Genome-wide analysis of BHLH and BZIP transcription factors and their temporal expression under abiotic stress conditions in groundnut (Arachis hypogaea L.)
Автор: Suchithra B., Shafia Hoor F., Nagesh Babu R.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 1 т.18, 2022 года.
Бесплатный доступ
Groundnut ( Arachis hypogaea L.), is an important subsistence oil yielding crop of the semi-arid tropics and often exposed to several environmental cues (high temperature, drought & heavy metal). Transcription factors can control the expression of many target genes through specific binding to the cis-acting elements in the promoters of the target genes. The basic leucine zipper (bZIP) and basic helix-loop-helix (bHLH) represents one of the largest as well as most diverse transcription factor (TFs) families. They are known to play role in both stress as well as in various plant developmental processes. In this study, a comprehensive phylogeny, chromosomal location, conserved motif identification and expression profiles under high temperature and drought stress. of bZIP and bhLH TF gene family was carried in groundnut. A total of 151 bZIP and 39 bHLH transcription factors have been identified from groundnut. Expression analysis during high temperature and heavy metal stress conditions. Gene expression studies revealed differential expressions of bZIP and bhLH TFs suggesting the possible role in various stress mitigation and can serve as a candidate genes for improving abiotic stress tolerance and can be helpful in enhancing the crop productivity under stress conditions.
Groundnut, bzip, bhlh, abiotic stress
Короткий адрес: https://sciup.org/143178338
IDR: 143178338
Текст научной статьи Genome-wide analysis of BHLH and BZIP transcription factors and their temporal expression under abiotic stress conditions in groundnut (Arachis hypogaea L.)
Plants are frequently being exposed to abiotic stresses such as drought, high salinity, high osmolarity, nutrient deficiency etc. These environmental factors negatively affect the plants leading to reduced growth and yield. Plants have evolved several defence mechanisms start from the alteration of gene expression and cellular metabolism to changes in plant growth, development, and crop yield (Akula Ramakrishna et al. , 2011). Following exposure to abiotic stress specific ion channels and kinase cascades are activated, reactive oxygen species (ROS), phytohormones like abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) accumulate, and a reprogramming of the genetic machinery results in adequate defense reactions and an increase in plant tolerance in order to minimize the biological damage caused by the stress (Ines Ben Rejeb et al. , 2014). Under stress conditions, plants synthesize ABA in various organs and initiate defense mechanisms, such as the regulation of stomatal aperture and expression of defense related genes conferring resistance to environmental stresses. Expression of functional proteins is largely controlled by specific transcription factors (TFs). Recent studies demonstrated that DREB1/ CBF, DREB2, AREB/ABF, and NAC have important roles in response to abiotic stresses in rice Kazuo Nakashima et al , 2009). TFs like MYB, AP2/ERF, NAC, bZIP, bHLH and WRKY families act as the early responders to environmental signals and trigger the expression of stress-induced genes that are important for plants to be tolerant to abiotic stress.
Groundnut is one of the important legume crops of tropical and semiarid tropical countries (annual production of ~ 46 million tons) where it provides a major source of edible oil and protein. Groundnut kernels contain 47-53% oil and 25-36% protein. The genus Arachis belongs to family Fabaceae, sub family Papilionaceae, Tribe Aeschynomeneae, Subtribe Stylosanthinae. The genus Arachis has more than 70 wild species, of which only Arachis hypogaea L is domesticated and commonly cultivated. The Arachis genus is composed mostly of diploid species (2n = 2x = 20). A. hypogaea is an allotetraploid (AABB-type genome; 2n = 4x = 40), derived from a hybridization event between two diploid species and polyploidization. Chromosomes are of mostly similar size and divided into A and B sub-genomes. Cytogenetic, phylogeographic and molecular evidence indicate A. duranensis and A. ipaensis as the donors of the A and B sub-genomes, respectively. In plant genomes approximately 7% of the coding sequences are assigned to transcription factors (TFs) (Soren Lindemose et al., 2013), and many of these are immediate-early abiotic stress-responsive genes (Kilian et al., 2012). A TF can control the expression of many target genes through specific binding to the cis-acting elements in the promoters of the target genes.
The basic leucine zipper (bZIP) transcription factor family is one of the largest and most conserved families, named according to the conserved bZIP domain that is composed of 60-80 amino acids and contains two functional regions: a basic region and a leucine zipper. The basic region is conserved and responsible for nuclear localization and DNA binding. The leucine zipper motif that consists of several repeats of leucine or other hydrophobic amino acids is involved in recognition and dimerization of bZIPs (Wei Hu et al. , 2016). Recent studies show that bZIP TFs play crucial roles in various aspects of biological processes, including organ differentiation, embryogenesis, seed maturation, flower and vascular development. Increasing evidences have also indicated that bZIP TFs take part in the regulation of plants’ response to biotic and abiotic stress. The basic helix–loop–helix (bHLH) proteins are a large superfamily of eukaryotic transcription factors, and play a central role in a wide range of metabolic, physiological, and developmental processes (Sonnenfeld et al. , 2005). Their bHLH domain contains approximately 60 amino acids, including a basic region and a HLH region (Murre et al. , 1989). The basic region, which consists of approximately 17 amino acids and is located at the N-terminus of the domain, is a DNA-binding region that allows HLH proteins to bind to a consensus hexanucleotide E-box (CANNTG) (Mark Eben Massari et al. , 2000). The HLH region is composed of two amphipathic helices consisting of hydrophobic residues linked by a divergent (both in length and primary sequence) loop, and functions as a dimerization domain
(Ferré D'Amaré et al. , 1994). The HLH domain promotes protein–protein interactions and allows for the formation of homodimeric or heterodimeric complexes. Several previous studies showed that bHLH plays an important role in protecting plants from abiotic stresses. A novel bHLH transcription factor, PebHLH35, enhanced the drought tolerance of Populus euphratica (Dong et al. , 2014). BrabHLH from Chinese cabbage participated in cold stress (Song et al. , 2014), and the grapevine bHLH transcription factor confers tolerance to cold stress in Arabidopsis (Xu et al. , 2014). Thus, bHLH TFs play an important role in various abiotic stresses. Agricultural production and quality are adversely affected by various abiotic stresses world-wide and this will be exacerbated by the deterioration of global climate. To feed a growing world population, it is very urgent to breed stress-tolerant crops with higher yields and improved qualities against multiple environmental stresses. Our study provides detailed characterization of bZIP and bHLH TFs which can be used as candidate genes to develop stress tolerant varieties in groundnut.
MATERIALS AND METHODS
Plant materials and stress treatment
Seeds of groundnut (ICG 1119) were surface sterilized and grown under controlled conditions at 28 °C day/25 °C night with a 12-h light/12-h dark photo period. After 10 days of germination, heavy metal stress was imposed hydroponically for 3 days with 300μM CdCl 2 and for high temperature stress seedlings were exposed to high-temperature [42 °C for 2h (induction) followed by 48 °C for 6h]. After the stress treatment, control and stress exposed tissues were harvested immediately and stored at -80 °C for further analysis.
Identification, characterization and sub-cellular localization of bHLH and bZIP proteins
The bHLH and bZIP domain containing protein sequences of groundnut were retrieved from the Plant Transcription Factor Database ver. 2.0. and Arachis genome (Peanut Base) for the hidden Markov model (HMM) profile of the bHLH and bZIP domain downloaded from the Peanut database using HAMMER (ver. 3.0). All redundant sequences were removed and the collected data were further curated by examining the presence of the conserved bHLH and bZIP domain with the help of Pfam , SMART and InterProScan web server. The length, molecular weight and pI of each deduced polypeptide were calculated using ExpasyProtParam tool . Further, WOLF PSORT tool was used to predict the subcellular localizations.
Multiple Sequence Alignment and Phylogenetic Analysis
Amino acid sequences of bHLH and bZIP TFs belonging to groundnut were imported to BioEdit v7.2.5 (Hall 1999) and multiple sequence alignment was performed with bHLH and bZIP protein sequences using ClustalW with default parameters. The bHLH and bZIP sequences were imported into MEGA v6.06 (Tamura et al , 2013) to construct a phylogenetic tree.
Genome wide distribution, Gene structure and Conserved Motif analysis
The chromosomal location of bHLH and bZIP genes were obtained from Peanut base website and the map was generated using MapInspect . Gene Structure Display Server from Centre for Bioinformatics, Peking University, was used to display the intron exon junctions . The genomic and mRNA sequences of bHLH and bZIP these were downloaded and used as query for generating its gene structure. A number of introns and exons were estimated based on this alignment and confirmed by the coordinates given in the sequences. The MEME Suite tool v4.9.1 was utilized for analysis of the conserved motifs.
Total RNA isolation and cDNA Synthesis and PCR amplification of bHLH and bZIP genes
Total RNA was isolated from control and stress treated shoot tissues using Trizol reagent and cDNA was synthesized by reverse transcription with 500ng of total RNA using PrimeScript RT Reagent Kit (Takara) according to the manufacturer’s instructions. Gene specific primers for AdbHLH48, AibHLH22, AdbZIP12
and AibZIP15 are listed in Table 1. cDNA concentration was checked using Nanodrop 2000 (Thermo Scientific). PCR reactions were setup using Taq DNA Polymerase. Each PCR reaction included 2 μl cDNA (1μg), 1 unit Taq DNA Polymerase, 10mM dNTPs, 2.5 μl Taq Assay Buffer (10X), 0.5 μl gene specific forward primer (10 μM), 0.5 μl reverse primer (10 μM), and made upto 25 μl with sterile water. The reactions conditions were 95 °C for 5 min followed by 35 cycles of 95 °C for 30 s, 54 °C for 45s and 72 °C for 30s; 72 °C for 2 min.
Expression analysis of bHLH and bZIP genes
All RNA samples were quantified by Nanodrop 2000 (Thermo Scientific). cDNA was synthesized by reverse transcription with 500ng of total RNA using PrimeScript RT Reagent Kit (Takara) according to the manufacturer’s instructions. Gene specific primers for AdbHLH48, AibHLH22, AdbZIP12 and AibZIP15 were designed using Primer3 software (Table 1). qRT- PCR reactions were performed using SYBR Green PCR Master mix (Takara) on CFX96 Real Time PCR (Biorad). Each PCR reaction (10 μl) included 2 μl cDNA (100ng), 5µ1 1x SYBR Green Master mix, 0.5 μl gene specific forward primer (10 μM), 0.5 μl reverse primer (10 μM), and 2 μl sterile water. The bHLH and bZIP expression was normalized against actin as reference gene. The reactions conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 54 °C for 45s and 72 °C for 30s. All reactions were run with three technical and the data was analyzed using 2 -ΔΔCT method.
RESULTS AND DISCUSSION
Identification of bHLH, Protein features, multiple sequence alignment and Phylogenetic analysis
To identify all the bHLH transcription factors, we retrieved all the predicted bHLH genes from Plant TFDB and Peanut Base . The keyword, HMM profile and BLAST search predicted that the groundnut genome encodes about 151 bHLH proteins. A total of 151 bHLH genes were identified from both A. duranensis and A.ipaënsis. They were named as AdbHLH1 to AdbHLH79, and AibHLH1 to AibHLH72 respectively. Basic information like molecular weight and pI of AdbHLH are depicted in Table 2. The average polypeptide length was 351.21 residues with the length ranging from 181aa (AdbHLH 77) to 665 aa (AdbHLH 67). The pI values range from 4.69 to 9.76. The sub-cellular localization results revealed that majority of the proteins were localized to nucleus and 2/76 were predicted to be localized in chloroplast and 1 in cytoplasm. Basic information like molecular weight and pI of AibHLH are depicted in Table 3. The average polypeptide length was 362.90 residues with the length ranging from 168aa (AibHLH 70) to 663 aa (AibHLH 18). The pI values range from 4.61 to 9.77. The sub- cellular localization results revealed that majority of the proteins were localized to nucleus and 1/76 were predicted to be localized in chloroplast. The multiple alignment of AdbHLH and AibHLH, proteins indicated that they share a highly conserved 7-9 domains consisting of N-terminal DNA binding domain and a variable C-terminal transcriptional regulation domain (Fig 1 and 2).
To examine the structure and phylogenetic relationships of groundnut bHLH TFs identified in our study, a combined phylogenetic tree was constructed with the aligned bHLH domains from groundnut. The relationship among the 79 AdbHLH and 72 AibHLH TFs was investigated through constructing phylogenetic trees using Neighbour Joining method and the tree topology revealed several pairs of bHLH proteins with a high degree of homology in the terminal nodes of each subfamily Fig 3 and 4. Examination of the phylogenetic tree emphasis that the groundnut AdbHLH TFs can be classified into seven major groups: Group 1 (17) Group 2 (15), Group 3 (11), Group 4 (7), Group 5 (11), Group 6 (13), Group7 (5). AibHLH can be classified into nine major groups: Group 1(17) , Group 2 (11), Group 3 (18), Group 4(4), Group 5(3),Group 6(7), Group7(3), Group8(1), Group 9(8).
Identification of bZIP, Protein features, Multiple sequence alignment and Phylogenetic analysis
To identify all the bZIP transcription factors, we retrieved all the predicted bZIP genes from Plant TFDB and Peanut Base . The keyword, HMM profile and BLAST search predicted that the groundnut genome encodes about 39 bZIP proteins. A total of 39 bZIP genes were identified from both A. Duranensis and A.ipaënsis. They were named as AdbZIP1 to AdbZIP18 and AibZIP1 to AibZIP21
respectively. Basic information like molecular weight and pI of AdbZIP are depicted in Table 4. The average polypeptide length was 293.4 residues with the length ranging from 146aa (AdbZIP 15) to 495 aa (AdbZIP 1). The pI values range from 5.03 to 9.9. The sub- cellular localization results revealed that majority of the proteins were localized to nucleus and 1/76 were predicted to be localized in endoplasmic reticulum. Basic information like molecular weight and pI of AibHLH are depicted in Table 5. The average polypeptide length was 296.4 residues with the length ranging from 145aa (AibZIP 18) to 800aa (AibZIP 8). The pI values range from 4.86 to 9.36. The sub- cellular localization results revealed that majority of the proteins were localized to nucleus and 2/76 were predicted to be localized in endoplasmic reticulum. The multiple alignment of AdbZIP and AibZIP indicated that they share 5 to 6 highly conserved domains consisting of N-terminal DNA binding domain and a variable C-terminal transcriptional regulation domain (Fig 5 and 6).
To examine the structure and phylogenetic relationships of groundnut bZIP TFs identified in our study, a combined phylogenetic tree was constructed with the aligned bZIP domains from groundnut. The relationship among the 18AdbZIP and 21AibZIP TFs was investigated through constructing phylogenetic trees using Neighbour Joining method and the tree topology revealed several pairs of bZIP proteins with a high degree of homology in the terminal nodes of each subfamily Fig 7 and 8. Examination of the phylogenetic tree emphasis that the groundnut AdbZIP is classified into 8 groups: Group1 (4), Group2 (1), Group3 (3), Group4 (2), Group5 (1), Group6 (1), Group7 (1), Group8 (5). AibZIP TFs are classified into 8 groups: Group1 (4), Group2 (2), Group3 (1), Group4 (4), Group5 (2), Group6 (1), Group7 (1), Group8 (6).
Chromosomal distribution and gene structure of bHLH and bZIP members
The genome of groundnut comprises of 20 chromosomes (10 from duranensis and 10 from ipaensis) varying in their length in which shortest being chromosome 8 and longest is the chromosome 3 in A. duranensis while in A.ipaënsis, shortest being chromosome 4 and longest being chromosome 9. In silico mapping of bHLH and bZIP indicated an uneven distribution of the genes on all the chromosomes. (Fig 9, 10, 11 and 12). The exact position (in bp) of each bHLH and bZIP genes on groundnut chromosomes is given in Table 2, 3 and 4. The gene structures were investigated through genomic annotation to determine the structural diversity. All bHLH and bZIP genes harbored at least two exons except few being the shortest not having intron. In addition, a separate phylogenetic tree was generated from the complete protein sequences of all the bHLH and bZIP genes (Fig 13, 14, 15 and 16).
Identification of conserved motifs
The MEME (Multiple Expectation Maximization for Motif Elicitation) server was used for exploring motif distribution in 79 AdbHLH, 72 AibHLH, 18 AdbZIP, and 21 AibZIP (Fig 17, 18 and 19). Five different conserved motifs were identified, of which most of them had at least three highly conserved motifs. The motif sequence logos are depicted in the Table 6, 7, 8 and 9. Some of these motifs have been characterized in animals regarding specificity in the DNA-binding sequence recognition and dimerization activities responsible for the activation or repression of target genes or for binding to small molecules. Multiple sequence alignment and identification of conserved motifs using MEME tool indicates that most of the bHLH and bZIP proteins possessed 5 to 6 sub-domains in the N termini that conferred the DNA-binding activities. The motif composition of these TF sequences may provide clues for further functional analysis of these TFs. However, the biological significance of most of the putative motifs remains to be elucidated.
PCR amplification of bHLH and bZIP genes
Total RNA was isolated from stress treated tissues (Fig 20 and 21). The PCR reaction mixtures were run on 1.5% Agarose gel prepared using 1X TAE buffer along with 100 bp ladder. A single band of AibZIP15 (704bp) and AdbZIP12 (709bp) was observed on gel. There was no PCR product for AdbHLH48 and AibHLH22 under high temperature stress (Fig 22b). Under heavy metal stress, AdbHLH48 (450bp) and AibZIP15 (709bp) were amplified and single band was observed on gel (Fig 22a). The band pattern is comparatively similar to the results obtained by quantitative PCR analysis.
Expression profiles bHLH genes during high temperature and high metal stress
The plant specific bHLH TFs play important role in regulation of diverse biological processes, including development, growth, cell division and responses to environmental stimuli. To cope with these stresses, plants have evolved a range of physiological and biochemical responses and a complex of signalling transduction pathways Rosa M. Pérez-Clemente et al., 2013). bHLH proteins are plant-specific TFs that have been shown to function in abiotic stress responses (Marie Pireyre et al., 2015). To investigate the responses of bHLH genes to high metal and high temperature stress, we analysed the expression profiles of one bHLH gene from each genome and expressed the results as fold changes with respect to the control. During heavy metal stress, bHLH belonging to Group 5 and Group 1 such as AdbHLH48 and AibHLH22 genes were down regulated with folds 4.49 and 1.56 respectively (Fig 23). During high temperature, bHLH genes were up-regulated and AdbHLH48 and AibHLH22 were found to be induced by 16.9 and 0.93 folds respectively (Fig 24). During heavy metal stress, bZIP genes belonging to Group 4 and Group 1 such as AdbZIP12 and AibZIP15 showed up regulation by 9.25 and 10.01 folds respectively. Out of 2 bZIP genes AdbZIP12 was down regulated by 12.9 fold and AibZIP15 was up-regulated by 10.68 fold (Fig 24). AibZIP showed increased expression under both drought and high temperature stress.
Table 1. List of primers for RT-qPCR
|
Gene Name |
Forward Primer |
Reverse Primer |
Length (bp) |
Tm CQ |
|
AdbHLH4S |
ACGGATCCTGACCTGTTCCAACTGCTTG |
TACTCGAGTCTATGAGCTCCGGGATGAG |
28 |
63.0 |
|
AibHLH22 |
ACGGATCCTCTCTTGCAGAGGGGAAAGA |
TACTCGAGC.A-AGTCnGGGHCACAGCA |
28 |
63.0 |
|
AdbZIP12 |
ACGGATCCACCACCAGCAAATGTTCTCC |
TACTCGAGAGGCGCAAGAATTAGGAACA |
28 |
63.0 |
|
AibZIPl? |
ACGGATCCGGCGTCTTCAAGTGGAACAT |
TACTCGAGAACTGGCTCCATGAATGACC |
28 |
63.0 |
Table 2. AdbHLH genes identified in Peanut, Chromosomal location, protein features and its localization prediction.
|
Protein |
Chromosome Number |
Chromosomal Location (bp) |
Deduced Polypeptide |
Subcellular Localization |
|||
|
Start |
End |
Length |
pI |
MW |
|||
|
AdbHLH1 |
A01 |
11180803 |
11184851 |
450 |
5.9 |
56349.39 |
nucleus |
|
AdbHLH2 |
A01 |
23818826 |
23821600 |
387 |
7.18 |
42810.87 |
Nucleus |
|
AdbHLH3 |
A01 |
24107575 |
24110292 |
268 |
7.03 |
29696.69 |
Nucleus |
|
AdbHLH4 |
A01 |
33747530 |
33755680 |
297 |
4.91 |
33725.91 |
Nucleus |
|
AdbHLH5 |
A01 |
76355573 |
76358681 |
255 |
8.73 |
27535.72 |
Nucleus |
|
AdbHLH6 |
A01 |
92963020 |
92966444 |
405 |
5.7 |
43265.18 |
Nucleus |
|
AdbHLH7 |
A02 |
4990418 |
4992478 |
447 |
6.04 |
50189.76 |
Nucleus |
|
AdbHLH8 |
A02 |
11490442 |
11491724 |
516 |
5.45 |
55840.51 |
Nucleus |
|
AdbHLH9 |
A02 |
65874875 |
65881124 |
338 |
6.42 |
37356.38 |
Nucleus |
|
AdbHLH10 |
A02 |
65875202 |
65880387 |
345 |
7.7 |
37894.06 |
Nucleus |
|
AdbHLH11 |
A02 |
66678616 |
66681794 |
407 |
9.24 |
44298.24 |
Nucleus |
|
AdbHLH12 |
A02 |
89000583 |
89007871 |
694 |
5.1 |
77631.15 |
Nucleus |
|
AdbHLH13 |
A03 |
3170735 |
3173105 |
465 |
6.38 |
51017.44 |
Nucleus |
|
AdbHLH14 |
A03 |
4396954 |
4399636 |
349 |
6.1 |
38798.89 |
Nucleus |
|
AdbHLH15 |
A03 |
12136351 |
12138613 |
216 |
6.84 |
24041.54 |
Nucleus |
|
AdbHLH16 |
A03 |
107170665 |
107172739 |
406 |
6.43 |
44670.22 |
Nucleus |
|
AdbHLH17 |
A03 |
117229922 |
117230891 |
338 |
7.14 |
38190.66 |
Nucleus |
|
AdbHLH18 |
A03 |
120673179 |
120676797 |
258 |
7 |
29055.83 |
Nucleus |
|
AdbHLH19 |
A03 |
123213398 |
123215577 |
337 |
4.69 |
37951.67 |
Nucleus |
|
AdbHLH20 |
A03 |
131184949 |
131187253 |
471 |
5.52 |
52619.98 |
Nucleus |
|
AdbHLH21 |
A04 |
29987499 |
29997525 |
247 |
5.34 |
28285.34 |
Nucleus |
|
AdbHLH22 |
A05 |
757006 |
758358 |
349 |
5.16 |
39414.5 |
Nucleus |
|
AdbHLH23 |
A05 |
4471852 |
4474149 |
334 |
6.9 |
36880.17 |
Nucleus |
|
AdbHLH24 |
A05 |
86030089 |
86031635 |
220 |
9.53 |
24900.43 |
Nucleus |
|
AdbHLH25 |
A05 |
104078105 |
104082033 |
367 |
6.07 |
40422.45 |
Nucleus |
|
AdbHLH26 |
A05 |
105232729 |
105237613 |
539 |
5.16 |
58266.45 |
Nucleus |
|
AdbHLH27 |
A06 |
11815778 |
11820126 |
357 |
5.69 |
39118.08 |
Nucleus |
|
AdbHLH28 |
A06 |
109557765 |
109560470 |
419 |
5.65 |
45509.36 |
Nucleus |
|
AdbHLH29 |
A06 |
110137735 |
110139757 |
277 |
5.85 |
31321.61 |
Nucleus |
|
AdbHLH30 |
A07 |
9293133 |
9294428 |
256 |
8.76 |
28456.33 |
Nucleus |
|
AdbHLH31 |
A07 |
55234108 |
55235523 |
417 |
6.44 |
52504.98 |
Nucleus |
|
AdbHLH32 |
A07 |
57713413 |
57715466 |
397 |
7.08 |
43397.54 |
Nucleus |
|
AdbHLH33 |
A07 |
68460163 |
68461453 |
321 |
4.83 |
36494.42 |
Nucleus |
|
AdbHLH34 |
A07 |
70853348 |
70871615 |
350 |
4.88 |
39768.86 |
Nucleus |
|
AdbHLH35 |
A07 |
75244942 |
75246494 |
313 |
9.18 |
34370.98 |
Nucleus |
|
AdbHLH36 |
A08 |
15304566 |
15307528 |
343 |
4.86 |
39261.21 |
Nucleus |
|
AdbHLH37 |
A08 |
17382662 |
17385316 |
393 |
5.85 |
42849.76 |
Nucleus |
|
AdbHLH38 |
A08 |
23771934 |
23774264 |
328 |
5.57 |
37212.97 |
Nucleus |
|
AdbHLH39 |
A08 |
24276717 |
24279598 |
401 |
5.68 |
44863.38 |
Nucleus |
|
AdbHLH40 |
A08 |
29959576 |
29960976 |
236 |
6.01 |
26574.31 |
Nucleus |
|
AdbHLH41 |
A08 |
43112521 |
43115242 |
400 |
8.88 |
44306.71 |
Nucleus |
|
AdbHLH42 |
A09 |
1141726 |
1145405 |
357 |
6.77 |
40041.13 |
Nucleus |
|
AdbHLH43 |
A09 |
4405214 |
4406928 |
366 |
5.31 |
41366.62 |
Nucleus |
|
AdbHLH44 |
A09 |
9616546 |
9619558 |
580 |
6.45 |
63460.81 |
Nucleus |
|
AdbHLH45 |
A09 |
116722167 |
116723907 |
291 |
7.76 |
31338.87 |
Chloroplast |
|
AdbHLH46 |
A10 |
4762764 |
4765178 |
368 |
7.66 |
41676.67 |
Nucleus |
|
AdbHLH47 |
A10 |
22120099 |
22122002 |
236 |
9.28 |
26260.28 |
Nucleus |
|
AdbHLH48 |
A10 |
104379283 |
104380857 |
238 |
7.86 |
26566 |
Nucleus |
|
AdbHLH49 |
A01 |
90749358 |
90752534 |
256 |
4.87 |
29223.97 |
Nucleus |
|
AdbHLH50 |
A03 |
19592294 |
19594435 |
221 |
6.67 |
25350.15 |
Nucleus |
|
AdbHLH51 |
A03 |
38984268 |
38987093 |
528 |
9.04 |
59651.37 |
Nucleus |
|
AdbHLH52 |
A03 |
121854601 |
121856521 |
327 |
6.41 |
36595.78 |
Nucleus |
|
AdbHLH53 |
A04 |
20876962 |
20880191 |
487 |
5.13 |
54204.89 |
Nucleus |
|
AdbHLH54 |
A05 |
1099767 |
1101296 |
336 |
6.28 |
37941.85 |
Nucleus |
|
AdbHLH55 |
A05 |
5650995 |
5652478 |
335 |
4.96 |
37474.88 |
Nucleus |
|
AdbHLH56 |
A05 |
5676022 |
5677733 |
322 |
6.57 |
36193.44 |
Nucleus |
|
AdbHLH57 |
A06 |
1704105 |
1705517 |
307 |
5.86 |
35063.41 |
Nucleus |
|
AdbHLH58 |
A06 |
4585188 |
4586562 |
278 |
5.36 |
31533.59 |
Nucleus |
|
AdbHLH59 |
A07 |
63884568 |
63887247 |
332 |
6.06 |
36849.39 |
Nucleus |
|
AdbHLH60 |
A07 |
70852467 |
70853604 |
193 |
9.76 |
21638.45 |
Nucleus |
|
AdbHLH61 |
A08 |
13571762 |
13573816 |
325 |
7.22 |
35758.02 |
Nucleus |
|
AdbHLH62 |
A08 |
31926538 |
31927699 |
228 |
7.07 |
26186.85 |
Nucleus |
|
AdbHLH63 |
A09 |
36737760 |
36739645 |
303 |
9.3 |
34193.65 |
Nucleus |
|
AdbHLH64 |
A10 |
57409973 |
57414243 |
311 |
6.38 |
35382.97 |
Nucleus |
|
AdbHLH65 |
A02 |
4340844 |
4342187 |
473 |
5.51 |
53353.28 |
Nucleus |
|
AdbHLH66 |
A03 |
6936674 |
6945870 |
279 |
7.7 |
31084.6 |
Nucleus |
|
AdbHLH67 |
A06 |
2305528 |
2307525 |
665 |
6.22 |
72541.1 |
Nucleus |
|
AdbHLH68 |
A07 |
66084895 |
66087281 |
262 |
8.84 |
28851.52 |
Nucleus |
|
AdbHLH69 |
A08 |
32827123 |
32829163 |
272 |
8.61 |
30093.18 |
Nucleus |
|
AdbHLH70 |
A09 |
96870471 |
96872697 |
223 |
6.33 |
25042.36 |
Nucleus |
|
AdbHLH71 |
A09 |
112385877 |
112387278 |
311 |
5.3 |
34671.55 |
Nucleus |
|
AdbHLH72 |
A09 |
112389260 |
112390463 |
302 |
5.2 |
33731.5 |
Nucleus |
|
AdbHLH73 |
A09 |
120376856 |
120378791 |
361 |
6.31 |
40610.33 |
Nucleus |
|
AdbHLH74 |
A03 |
19352315 |
19353746 |
208 |
5.69 |
23649.96 |
Cytoplasm |
|
AdbHLH75 |
A05 |
105520640 |
105522451 |
262 |
6.01 |
29705.07 |
Nucleus |
|
AdbHLH76 |
A09 |
110546064 |
110547184 |
275 |
6.99 |
31371.18 |
Nucleus |
|
AdbHLH77 |
A10 |
2767037 |
2768803 |
181 |
9.26 |
20830.97 |
Nucleus |
|
AdbHLH78 |
A04 |
62752020 |
62756877 |
662 |
5.54 |
74386.17 |
chloroplast |
|
AdbHLH79 |
A06 |
14352907 |
14359399 |
572 |
8.25 |
64663.36 |
Nucleus |
Table 3 : AibHLH genes identified in Peanut, Chromosomal location, protein features and its localization prediction.
|
Protein |
Chromosome Number |
Chromosomal Location |
Deduced Polypeptide |
Subcellular Localization |
|||
|
Start |
End |
Length |
pI |
MW |
|||
|
AibHLH1 |
B01 |
636283 |
640342 |
520 |
5.9 |
56387.44 |
nucleus |
|
AibHLH2 |
B01 |
30217938 |
30223799 |
354 |
4.75 |
39553.40 |
nucleus |
|
AibHLH3 |
B01 |
107608827 |
107611906 |
255 |
8.73 |
27506.73 |
nucleus |
|
AibHLH4 |
B01 |
136640565 |
136641988 |
255 |
4.87 |
29061.79 |
nucleus |
|
AibHLH5 |
B02 |
77755308 |
77758153 |
413 |
9.27 |
44795.72 |
nucleus |
|
AibHLH6 |
B02 |
94625362 |
94627028 |
189 |
6.5 |
21513.15 |
nucleus |
|
AibHLH7 |
B03 |
108120840 |
108123374 |
271 |
8.77 |
29901.1 |
nucleus |
|
AibHLH8 |
B03 |
121291052 |
121294003 |
258 |
7.04 |
28944.73 |
nucleus |
|
AibHLH9 |
B03 |
123867236 |
123869445 |
347 |
4.7 |
39115.85 |
nucleus |
|
AibHLH10 |
B03 |
132161677 |
132163387 |
508 |
5.45 |
56536.24 |
nucleus |
|
AibHLH11 |
B04 |
28150372 |
28160597 |
219 |
6.86 |
25266.9 |
nucleus |
|
AibHLH12 |
B05 |
746211 |
747857 |
350 |
5.06 |
39482.53 |
nucleus |
|
AibHLH13 |
B05 |
4501469 |
4503100 |
304 |
7.2 |
33763.84 |
nucleus |
|
AibHLH14 |
B05 |
98520553 |
98525231 |
536 |
5.15 |
57911.12 |
nucleus |
|
AibHLH15 |
B05 |
109099855 |
109104474 |
367 |
6.07 |
40381.4 |
nucleus |
|
AibHLH16 |
B06 |
3996822 |
4000058 |
209 |
9.69 |
23867 |
nucleus |
|
AibHLH17 |
B06 |
4110261 |
4113865 |
362 |
5.85 |
39529.32 |
nucleus |
|
AibHLH18 |
B06 |
18357835 |
18358780 |
663 |
6.22 |
72369.88 |
nucleus |
|
AibHLH19 |
B06 |
134206069 |
134209265 |
420 |
5.65 |
45537.41 |
nucleus |
|
AibHLH20 |
B07 |
9285262 |
9287119 |
256 |
8.6 |
28370.24 |
nucleus |
|
AibHLH21 |
B07 |
33652692 |
33654351 |
357 |
4.71 |
40780.16 |
nucleus |
|
AibHLH22 |
B07 |
62509004 |
62511083 |
395 |
7.07 |
43147.23 |
nucleus |
|
AibHLH23 |
B07 |
123477966 |
123480961 |
311 |
4.88 |
35544.09 |
nucleus |
|
AibHLH24 |
B07 |
125315732 |
125318601 |
380 |
5.74 |
41449.09 |
nucleus |
|
AibHLH25 |
B08 |
990433 |
991857 |
405 |
6.35 |
52694.21 |
nucleus |
|
AibHLH26 |
B08 |
1799227 |
1801832 |
330 |
5.44 |
37429.29 |
nucleus |
|
AibHLH27 |
B08 |
2084005 |
2086453 |
402 |
5.68 |
44927.41 |
nucleus |
|
AibHLH28 |
B08 |
7512500 |
7513857 |
180 |
8.8 |
20429.6 |
nucleus |
|
AibHLH29 |
B08 |
89807832 |
89809286 |
329 |
4.91 |
37237.77 |
nucleus |
|
AibHLH30 |
B08 |
128695301 |
128698133 |
369 |
8.19 |
41646.29 |
nucleus |
|
AibHLH31 |
B09 |
276697 |
278046 |
344 |
4.61 |
39789.54 |
nucleus |
|
AibHLH32 |
B09 |
1342273 |
1346258 |
242 |
9.2 |
27208.04 |
nucleus |
|
AibHLH33 |
B09 |
139937689 |
139941958 |
327 |
8.82 |
34926.71 |
nucleus |
|
AibHLH34 |
B09 |
6839044 |
6841764 |
354 |
6.97 |
39996.66 |
nucleus |
|
AibHLH35 |
B10 |
131077378 |
131078628 |
238 |
8.46 |
26579.05 |
nucleus |
|
AibHLH36 |
B01 |
636822 |
639231 |
520 |
5.9 |
56387.44 |
nucleus |
|
AibHLH37 |
B01 |
773401 |
775837 |
451 |
5.37 |
51213.51 |
nucleus |
|
AibHLH38 |
B01 |
29853579 |
29855816 |
365 |
7.18 |
40438.28 |
nucleus |
|
AibHLH39 |
B01 |
134857606 |
134860895 |
407 |
5.7 |
43503.43 |
nucleus |
|
AibHLH40 |
B01 |
137028861 |
137032298 |
416 |
9.75 |
46883.57 |
chloroplast |
|
AibHLH41 |
B02 |
6253314 |
6255779 |
446 |
6.13 |
50118.64 |
nucleus |
|
AibHLH42 |
B02 |
102553396 |
102557165 |
661 |
4.89 |
73703.12 |
nucleus |
|
AibHLH43 |
B02 |
105759050 |
105761496 |
338 |
6.81 |
38166.63 |
nucleus |
|
AibHLH44 |
B03 |
5875560 |
5878134 |
463 |
6.38 |
50834.15 |
nucleus |
|
AibHLH45 |
B03 |
10090832 |
10092450 |
279 |
7.7 |
30985.46 |
nucleus |
|
AibHLH46 |
B03 |
14817318 |
14819570 |
217 |
5.99 |
24186.71 |
nucleus |
|
AibHLH47 |
B03 |
41339015 |
41343427 |
539 |
8.88 |
60474.21 |
nucleus |
|
AibHLH48 |
B03 |
122440911 |
122442823 |
327 |
6.03 |
36538.68 |
nucleus |
|
AibHLH49 |
B04 |
20530498 |
20533734 |
487 |
5.13 |
54204.89 |
nucleus |
|
AibHLH50 |
B05 |
1081535 |
1082506 |
342 |
6.24 |
38412.28 |
nucleus |
|
AibHLH51 |
B05 |
5834411 |
5835652 |
335 |
5 |
37457.8 |
nucleus |
|
AibHLH52 |
B06 |
13767672 |
13769322 |
289 |
5.2 |
32933.15 |
nucleus |
|
AibHLH53 |
B06 |
20280531 |
20282138 |
530 |
6.11 |
58644.05 |
nucleus |
|
AibHLH54 |
B06 |
134873226 |
134875351 |
362 |
4.9 |
41028.16 |
nucleus |
|
AibHLH55 |
B07 |
38152793 |
38153907 |
266 |
8.24 |
29235.88 |
nucleus |
|
AibHLH56 |
B07 |
42703139 |
42704374 |
332 |
6.06 |
36819.32 |
nucleus |
|
AibHLH57 |
B07 |
121797374 |
121799212 |
310 |
8.77 |
34239.27 |
nucleus |
|
AibHLH58 |
B07 |
125783661 |
125785901 |
272 |
4.79 |
30831.18 |
nucleus |
|
AibHLH59 |
B09 |
269215 |
270201 |
192 |
9.77 |
21471.28 |
nucleus |
|
AibHLH60 |
B09 |
44277247 |
44279136 |
303 |
9.15 |
34152.55 |
nucleus |
|
AibHLH61 |
B09 |
131488064 |
131489865 |
369 |
6.16 |
41501.24 |
nucleus |
|
AibHLH62 |
B09 |
146220285 |
146221793 |
272 |
6.53 |
31041.7 |
nucleus |
|
AibHLH63 |
B10 |
72699993 |
72703002 |
305 |
6.56 |
34721.31 |
nucleus |
|
AibHLH64 |
B02 |
4649893 |
4651764 |
656 |
6.38 |
72947.22 |
nucleus |
|
AibHLH65 |
B02 |
14875849 |
14877865 |
516 |
5.45 |
55794.42 |
nucleus |
|
AibHLH66 |
B04 |
76925964 |
76930950 |
662 |
5.59 |
74255 |
nucleus |
|
AibHLH67 |
B06 |
2329181 |
2336236 |
569 |
7.11 |
64242.7 |
nucleus |
|
AibHLH68 |
B08 |
7512297 |
7514586 |
180 |
8.8 |
20429.6 |
nucleus |
|
AibHLH69 |
B09 |
118317111 |
118319428 |
220 |
6.4 |
24837.11 |
nucleus |
|
AibHLH70 |
B03 |
21876207 |
21877320 |
168 |
8.31 |
19027.77 |
nucleus |
|
AibHLH71 |
B05 |
145697971 |
145699460 |
264 |
5.75 |
29877.09 |
nucleus |
|
AibHLH72 |
B03 |
125089589 |
125093842 |
480 |
5.93 |
54008.05 |
nucleus |
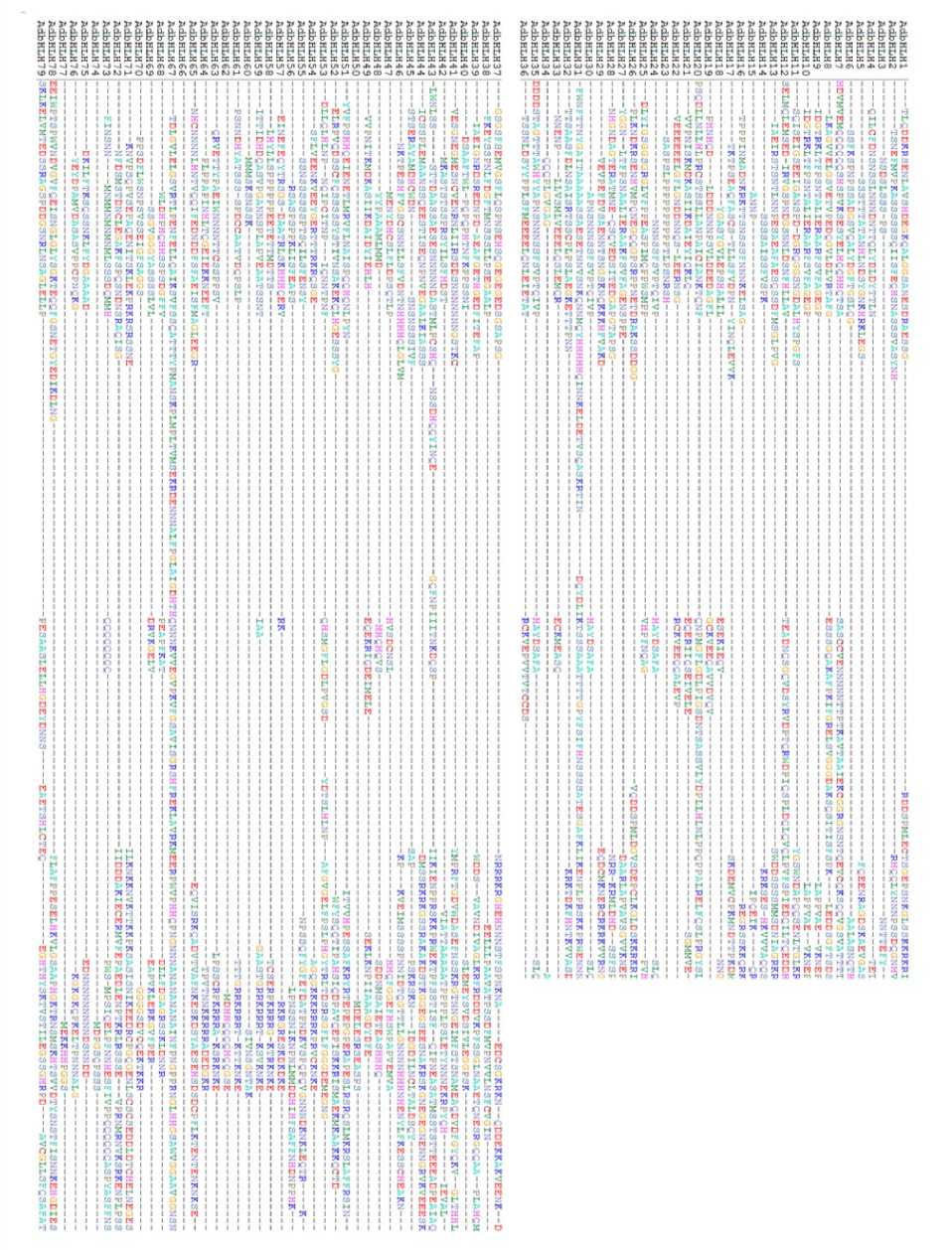
Figure 1. Multiple alignment of 79 AdbHLH TFs of groundnut

Figure 1. Continued

Figure 1. Continued
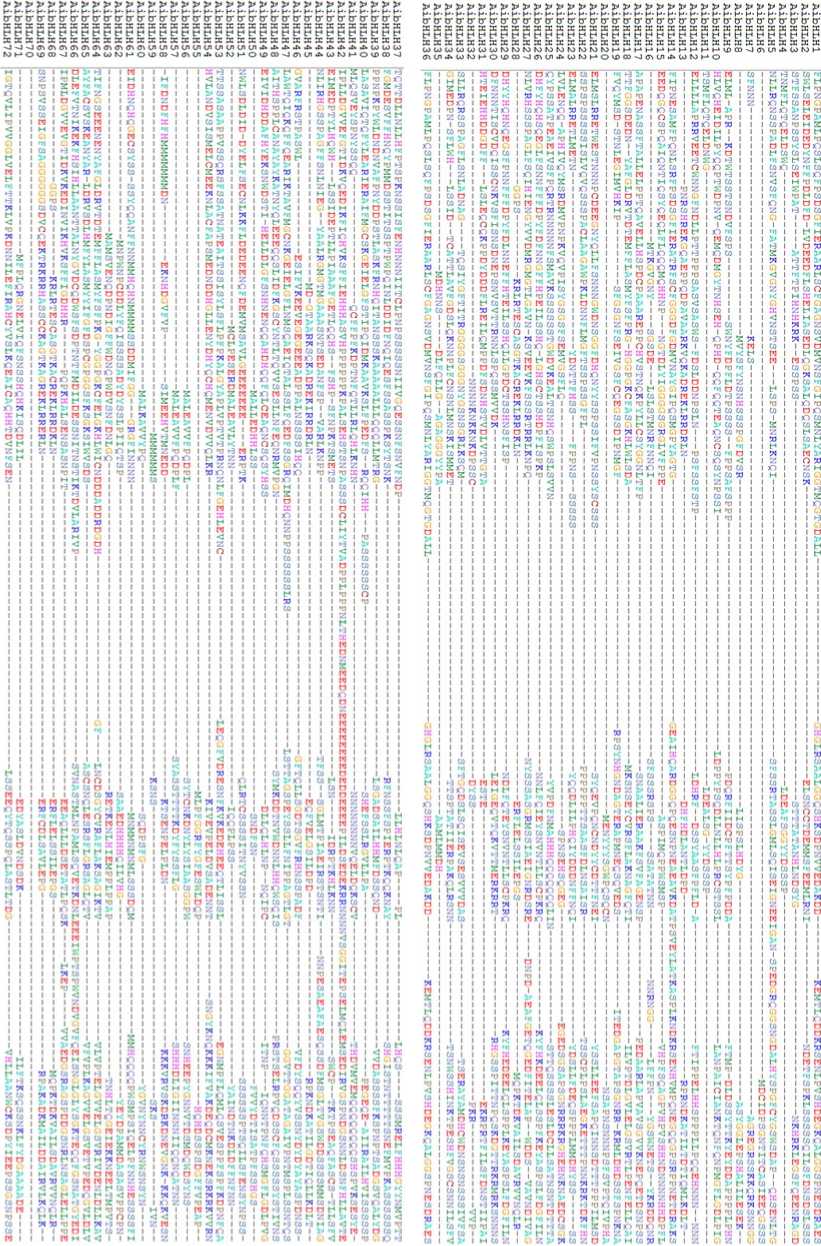
Figure 2. Multiple alignment of 72 AibHLH TFs of groundnut
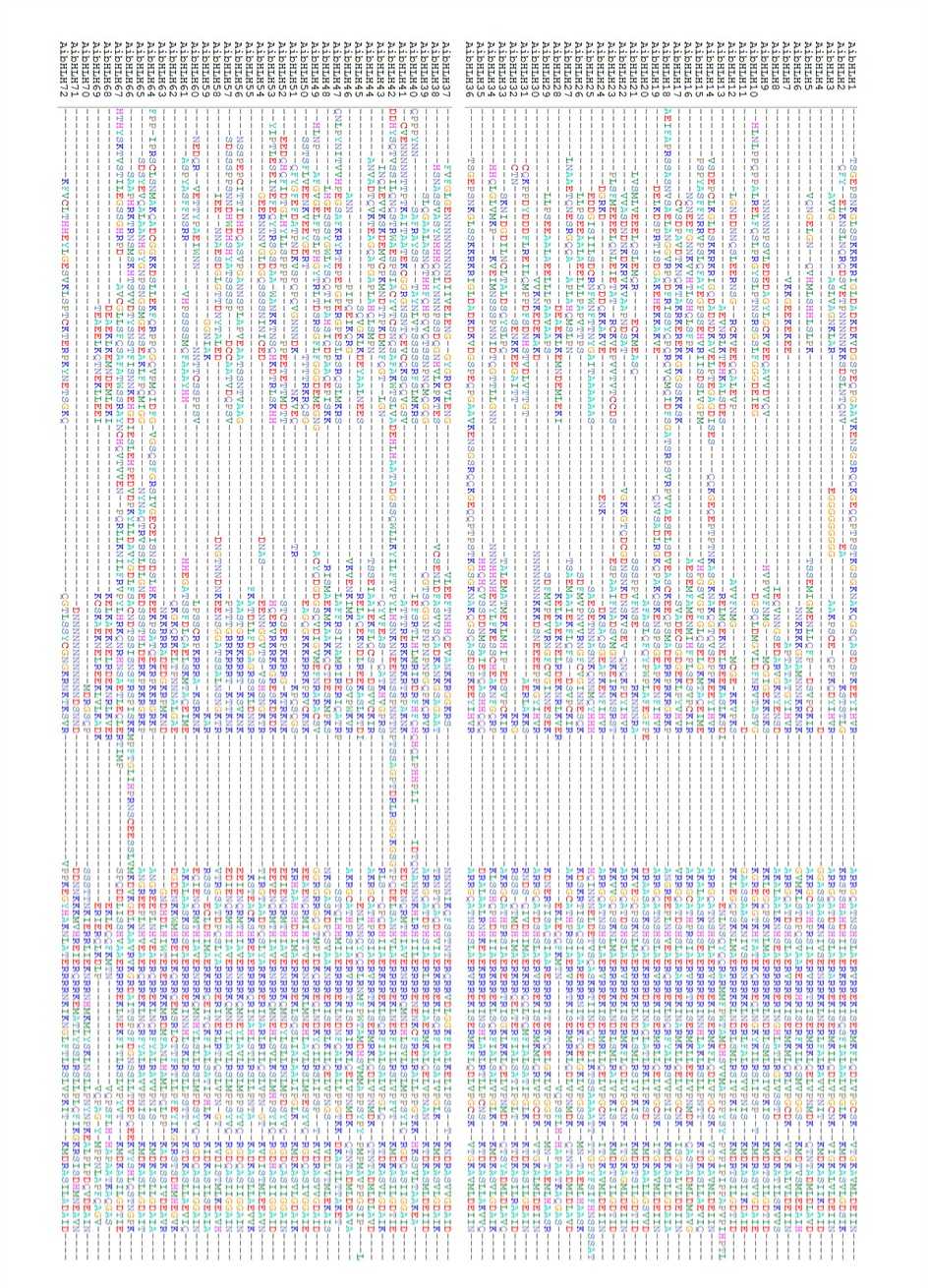
Figure 2. Continued

Figure 2. Continued
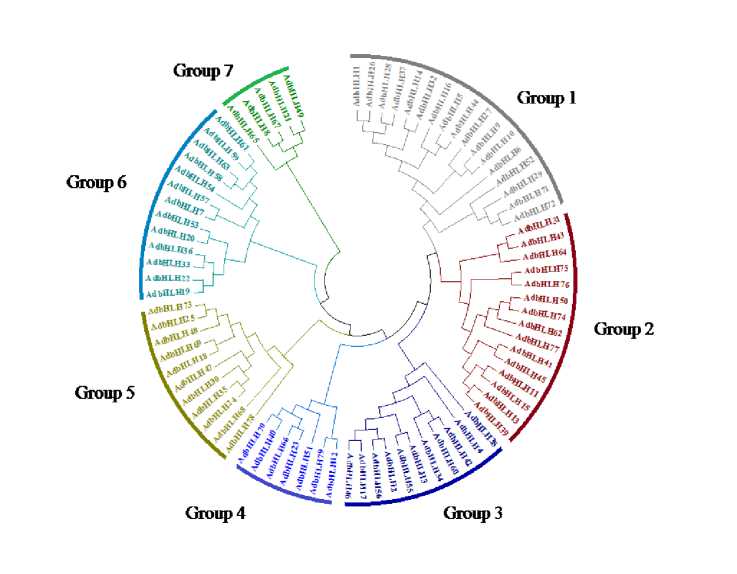
Figure 3. Phylogenetic relationship and gene structure of the bHLH genes. Phylogenetic tree was constructed with MEGA 6.0 on a multiple alignment of 79 amino acid sequences of bHLH genes from Arachis duranensis Exon/ intron structure of bHLH genes are represented by boxes and black lines, respectively.
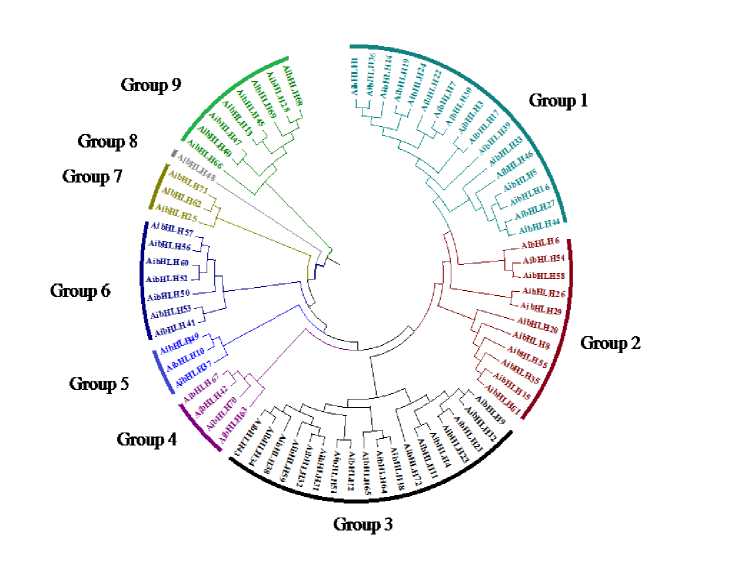
Figure 4. Phylogenetic relationship and gene structure of the bHLH genes. Phylogenetic tree was constructed with MEGA6.0 on a multiple alignment of 72 amino acid sequences of bHLH genes from Arachis ipaënsis.
Table 4. AdbZIP and AibZIP proteins identified in Peanut, Chromosomal location, protein features and its localization prediction.
|
Protein |
Chromosome Number |
Chromosomal Location (bp) |
Deduced Polypeptide |
Subcellular Localization |
|||
|
Start |
End |
Length (aa) |
pI |
MW |
|||
|
AdbZIP1 |
A06 |
67093845 |
67094858 |
495 |
7.07 |
54713.19 |
Nucleus |
|
AdbZIP2 |
A07 |
72015371 |
72018639 |
322 |
5.94 |
35914.02 |
Nucleus |
|
AdbZIP3 |
A10 |
95293270 |
95294273 |
316 |
5.03 |
33962.22 |
Nucleus |
|
AdbZIP4 |
A10 |
104633025 |
104633360 |
212 |
5.7 |
24810.41 |
Nucleus |
|
AdbZIP5 |
A01 |
104038428 |
104038856 |
217 |
8.48 |
24519.65 |
Nucleus |
|
AdbZIP6 |
A02 |
45495034 |
45497618 |
443 |
6.06 |
48807.58 |
Nucleus |
|
AdbZIP7 |
A10 |
104137408 |
104138555 |
800 |
5.84 |
86930.94 |
E.R |
|
AdbZIP8 |
A03 |
21151650 |
21152954 |
304 |
5.65 |
33778.3551 |
Nucleus |
|
AdbZIP9 |
A08 |
1090955 |
1091461 |
168 |
7.1 |
19345.84 |
Nucleus |
|
AdbZIP10 |
A08 |
32533691 |
32536893 |
332 |
5.3 |
37390.56 |
Nucleus |
|
AdbZIP11 |
A05 |
4161574 |
4162581 |
224 |
9.9 |
24845.75 |
Nucleus |
|
AdbZIP12 |
A06 |
104086507 |
104089154 |
373 |
5.18 |
40440.98 |
Nucleus |
|
AdbZIP13 |
A07 |
23354880 |
23357810 |
225 |
9.13 |
25954.56 |
Nucleus |
|
AdbZIP14 |
A03 |
4,793,469 |
4,793,906 |
163 |
6.12 |
18187.23 |
Nucleus |
|
AdbZIP15 |
A06 |
7,343,727 |
7,344,167 |
146 |
5.78 |
16384.81 |
Nucleus |
|
AdbZIP16 |
A03 |
134,132,232 |
134,132,612 |
160 |
6.14 |
17947.95 |
Nucleus |
|
AdbZIP17 |
A05 |
4,161,574 |
.4,162,581 |
224 |
9.9 |
24845.75 |
Nucleus |
|
AdbZIP18 |
A06 |
16,421,562 |
16,422,038 |
158 |
8.91 |
18493.87 |
Nucleus |
|
AibZIP1 |
B03 |
135171129 |
135171510 |
155 |
6.22 |
17528.52 |
Nucleus |
|
AibZIP2 |
B04 |
130023980 |
130024291 |
224 |
4.98 |
26262.69 |
Nucleus |
|
AibZIP3 |
B05 |
101761857 |
101762086 |
234 |
4.86 |
26920.53 |
Nucleus |
|
AibZIP4 |
B05 |
1567450 |
1572508 |
388 |
5.72 |
41074.03 |
Nucleus |
|
AibZIP5 |
B07 |
106034150 |
106034580 |
164 |
7.1 |
18889.34 |
Nucleus |
|
AibZIP6 |
B08 |
25788300 |
25790824 |
307 |
7.21 |
34005.71 |
Nucleus |
|
AibZIP7 |
B10 |
119022617 |
119023360 |
327 |
5.11 |
35139.44 |
Nucleus |
|
AibZIP8 |
B10 |
130798698 |
130800592 |
800 |
5.89 |
86979.07 |
E .R |
|
AibZIP9 |
B10 |
131256375 |
131256710 |
216 |
6.04 |
25239.89 |
Nucleus |
|
AibZIP10 |
B03 |
8405762 |
8407205 |
271 |
6.21 |
29420.98 |
Nucleus |
|
AibZIP11 |
B10 |
130798698 |
130799838 |
800 |
5.89 |
86979.07 |
E .R |
|
AibZIP12 |
B01 |
704446 |
705030 |
194 |
5.85 |
22774.32 |
Nucleus |
|
AibZIP13 |
B01 |
26679598 |
26679918 |
183 |
9.36 |
21497.39 |
Nucleus |
|
AibZIP14 |
B02 |
54142154 |
54144616 |
296 |
5.72 |
33288.03 |
Nucleus |
|
AibZIP15 |
B03 |
7502032 |
7502526 |
164 |
6.12 |
18274.30 |
Nucleus |
|
AibZIP16 |
B03 |
23484533 |
23487428 |
344 |
5.66 |
39240.76 |
Nucleus |
|
AibZIP17 |
B08 |
11006331 |
11009532 |
331 |
5.38 |
37397.60 |
Nucleus |
|
AibZIP18 |
B09 |
21764782 |
21765285 |
145 |
5.61 |
16555.98 |
Nucleus |
|
AibZIP19 |
B06 |
128409215 |
128411175 |
372 |
5.18 |
40414.98 |
Nucleus |
|
AibZIP20 |
B06 |
:9,088,845 |
9,089,285 |
146 |
5.78 |
16384.81 |
Nucleus |
|
AibZIP21 |
B07 |
:106,033,813 |
106,034,307 |
164 |
7.10 |
18889.34 |
Nucleus |

Figure 5. Multiple alignment of 18 AdbZIP TFs of groundnut

Figure 6. Multiple alignment of 21 AibZIP TFs of groundnut ukf»
MSIP1S
UK IP 14
MEF16
UK Fl
UKF4 J
UKFi "
UK FIS
Group 1
Group 2
Group 3
Group 4
Group 8
Group 5
Group 6
Group 7
MEH ] .UK FT ] UK Fil '
MEMO
Figure 7. Phylogenetic relationship and gene structure of the bZIP genes. Phylogenetic tree was constructed with MEGA6.0 on a multiple alignment of 18 amino acid sequences of bZIP genes from Arachis duranensis Exon/intron structure of bZIP genes are represented by boxes and black lines, respectively.
UKF13
UEFI’
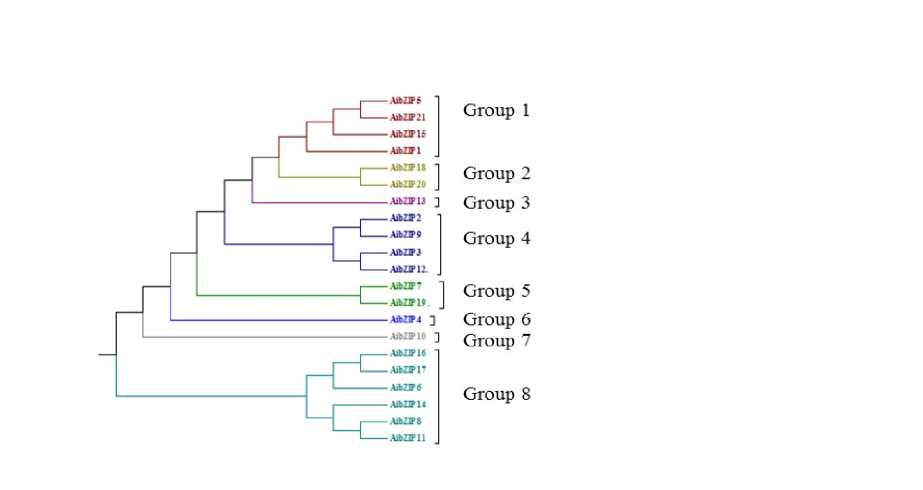
Figure 8. Phylogenetic relationship and gene structure of the bZIP genes. Phylogenetic tree was constructed with
MEGA6.0 on a multiple alignment of 21 amino acid sequences of bZIP genes from Arachis ipaënsis

Figure 9. Distribution of 79 bHLH genes from Arachis duranensis on peanut chromosomes and physical location of each bHLH gene on the ten chromosomes from each species (positions in cM).

Figure 10. Distribution of 72 bHLH genes from Arachis ipaensis on Peanut chromosomes and physical location of each bHLH gene on the ten chromosomes from each species (positions in cM).
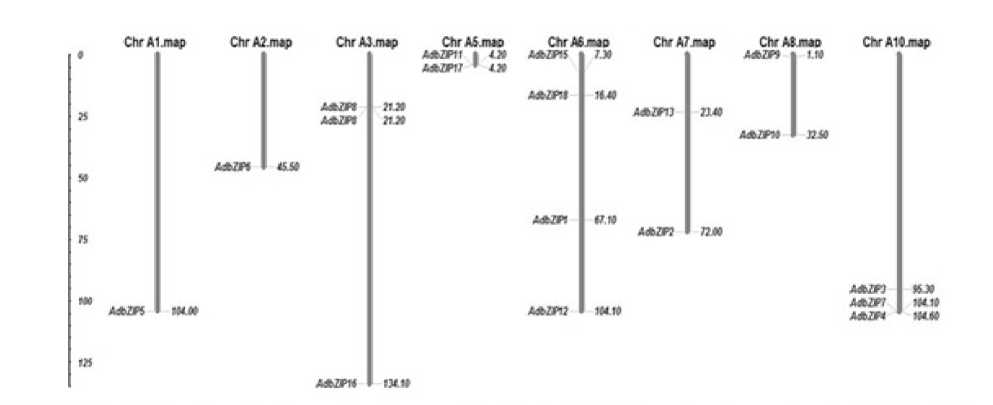
Figure 11. Distribution of 18 bZIP genes from Arachis duranensis on Peanut chromosomes and physical location of each bZIP gene on the ten chromosomes from each species (positions in cM).
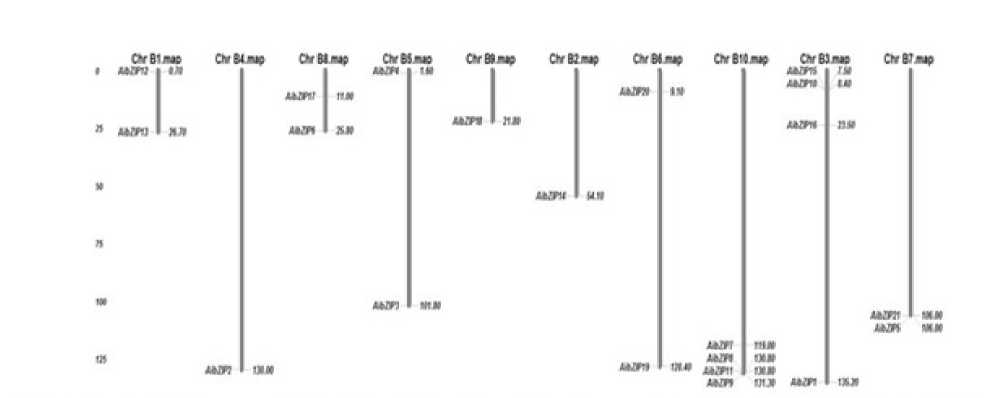
bZIP gene on the ten chromosomes from each species (positions in cM).
Figure 12. Distribution of 21 bZIP genes from Arachis ipaensis on Peanut chromosomes and physical location of each

Figure 13. Phylogenetic relationship and gene structure of the bHLH genes. Phylogenetic tree was constructed with MEGA6.0 on a multiple alignment of 79 amino acid sequences of bHLH genes from Arachis duranensis . Exon/ intron structure of bHLH genes are represented by boxes and black lines, respectively.
Figure 14. Phylogenetic relationship and gene structure of the bHLH genes. Phylogenetic tree was constructed with MEGA6.0 on a multiple alignment of 72 amino acid sequences of bHLH genes from Arachis ipaënsis Exon/intron structure of bHLH genes are represented by boxes and black lines, respectively
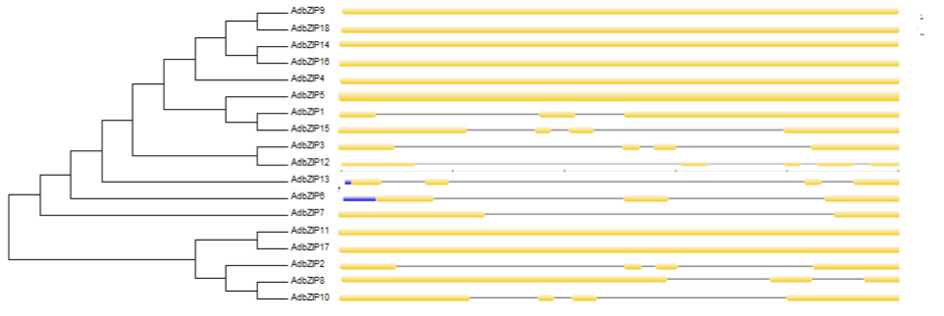
Figure 15. Phylogenetic relationship and gene structure of the bZIP genes. Phylogenetic tree was constructed with MEGA6.0 on a multiple alignment of 18 amino acid sequences of bZIP genes from Arachis duranensis Exon/intron structure of bZIP genes are represented by boxes and black lines, respectively.
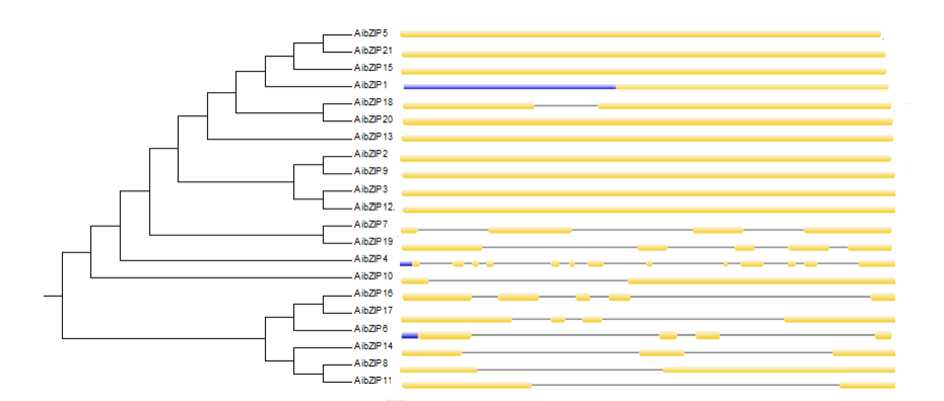
Figure 16. Phylogenetic relationship and gene structure of the bZIP genes. Phylogenetic tree was constructed with MEGA6.0 on a multiple alignment of 21 amino acid sequences of bZIP genes from Arachis ipaënsis Exon/intron structure of bZIP genes are represented by boxes and black lines, respectively
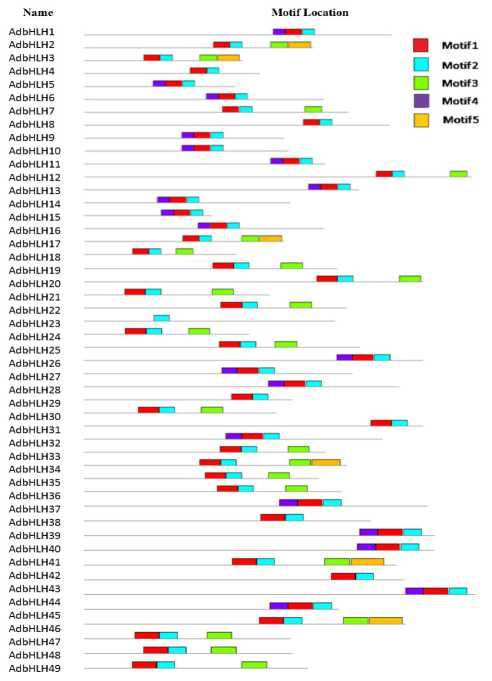
Figure 17. Schematic representation of conserved motifs in the AdbHLH proteins predicted by MEME. Each motif is represented by a number in the colored box. The black lines represent non-conserved sequences.
Name
AibHLHl
AibHLH2
AibHLH3
AibHLH4
AibHLH5
AibHLH6
AibHLH7
AibHLH8
AibHLH9 AibHLHIO AibHLHll AibHLH12 AibHLH13 AibHLH14 AibHLH15 AibHLHIG AibHLH17 AibHLH18 AibHLH19 AibHLH2O AibHLH21 AibHLH22 AibHLH23 AibHLH24 AibHLH25 AibHLH26 AibHLH27 AibHLH28 AibHLH29 AibHLH30 AibHLH31 AibHLH32 AibHLH33 AibHLH34 AibHLH35 AibHLH36 AibHLH37 AibHLH38 AibHLH39 AibHLH40 AibHLH41 AibHLH42 AibHLH43 AibHLH44 AibHLH45 AibHLH46 AibHLH47 AibHLH48 AibHLH49 AibHLH5O
Motif Location
AibHLH51 AibHLH52 AibHLH53 AibHLH54 AibHLH55 AibHLH56 AibHLH57 AibHLH58 AibHLH59 AibHLH60 AibHLH61 AibHLH62 AibHLH63 AibHLH64
AibHLH65 AibHLH66 AibHLH67 AibHLH68 AibHLH69 AibHLH7O AibHLH71 AibHLH72
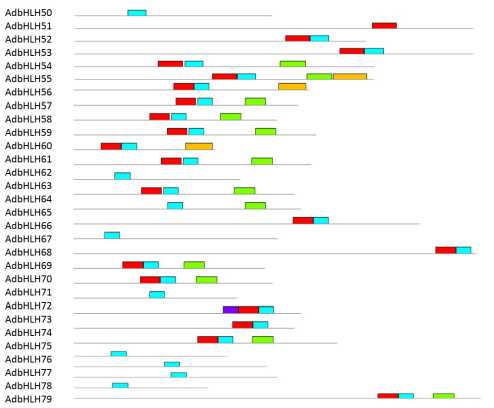
Figure
18. Schematic representation of conserved motifs AibHLH proteins predicted by MEME. Each motif is represented by a number in the colored box. The black lines represent non-conserved sequences.

Figure 19. Schematic representation of conserved motifs in the AdbZIP and AibZIP proteins predicted by MEME. Each motif is represented by a number in the colored box. The black lines represent non-conserved sequences.
Table 6 : Conserved motif logos identified in AdbHLH proteins using MEME tool
|
Sl.No |
Motif Logo |
|
1 |
1a JgRIfejH^J&u «ate о-1 _ ем „ _ „, «, г- со <п а ^ см со ? ^ JO г- "^ЯЯ^ЯЯЯЯКЙ |
|
2 |
’^^L^aW^X^bL^”^ Ь |
|
3 |
|
|
4 |
I^s^^yR^bR1 ^АТв®^ |
|
5 |
' §SAtF tedD J ^Q^mI^^^^ |
Table 7 : Conserved motif logos identified in AibHLH proteins using MEME tool
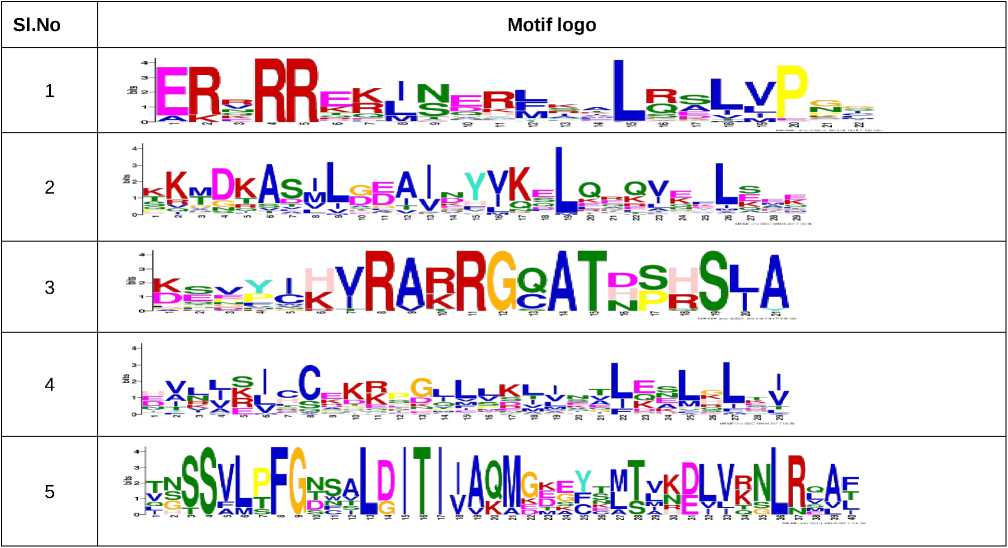
Table 8 : Conserved motif logos identified in AdbZIP proteins using MEME tool
|
Name |
Motif Logo |
|
1 |
•^ KK^R^NRiSAgRSRgRKsM .^HiUeeXe- U=|y |
|
2 |
'к:?Хйй29в№вЬ1^^|8^е^ |
|
3 |
<Ыь^е*1^^М^ |
|
4 |
•■Ш SIR SSSSW FQCFUR FT И |
|
5 |
QQQKLCEAAVLN1QQKKKSALVRIFTA F |
Table 9 : Conserved motif logos identified in AibZIP proteins using MEME tool
Name
Motif Logo






Figure 20 : Control and heavy metal treated seedlings

Figure 21 : Control and high temperature stress treated seedlings
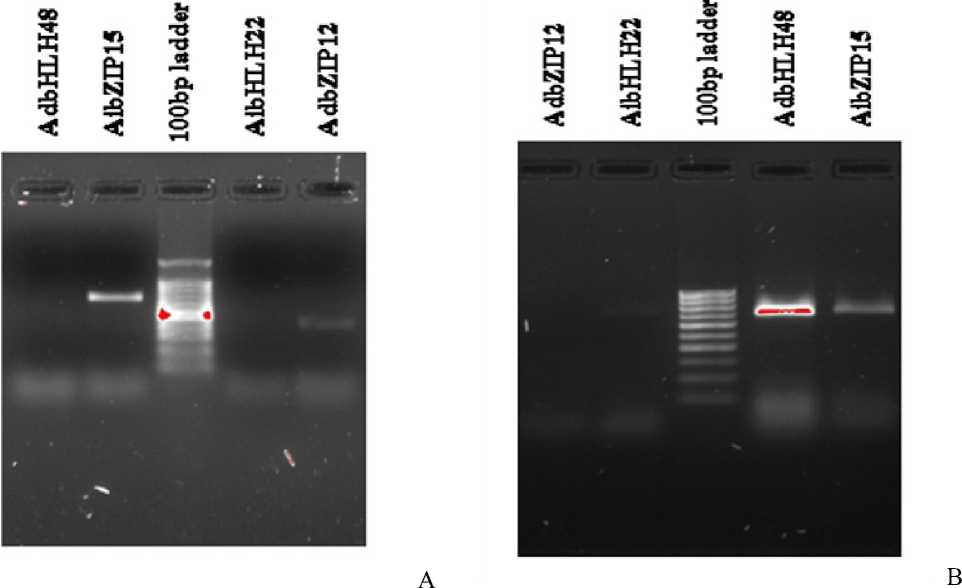
Figure 22 : A) Agarose gel electrophoresis of PCR amplified products under high temperature stress. B) Agarose gel electrophoresis of PCR amplified products under heavy metal stress
Cadmium Stress
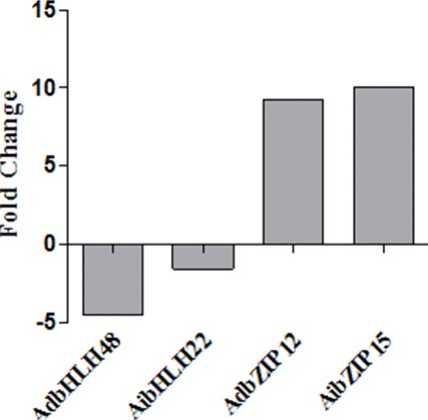
Figure 23: Expression profile of AdbHLH and AibHLH genes obtained by RT-qPCR of Cadmium chloride treated shoot samples.
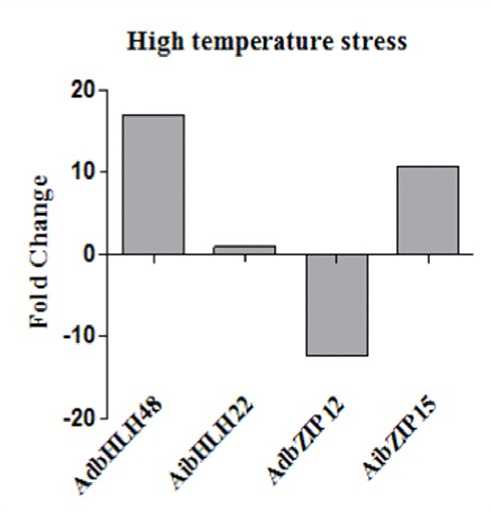
Figure 24: Expression profile of AdbZIP and AibZIP genes obtained by RT-qPCR of high temperature treated shoot samples.
CONCLUSION
Plants growing in their natural habitats are often challenged simultaneously by multiple stress factors, both abiotic and biotic. Several families of plant TFs play significant roles in translating abiotic stress signals into changes in gene expression. So far, research into TFs that regulate abiotic stress responses has mainly focused on single TFs and their isolated function. The present study comprising genome- wide analysis of bHLH and bZIP TFs, detailed protein features, motif composition, multiple sequence alignment, phylogenetic analysis, gene structure, chromosomal location and expression analysis under high temperature and heavy metal stress provides valuable data for further functional analysis to develop multi stress tolerant varieties in groundnut.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.
Список литературы Genome-wide analysis of BHLH and BZIP transcription factors and their temporal expression under abiotic stress conditions in groundnut (Arachis hypogaea L.)
- Agati, G., Biricolti, S., Guidi, L., Ferrini, F., Fini, A., & Tattini, M. (2011). The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. Journal of plant physiology, 168(3), 204-212
- Atchley, W. R., Terhalle, W., & Dress, A. (1999). Positional dependence, cliques, and predictive motifs in the bHLH protein domain. Journal of molecular evolution, 48(5), 501-516.
- Dong, Y., Wang, C., Han, X., Tang, S., Liu, S ., Xia, X., & Yin, W. (2014). A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochemical and biophysical research communications, 450(1), 453-458.
- Ferre-D'Amare, A. R., Pognonec, P., Roeder, R. G., & Burley, S. K. (1994). Structure and function of the b/HLH/Z domain of USF. The EMBO journal, 13(1), 180-189.
- Goding, C. R. (2000). Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes & development, 14(14), 1712-1728.
- Hall, T., Biosciences, I., & Carlsbad, C. (2011). BioEdit: an important software for molecular biology. GERF Bull Biosci, 2(1), 60-61.
- Hu, W., Yang, H., Yan, Y., Wei, Y., Tie, W., Ding, Z., & Li, K. (2016). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Scientific reports, 6(1), 1-12.
- Kilian, J., Peschke, F., Berendzen, K. W., Harter, K., & Wanke, D. (2012). Prerequisites, performance and profits of transcriptional profiling the abiotic stress response. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 1819(2), 166-175.
- Luo, X. P., Zhao, H. X., Xue, J., Li, C. L., Chen, H., Park, S. U., & Wu, Q. (2016). Cloning of two basic helix-loop-helix transcription factor genes from Tartary buckwheat (Fagopyrum tataricum) and their proteins: regulators of transcription in eucaryotic organisms. Molecular and cellular biology, 20(2), 429-440.
- Nakashima, K., Ito, Y., & Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant physiology, 149(1), 88-95.
- Rejeb, I. B., Pastor, V., & Mauch-Mani, B. (2014). Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants, 3(4), 458-475.
- Simionato, E., Ledent, V., Richards, G., Thomas-Chollier, M., Kerner, P., Coornaert, D..... & Vervoort, M. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC evolutionary biology, 7(1), 1-18.
- Song, X. M., Huang, Z. N., Duan, W. K., Ren, J., Liu, T. K., Li, Y., & Hou, X. L. (2014). Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Molecular genetics and genomics, 289(1), 77-91.
- Sonnenfeld, M. J., Delvecchio, C., & Sun, X. (2005). Analysis of the transcriptional activation domain of the Drosophila tango bHLH-PAS transcription factor. Development genes and evolution, 215(5), 221-229.
- Sun, X. H., Copeland, N. G., Jenkins, N. A., & Baltimore, D. (1991). Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Molecular and cellular biology, 11(11), 5603-5611.
- Xu, P., Jiang, L., Wu, J., Li, W., Fan, S., & Zhang, S. (2014). Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Molecular biology reports, 41(8), 4899-4909.
- Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: evolution, 33(7), 1870-1874. Ledent, V., & Vervoort, M. (2001). The basic helix-loop-754-770.
- Lindemose, S., O'Shea, C., Jensen, M. K., & Skriver, K. expression under abiotic stress. Turkish Journal of Biology, 40(6), 1192-1201. Massari, M. E., & Murre, C. (2000). Helix-loop-helix


