest strings which are substrings of both S and T .
A generalization is the k-common substring problem [16].
Given the set of strings = {S1S2,, ... ,Sk }, where |S, | = n , and £ n , = N. Find for each 2 < к < К, the longest strings which occur as substrings of at least strings.
The algorithm suggested in this paper aims to minimize both the time of processing and the size of allocated memory. The algorithm detects not only the longest common string but finds exactly all possible common substrings. It is based on the convolution between the two documents (named major sequence ( X ) which is represented as array and the minor one ( Y ) which is represented as circular linked list. An array ( Z ) of linked lists is established and a new node is created for each match between two substrings. The first string is kept fixed while the another one is rotated, using pointer, to left in one character step. If two or more matches in different locations in string Y share the same location in string X , the corresponding nodes will construct a unique linked-list. Accordingly, by the end of processing, we obtain a group of linked-lists containing nodes arranged in certain manner representing all possible matches between sequences X and Y . The array Z of linked lists is then traversed horizontally and vertically looking for the matches between string X and Y .
The rest of the paper is organized as follows. Section II discusses the suggested algorithm, and the implementation and experimental results are discussed in Section III. We conclude the paper in Section IV.
-
II. The Suggested Algorithm
The suggested algorithm compares two documents to find all possible identical matches. The data structure required for the suggested algorithm is shown in Fig.1. The major string is represented in array X while the second one (the minor string) is represented in circular linked-list Y . Each node in the circular linked list Y consists of two fields: the first field holds character value while the second one points to the next node in a circular manner. The initialization process (shown in Fig.2) reads the major document (the longer one) character by character in array X (each array cell holds one character value) and the minor document is read into the circular linked-list Y respectively. In addition, it establishes an empty linked-list array Z , where each cell of array Z is a separate linked-list. The data structures X , Y and Z are all of the same size as the major document (measured in characters). The major document refers to the longer one. If the major document is longer than the minor one, the rest of the circular linked-list Y will be filled with a not used special character (i.e. Null ).
The process of matching is described in the flowchart shown in Fig.3; where it begins by creating two counters: I for loop number and shift_no for the shifting number. Two nested loops will be used, one for left-shifting the sequence represented in the circular linked-list Y, and the inner loop for comparing every character in X with the corresponding one in Y . If there is a match between the the character stored in cell number n in X and the character stored in node number n in the shifted circular linked-list Y , then a new node will be added to the linked-list stored in cell number n in array Z . The new node consists of two fields; the first one contains the shift number ( shift_no counter) and the second one contains a pointer to the next node in the same linked-list (it must be null until adding new node).
The last process scans and inspects the contents of the linked-list array Z looking for the contiguous nodes which contains the same shift_no in its data fields. The result of this process is added to a list yielding the longest contiguous nodes sequence.
The following example illustrate the matching process of the suggested algorithm:
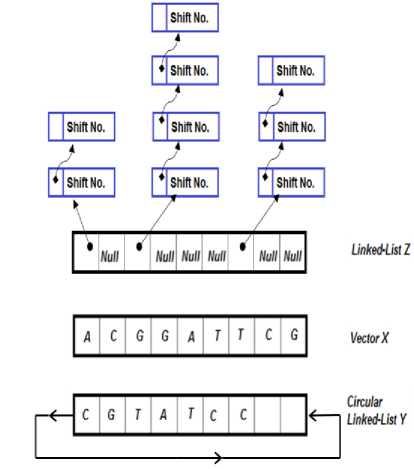
Fig.1. The data structures used in the suggested approach
Input: two strings
Major string = “ please let me learn better”
Minor string = “ release letter”
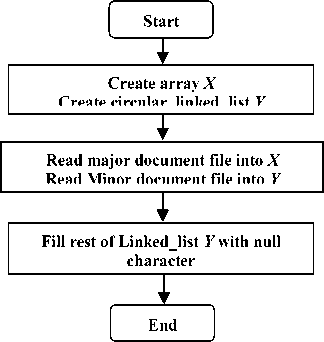
Fig.2. Flow chart of the initialization process
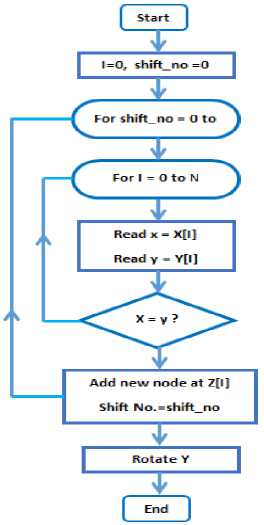
Fig.3. Flow chart of the matching process
As illustrated in Fig.5 B, the first two rows at top of the right table contain the two input strings X and Y respectively. The minor string is rotated to left one character at a time yielding to the successive strings shown in the rest of the same table. The convolution process is performed every time the minor string is rotated, the contents of the minor string is compared, character by character with the corresponding characters of the major string yielding to the corresponding row in the left table
(Fig.5 A) in the figure. If two characters are equal, a new node is added to the linked list related to this position in the major string. The new node contains two fields: the first one contains the shift_no and the other contains a pointer to the next node (null until adding the next node). The new node is represented by “ 1 ” in the left table. The two linked-lists shown in Fig.4 represent the added nodes in the second and third columns of the left table (Fig. 5 A) respectively and they are corresponding to characters “ l ” and “ e ” of the major string. Inspecting the rows contained in the left table, it is noticed that each row represents the result of comparing the major string with the minor one at certain rotation number. The “ ones ” in a row means that the corresponding characters in both strings are equal. The contiguous “ ones ” means that the corresponding characters is common substring in both strings. For example the ones in the row number “ 1 ” in the left table represents the substrings: “ lease let ”, “ ” and “ r ” respectively.


Fig.4. the added nodes in the second and third columns of the left table corresponding to characters “l” and “e” of the major string
|
Shift No. |
Result of comparing major string with the rotated minor string |
|||||||||||||||||||||||||
|
0 |
1 |
1 |
||||||||||||||||||||||||
|
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
||||||||||||||||
|
2 |
1 |
1 |
||||||||||||||||||||||||
|
3 |
1 |
1 |
1 |
|||||||||||||||||||||||
|
4 |
1 |
1 |
1 |
1 |
1 |
1 |
||||||||||||||||||||
|
5 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
6 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
7 |
1 |
1 |
1 |
1 |
1 |
|||||||||||||||||||||
|
8 |
1 |
1 |
1 |
1 |
1 |
1 |
||||||||||||||||||||
|
9 |
1 |
1 |
1 |
1 |
1 |
|||||||||||||||||||||
|
10 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
11 |
1 |
1 |
1 |
1 |
1 |
1 |
||||||||||||||||||||
|
12 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
13 |
1 |
1 |
1 |
|||||||||||||||||||||||
|
14 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
|||||||||||||||||
|
15 |
1 |
1 |
1 |
1 |
1 |
|||||||||||||||||||||
|
16 |
1 |
|||||||||||||||||||||||||
|
17 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
18 |
1 |
|||||||||||||||||||||||||
|
19 |
1 |
1 |
||||||||||||||||||||||||
|
20 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
21 |
1 |
1 |
||||||||||||||||||||||||
|
22 |
1 |
1 |
1 |
|||||||||||||||||||||||
|
23 |
1 |
1 |
1 |
1 |
||||||||||||||||||||||
|
24 |
1 |
1 |
1 |
|||||||||||||||||||||||
|
25 |
1 |
1 |
||||||||||||||||||||||||
A)
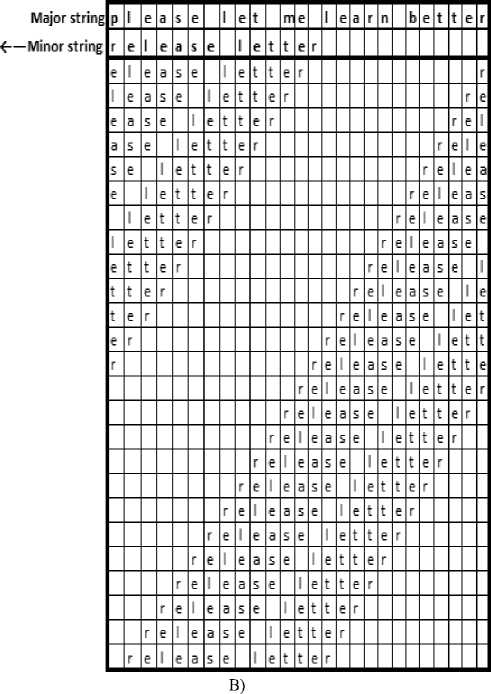
Fig.5. Illustration example to find matches between strings: ”please let me learn better” as major string and “release letter” as minor one
-
III. Implementation and Experimental Results
The suggested algorithm has been implemented using C# (Microsoft Visual Studio) language to perform comparison between two input long strings and display all possible common substrings in suitable manner. The program has been tested with different lengths strings and the time of processing is computed in each case. The snapshot in Fig.6 is taken after running the program for the example explained in Fig.5. The resulted common substrings between the two input strings are listed in the data view component as shown in the mentioned Fig. along with the locations of those substrings and their lengths in both input strings. The relation between the time of processing and its dependence on the ratio between the lengths of both minor and some major strings
(see Table-1) have been experimentally studied yielding to Table-2 which is presented and plotted in Fig.7. The major string was 1000 character and the minor string is varied gradually from 50 to 1000 characters length. It is noticed that this relation is almost linear. Moreover, the time of processing has been studied in more deep: the time of processing consists of two components; the first one is consumed in constructing the linked lists related to each position in the major string which contains the matches between the two strings, and the other component is consumed in traversing the these linked lists looking for continuous strings. It has been found that the first time component is trivial compared to the second one (Table-2). The relation between time of processing and its dependence on the length of the major string is considered and inspected yielding to Table-3 which is represented in Fig.8 respectively.
Table 1. LCS for different input strings
|
Major string |
Minor string |
LCS |
|
please let me learn better |
release letter |
Lease let |
|
A state diagram is a model of a reactive system. state diagrams are used to model complex logic. The model defines a finite set of states and behaviors and how the system transitions from one state to another when certain conditions are true. |
state diagrams are used to model complex logic in dynamic systems from one state to another |
state diagrams are used to model complex logic |
|
we can format hard disk |
information science |
format |
|
his computer is encoded |
the letter is enclosed |
ter is enc |
|
this manual machine is very old |
the computational machines |
Al machine |
Table 2. Relation between time of creating linked lists and traversing them (length of major string = 1000 characters).
|
Minor string length |
Time of creating linked list |
Time of traversing linked list |
Total time (seconds) |
|
50 |
16 |
1597 |
1613 |
|
100 |
24 |
1879 |
1903 |
|
150 |
29 |
2311 |
2340 |
|
200 |
30 |
2847 |
2877 |
|
250 |
34 |
3211 |
3245 |
|
300 |
35 |
3688 |
3723 |
|
350 |
36 |
4380 |
4416 |
|
400 |
36 |
5094 |
5130 |
|
450 |
37 |
5988 |
6025 |
|
500 |
37 |
6957 |
6994 |
|
550 |
37 |
8649 |
8686 |
|
600 |
37 |
10400 |
10437 |
|
650 |
37 |
11505 |
11542 |
|
700 |
37 |
13042 |
13079 |
|
750 |
37 |
15264 |
15301 |
|
800 |
37 |
16389 |
16426 |
|
850 |
38 |
17689 |
17727 |
|
900 |
38 |
18770 |
18808 |
|
950 |
38 |
20349 |
20387 |
|
1000 |
40 |
21422 |
21462 |
Table 3. Relation between total processing time and length of string (both minor and major strings are of the same size).
|
Minor string length |
Time of creating linked list |
Time of traversing linked list |
Total time (seconds) |
|
200 |
4 |
277 |
281 |
|
400 |
8 |
966 |
974 |
|
500 |
16 |
1710 |
1726 |
|
600 |
22 |
2705 |
2727 |
|
700 |
26 |
4197 |
4223 |
|
800 |
29 |
7596 |
7625 |
|
1000 |
37 |
19188 |
19225 |
|
1200 |
75 |
34815 |
34890 |
|
1500 |
119 |
64296 |
64415 |
|
2000 |
300 |
155075 |
155375 |
|
2250 |
442 |
224355 |
224797 |
|
2500 |
576 |
310108 |
310684 |
|
3000 |
976 |
530132 |
531108 |
|
4000 |
1779 |
1340481 |
1342260 |
|
4500 |
2111 |
1919311 |
1921422 |
|
5000 |
2695 |
2656767 |
2659462 |
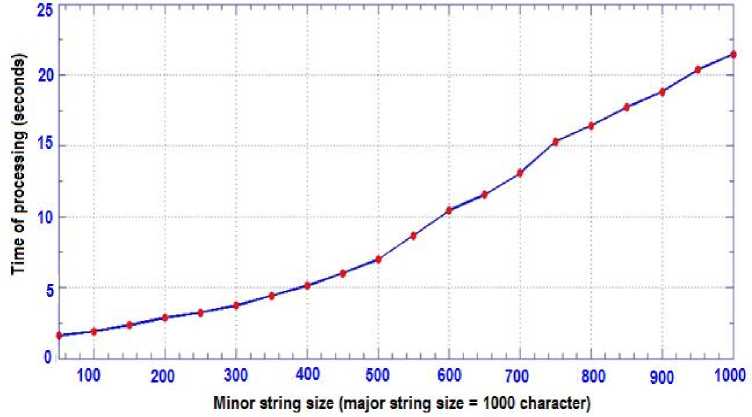
Fig.6. The snapshot of the implemented program running for the illustration example.
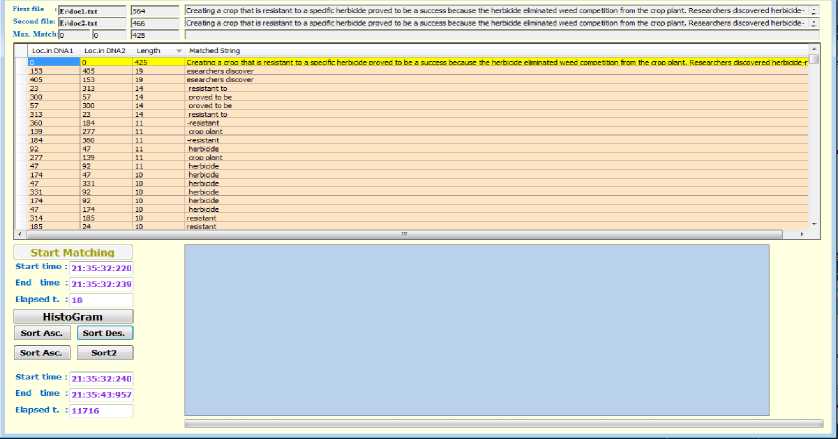
Fig.7. The relation between the time of processing and its dependence on the ratio between the lengths of both minor and some major strings
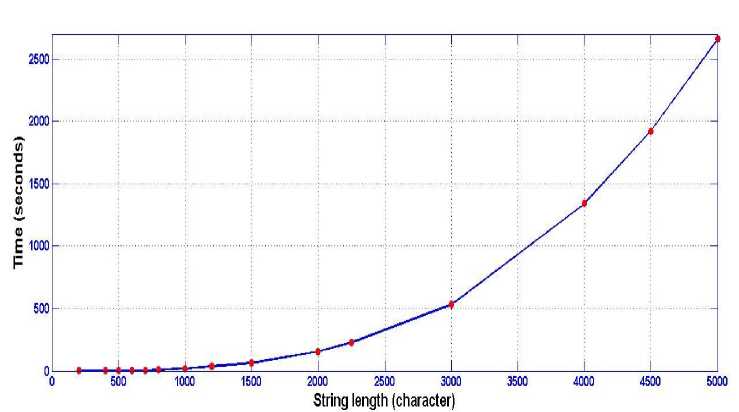
Fig.8. Relation between time consumed in constructing linked lists and time consumed in traversing them
-
IV. Conclusion
Documents matching is a fundamental and upcoming area in data processing and internet search. The algorithm proposed in this paper addressed the problem of locating the longest common substring (LCS) in two different sequences. It is based on the convolution between the two strings (named major sequence X and minor one Y ) and creating a node to the linked-list in the corresponding cell of array Z for each match between two substrings. If two or more matches share the same location in string X , the corresponding nodes will construct a single linked-list yielding to a group of linked-lists containing nodes arranged in certain manner representing all possible matches between sequences X and Y . The obtained results presented very good speedups and indicate that impressive improvements has been achieved. basing on this algorithm, an application can be developed to find the longest common substring between two or more DNA sequences. The prposed algorithm can be developed to locate the longest common strings between the major document and several minor strings. Moreover, the algorithm needs to be developed to be able to run in parallel processing manner to cope with the long time processing problem.
Список литературы Finding Longest Common Substrings in Documents
- Gusfield, D.: Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge University Press (1997).
- Hui, L.C.K.: Color Set Size Problem with Applications to String Matching. In: Proc. 3rd CPM (LNCS 644). pp. 230–243 (1992).
- Weiner, P.: Linear Pattern Matching Algorithms. In: Proc. 14th FOCS (SWAT). pp. 1–11 (1973).
- J. A. W. Faidhi and S. K. Robinson, "An empirical approach for detecting program similarity within a university programming environment", Computers & Education, vol. 11, no. 1, (1987), pp. 11-19.
- U. Manber, "Finding similar files in a large file system[C/OL]", In: Proceedings of the Winter USENIX Conference, (1994), pp. 1-10.
- BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi, (2011) September.
- Weiner, P.: Linear Pattern Matching Algorithms. In: Proc. 14th FOCS (SWAT). pp. 1–11 (1973).
- Hui, L.C.K.: Color Set Size Problem with Applications to String Matching. In: Proc. 3rd CPM (LNCS 644). pp. 230–243 (1992).
- S. Grier, "A tool that detects plagiarism in Pascal programs", ACM SIGCSE Bulletin, vol. 13, no. 1, (1981), pp. 15-20.
- J. K. Me, M. R. Panigrahi, G. N. Dash and P. K. Meher, Wavelet Based Lossless DNA Sequence Compression for Faster Detection of Eukaryotic Protein Coding Regions, IJIGSP Vol.4, No.7, July 2012.
- Kamta Nath Mishra, P. C. Srivastava, Anupam Agrawal, Vivek Tripathi, Rishu Garg, Minutiae Distances and Orientation Fields Based Thumbprint Identification of Identical Twins, IJIGSP Vol.5, No.2, February 2013.
- Murugan. A. and Lavanya. B. DNA algorithmic approach to solve GCS problem. Journal of Computational Intelligence in Bioinformatics,3(2):239-247, 2010.
- Murugan. A., Lavanya. B., and Shyamala. K.A novel programming approach for DNA computing. International Journal of Computational Intelligence Research, 7(2):199-209, 2011.
- Lavanya. B. and Murugan. A. Discovering sequence motifs of different patterns parallelly using DNA operations. International Journal of Computer Applications, 3(1):18-24, Nov 2011.
- Lavanya. B. and Murugan. A. A DNA based approach to _nd closed repetitive gapped subsequence from a sequence database. International Journal of Computer Applications, 29(5):45-49, Sep 2011.
- http://en.wikipedia.org/wiki/Longest_common_substring_problem


