Genome wide identification and in-silico transcriptional and expression analysis specific protease against biotic stress in plant species
Автор: Dutta Sanjukta, Sharma Arti
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 1 т.18, 2022 года.
Бесплатный доступ
The post-translational modification involves protein degradation which helps to degrade the overexpressed proteins in plants. This includes the process of ubiquitination and deubiquitination where the targets attached with the substrates are cleaved and can play a major role in plant-pathogen interactions. By 26S/proteasome pathway, the pathogens are targeted for the defense of the plant and thus the proteases involved in the process are taken for the study. The study involves the digital expression of the genes which helps to infer the gene expression among various important plant tissues which helps to manifest the responses against the pathogenic stresses. Ubiquitin-specific protease are seen to expressed maximum among the plants species taken for study and thus it can be generalized that these protease genes can help to combat against the biotic stresses in certain tissue specific regions as their own immune response.
Protease, biotic stress, deubiquitin enzymes, ubiquitin/26s proteasome
Короткий адрес: https://sciup.org/143178334
IDR: 143178334
Текст научной статьи Genome wide identification and in-silico transcriptional and expression analysis specific protease against biotic stress in plant species
The study of plant defense system is quite demanding in the field of research and agriculture. In India, scientists are constantly searching the root of defense mechanism in plants to increase the production of various economical crops and harvesting material. This has a huge effect on not only science but also in the field of economy, growth in production and good farming. In the last decade of research, various pieces of literature have been cited on the relationship between plant and pathogen, and thus new and advanced work is in progress in recent times. Plants being a sessile organism need to have more prominent and effectual defense system for their survival and thus the various mechanisms of defense in plants encompass through the complex steps of signaling and apoptotic pathways to overcome through it (CM Pickart, 2003). The defense genes get initiated after the recognition of the foreign pathogens which can harm the plants and invade themselves through various ways (Diaz I., 2018). Protease plays huge role in defense mechanisms as well as in the metabolic changes occurred in plants (Reyes-Turcu et al ., 2009).
Being a necessary and crucial component in several regulations of plants and its pathways, protease has the ability to combat the adverse effects done by the pathogens like bacteria, fungi and viruses. Various proteases have various functions and their own pathways to deal with different stresses (Doherty et al 2002). Pathogens like bacteria have various stages of attacking the target genes of the host plant like, recognizing suitable hosts, evading the barriers of the host organisms and spreading its effect in the specific parts of the organism, the development of the effect and causing bigger symptoms and then the disease gets transmitted to a new host (Donata et al., 2019). The virulence can be different in different pathogens, pests or microorganisms. The virulence generally invades the host defense pathways initially where the symptoms get produced (Dangel J., & J.J.,2001; Vorholt and A.,2012; Colwell and R.,2000). Thus protease helps in the cleavage of peptide bonds in the PTM, where the pathogens also get affected which targets the particular genes of the same plant. Recognition of pathogen, hypersensitive response or the releases of hormones are steps where protease plays important role (Van der Hoorn RAL, 2008).
Proteases in plants are the superfamily of proteins which has a total 2% of coding genes approximately. According to MEROPS peptidase database , Arabidopsis thaliana encodes over 800 proteases and Oryza sativa has about 600 approximately which are further divided into families, subfamilies and clan ( Rowling et al., 2018; Hou et al.,2018). There are two types of enzymes in protease which are endopeptidase an exopeptidase. Exopeptidase acts on its terminal end and can be grouped as aminopeptidase (Which act on N-terminus) and carboxypeptidase (which act on C-terminus). On the other hand, endopeptidase only acts in between the peptide chains and can be grouped on the basis of the active site residue. Thus proteases are categorized into four groups such as Cystine protease, Serine protease, Metalloprotease and Aspartic protease (Grudkowska and Zagdanska,2004). There are 15 families in the cysteine proteases such as PLCPs (C1), Calpains (C2), legumain (C13), USP (C19), etc and they are grouped into 5 clans (Grudkowska and Zagdanska,2004; Van der Hoorn RAL,2008). Serine protease has 14 families which again divided into a group of clans where subtilisin-like proteases are mostly found in plants. Aspartic proteases, on the other hand, have 5 families and further grouped into 2 clans. The most abundant aspartic proteases found in plants are PLAPs, which is an A1 family. And lastly, metalloprotease has 19 families and are categorized into 12 clans (Hou et al.,2018; Van der Hoorn RAL,2008; Moon et al, 2004). Most of these proteases are involved in the defense as they recognize and can combat against most of the stress that plants deal with.
Plants can perform proteolysis which can degrade some regulatory proteins and thus the proteolysis use a multisubunit protease complex known as 26S proteasome along with the covalent attachment of a protein called ubiquitin. This helps in the regulation in plants in the degradation or ubiquitinylation (Dielen et al., 2010). On the other hand, there are few groups of proteins and enzymes which helps in inhibiting the regulation or the process that is deubiquitinating enzymes and thus helps in deubiquitinylation (Hou et al.,2018).
So, the expression of Ubiquitin-specific protease is investigated to perform the comparative analysis for the plant defense from biotic stresses. The plants taken for studies are Arabidopsis thaliana, Chorchorus capsularis, Oryza sativa (indica) and Zea mays . Since these plants are both agronomically and economically important in India, so this study was conducted on these model plants.
MATERIALS AND METHODS
The experimental procedures carried out for the comparative analysis among the model plants ( Arabidopsis thaliana, Chorchorus capsularis, Oryza sativa (indica) and Zea mays ) are done with the help of dry lab techniques and online bioinformatics tools. The analysis study is mainly based on the data of genome sequences which are taken from plant databases. The genes which are taken for the analysis belong to the protease or peptidase family that acts as defense genes in plants against biotic stress like bacteria or fungi. Further methods for the study are being carried out by the following procedures.
Collection of data samples
The whole genome sequences are available on EnsemblPlants . Ubiquitin family gene sequences are searched for each respective plant species (Arabidopsis thaliana, Chorchorus capsularis, Oryza sativa (indica) and Zea mays) in EnsemblPlants. From there, the defense genes like ubiquitin-specific protease, cysteine protease and other defense gene sequences are then taken and the FASTA files were downloaded from Uniprot .
General pathway analysis
Since protease plays a major role in various mechanism of plant, thus the pathways of each plant species are checked with respect to the pathway against the biotic stimuli. The general biological pathways are studied in Plant Reactome Gramene Pathways
which is an open source and curated pathway database. The pathway event of recognition of fungal and bacterial pathogens and immunity responses are visualized in the pathway browser and thus analyzed accordingly. In pathway analysis, all the events along with the end products are observed and investigated in each family and further analysis can be done with it.
Gene expression of the data in the sample plants
The datasets of the downloaded genes of respective model plants are further taken for the gene expression analysis. The gene expression of Arabidopsis thaliana is checked in the TAIR database . The rest three species of plants have infrequent information in their respective databases, thus their gene expressions are further investigated in EST database (which consist of database of GenBank, EMBL, DDBJ sequences from EST divisions) and thus the gene expression information was enquired.
RESULTS
The dataset of genes includes 14 genes of Arabidopsis thaliana, 21 genes of Oryza sativa indica, 24 genes of Zea mays and 28 genes of Chorchorus capsularis (Table 1) . The genes from the protease families that are collected as datasets are analyzed through the pathway analysis and the digital expression in plants-pathogens interaction. In this study, the specific proteases mainly include the DUB genes which include the ubiquitin specific proteases, ubiquitin proteases, SUMO proteases, peptidase C19 and other genes from ubiquitin family and cysteine proteases in each plant species which are taken.
The bacterial and fungal recognition undergoes few events where it forms the chitin complex which activates CERK1 by forming another complex. Another bacterial and fungal recognition activates Rac1, which occurs in all these species. The cell then undergoes ubiquitination of SPIN6 (SPL11 interacting protein 6) which is a GTPase activating protein. SPIN6 interacts with SPL11 both in vivo and in vitro and thus can be observed in the event called Phosphorylation of SPL11 of the pathway. These events of interaction occur via 26S/Ubiquitin proteasome pathway. Meanwhile the recognition of the pathogen triggers an oxidative burst which generates superoxide dismutase. In this pathway, RBOHB (Respiratory burst oxidase homologs protein B) is responsible for the oxidative burst which helps in the defense related NADPH oxidase interaction with Rac1 component in the cell. As a result, oxygen and Hydrogen peroxide is released as Reactive Oxygen Species (ROS).
The pathway starts from the fungal and bacterial recognition by LYP4 and LYP6 followed by the activation of Rac1 protein. LYP6 and LYP4 are Lysin Motif Containing Proteins which is involved in the immunity process of plants. The Rac1 GTPase:GDP long with GTP gives Rac1GTPase:GDP complex and releases GDP
In this case, two types of ubiquitination can occur. One is ubiquitination of SPIN6 which takes place at nucleoplasm and ubiquitination of SDS (SPL11 cell death suppressor 2) which takes place in both plasma membrane and cytosol. In case of SDS2, the ubiquitination changes the homologues of phosphorylated SDS2 to homologues of ubiquitinated SDS2 after regulating and interacting with homologues of phosphorylated SPL11. This helps to regulate the immunity as well as in the phosphorylation of RBOHB (NADPH oxidase) to generate superoxide dismutase.
The pathway starts with chitin which is the first signal of the pathogen to cause the cell infection. Chitin is recognized by both LYP4 and LYP6 which forms chitin: LYP4 and chitin: LYP6 complex respectively. Fungal recognition also activates CERK1 (receptor like kinase) that helps in chitin elicitor signaling or chitin complexes in Arabidopsis thaliana . Then again the activation of Rac1 takes place via bacterial and fungal recognition forming Rac1GTPase: GTP and GDP. The event is then followed by Ubiquitination of SPIN6 which has the interacting protein (OsI_27182 gene) having a gene entity of BGIOSGA026315 with reference to EnsemblPlant database
ary?db=core;g=BGIOSGA02 6315;r=7:25704438-
25713192;t=BGIOSGA026315-TA) after interaction of phosphor-phosphorylated SPL11. Then the production of ROS is done by the RBOHB for further induction of defense gene expressions.
The pathway for Zea mays is similar to that of Oryza sativa indica where the pathogen recognition activates Rac1 and CERK1. But the entities for the Ubiquitination become different as the Homologues of SPIN6 after interaction with the phosphorylated SPL11 gives homologues of SPIN6 ubi. Then after the activation of Rac1, the phosphorylation of RBOHB is done by SDS2 complex which helps in releasing the product of ROS.
In case of Jute, the pathway does not follow any specific pattern for recognition of pathogen. So they are mainly involved in the pathway of Ubiquitination and ROS production. The phosphor-phosphorylated SPL11 helps in the ubiquitination of homologues of SPIN6 to homologues of SPIN6 ubi, in the nucleoplasm, and the phosphor-phosphorylated RBOHB helps in the generation of superoxide followed by the production of superoxide dismutase. The catalyst involves the superoxide dismutase copper chaperone activity of the gene CCACVL1_04695.
It can be seen that all four plant sample have shown different level of expression (Supplementary Table 1). In case of Arabidopsis thaliana, 8 genes are found to be expressed in root and flower, 7 genes are expressed in leaf, carpel and seeds, 6 genes are found in the inflorescence, 5 genes are found in petal, sepals and shoot and less than 5 genes are found in rest of the tissues like stem, endosperm and embryo (Fig. 1). Genes are mostly seen to be ubiquitin specific protease and cysteine proteases. In Zea mays, Chorchorus capsularis and Oryza sativa indica, the gene expression in specific tissue parts was less that other sample plants as most of the genes get expressed globally. In Zea mays, 3 genes have seen to be expressed in endosperm and 1 gene is expressed in inflorescence, root and embryo (Fig. 2). Seven genes has been expressed in leaf of Chorchorus capsularis, 4 genes get expressed in root and less than 4 genes are expressed in flower, callus, stem, shoot, and seed (Fig. 3). And lastly, in Oryza sativa indica , genes are expressed mostly in roots, and less than 4 genes are expressed in leaf, flower, endosperm and embryo (Fig. 4). Rice has minimum or no protease gene expression in parts like stem, shoot, and inflorescence.
Table. 1. Comparative pathway analysis of Arabidopsis thaliana, Oryza sativa indica, Zea mays and Chorchorus
|
capsularis. |
||||||||
|
Pathway Steps |
Arabidopsis |
Oryza sativa |
Zea Mays |
Chorchorus capsularis |
||||
|
Input |
Output |
Input |
Output |
Input |
Output |
Input |
Output |
|
|
Chitin |
Chitin |
|||||||
|
Fungal pathogen |
(infection) |
Chitin:LYP4 |
(infection) |
Chitin:LYP4 |
||||
|
rec step1 LYP4 |
LYP4 (protein) |
(complex) |
LYP4 (protein) |
(complex) |
||||
|
Chitin |
Chitin |
|||||||
|
Fungal pathogen |
(infection) |
Chitin:LYP6 |
(infection) |
Chitin:LYP6 |
||||
|
rec step1 LYP6 |
LYP6 (protein) |
(complex) |
LYP6 (protein) |
(complex) |
||||
|
Fungal recognition step2 LYP6 chitin |
CERK1: |
Chitin:LYP6 |
CERK1: |
Chitin:LYP6: |
||||
|
RacGEF1 (complex) |
: phospho-CERK1: RacGEF1 |
RacGEF1 (complex) |
phospho-CERK1: RacGEF1 |
|||||
|
activates CERK1 |
Chitin:LYP6 |
Chitin:LYP6 |
||||||
|
(Complex) |
(Complex) |
|||||||
|
Fungal |
Rac1GTPa |
Rac1GTPa |
Rac1GTPase: |
Rac1GTPa |
Rac1GTPas |
Rac1GTPase |
||
|
recognition by |
se: GDP |
se:GTP |
GDP |
se:GTP |
e: GDP |
:GTP |
||
|
LYP6 step 3 activation of |
(complex) |
(complex) |
(complex) |
(complex) |
(complex) |
(complex) |
||
|
Rac1 |
GTP |
GDP |
GTP |
GDP |
GTP |
GDP |
||
|
Bacterial |
Rac1GTPa |
Rac1GTPa |
Rac1GTPase: |
Rac1GTPa |
Rac1GTPas |
Rac1GTPase |
||
|
pathogen recognition by |
se: GDP |
se:GTP |
GDP |
se:GTP |
e: GDP |
:GTP |
||
|
LYP4 step 3 activation of |
GTP |
GDP |
GTP |
GDP |
GTP |
GDP |
||
|
Rac1 Bacterial |
Rac1GTPa |
Rac1GTPa |
Rac1GTPase: |
Rac1GTPa |
Rac1GTPas |
Rac1GTPase |
||
|
recognition by LYP6 |
se: GDP |
se:GTP |
GDP |
se:GTP |
e: GDP |
:GTP |
||
|
step3activation of Rac1 by LYP6 |
GTP |
GDP |
GTP |
GDP |
GTP |
GDP |
||
|
2xCEBiP: |
Chitin: |
2xCEBiP: |
Chitin: CEBiP: |
|||||
|
Fungal |
Chitin |
CEBiP: |
Chitin |
|||||
|
recognition step 2 CEBiPchitin activates CERK1 |
(complex) CERK1: |
PhosCERK 1: RacGEF1 |
(complex) CERK1: |
PhosCERK1: RacGEF1 |
||||
|
2XRac1GT Pase: |
RacGEF1 |
(complex) |
RacGEF1 |
(complex) |
||||
|
Fungal recognition Step 3 activation of Rac1 by Chitin |
GDP |
2XRac1GTPa se: GDP GTP |
GDP |
2XRac1GTP ase: GDP GTP |
GDP |
|||
|
GDP GTP |
||||||||
|
Rac1GTPa |
Rac1GTPa |
Rac1GTPase |
||||||
|
se: GTP |
se: GTP |
: GTP |
||||||
|
Fungal recognition by LYP4 step 3 activation of |
Rac1GTPa se: GDP GTP |
GDP |
Rac1GTPase: GDP GTP |
GDP |
Rac1GTPas e: GDP GTP |
GDP |
||
|
Rac1GTPa |
Rac1GTPa |
Rac1GTPase |
||||||
|
Rac1 |
se: GTP |
se: GTP |
: GTP |
Homol |
||||
|
Ubiquitination of SPIN6 |
Homologu es of |
Homologue s of SPIN6 |
BGIOSGA026 315 (Protein) |
BGIOSGA0 26315 |
Homologues |
Homologues |
ogues of |
Homologu es of |
|
SPIN6 |
ubi |
(SPIN) |
of SPIN6 |
of SPIN6 ubi |
SPIN6 |
SPIN6 |
||
|
Homologu |
Homologue |
|||||||
|
Ubiquitination of |
es of |
s of |
||||||
|
SDS2 |
phosphoryl |
ubiquitinate |
||||||
|
ated-SDS2 |
d-SDS2 |
|||||||
|
phosphorylation of SPL11 |
Homologu es of SPL11 |
Homologue s of phosphoryl |
SPL11 |
Phosphophosphoryl ated SPL11 |
Homologues of SPL11 |
Homologues of phosphorylat |
SPL11 |
Phosphophosphoryl ated |
|
ated SPL11 |
ed SPL11 |
SPL11 |
||||||
|
Dephosphorylati on of Rac1GTP |
Rac1GTPa |
Rac1GTPa |
Rac1GTPase: |
Rac1GTPa |
Rac1GTPas |
Rac1GTPase |
||
|
se:GTP |
se: GDP |
GTP |
se: GDP |
e:GTP |
: GDP |
|||
|
GDP |
GTP |
GDP |
GTP |
GDP |
GTP |
|||
|
Phospho- |
Homologues |
|||||||
|
Phosphorylation |
RBOHB |
phosphoryl |
RBOHB |
of |
||||
|
of RBOHB |
ated- |
phosphorylat |
||||||
|
RBOHB |
ed- RBOHB |
|||||||
|
RBOHB generate |
O2 |
O2- |
O2 |
O2- |
O2 |
O2- |
O2 |
O2- |
|
superoxide in plasma membrane in |
NADPH |
H + |
NADPH |
H + |
NADPH |
H + |
NADP H |
H + |
|
response to biotic stress |
NADP + |
NADP + |
NADP + |
NADP + |
||||
|
Superoxide dismutase |
H+ O2- |
O 2 |
H+ O2- |
O 2 |
H+ O2- |
O 2 |
H+ O2- |
O 2 |
|
cytosol |
H 2 O 2 |
H 2 O 2 |
H 2 O 2 |
H 2 O 2 |
||||
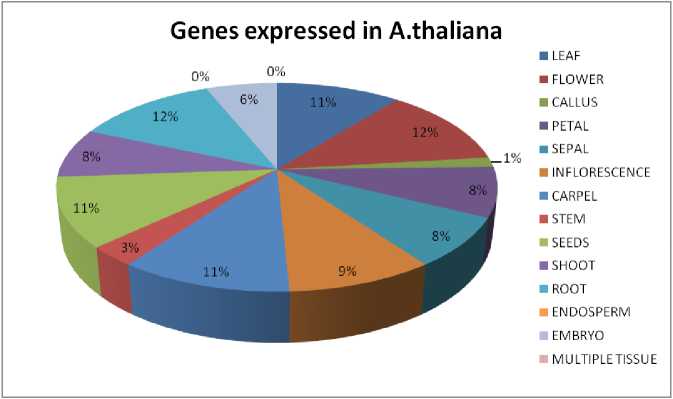
Figure 1 Gene expression in A.thaliana : The diagram shows the amount of gene expressed in the tissue specific region
of the respective plant.
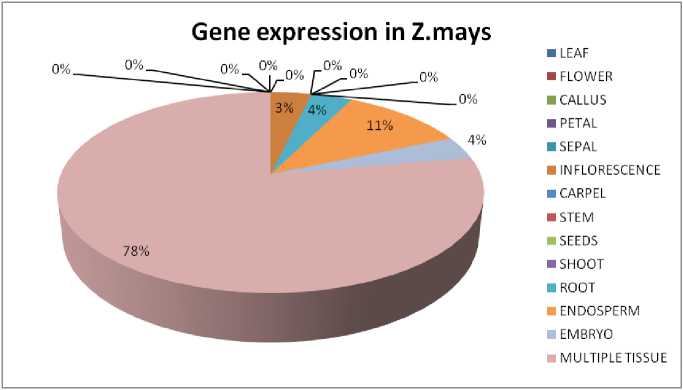
Figure 2 Gene expression in Z.mays : The diagram shows the amount of gene expressed in the tissue specific region of the respective plant.
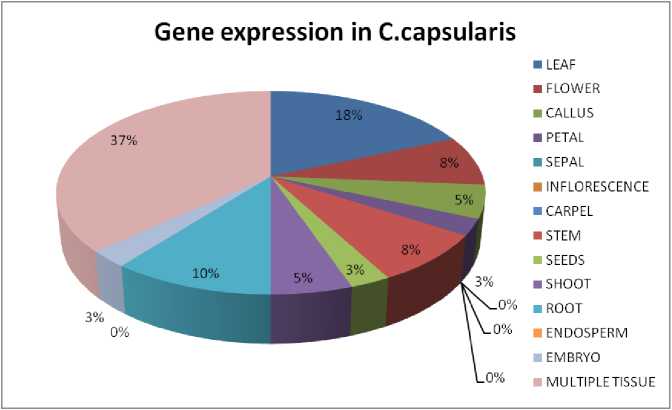
Figure 3 Gene expression in C.capsularis ; The diagram shows the amount of gene expressed in the tissue specific region of the respective plant.
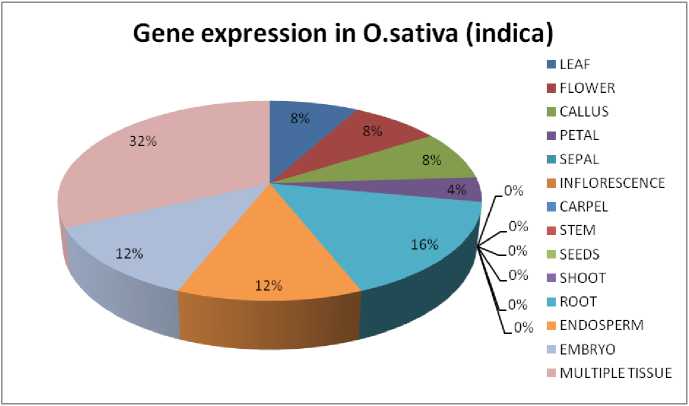
Figure 4 Gene expression in O.sativa (indica); the diagram shows the amount of gene expressed in the tissue specific region of the respective plant.
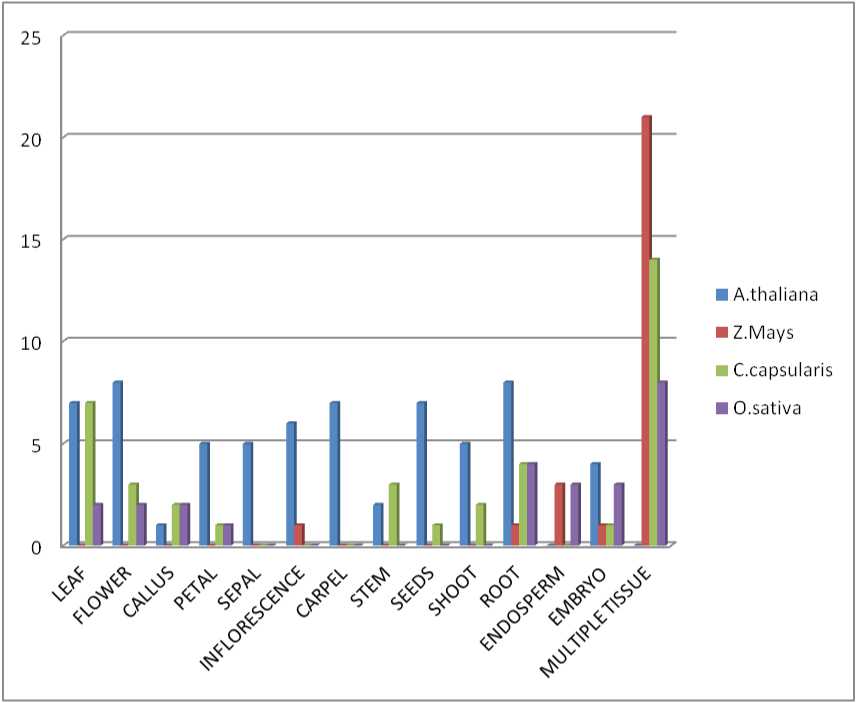
Figure 5 The bar graph represents the comparative gene expression level of proteases in different parts of the plants among the plant species under study.
Most of the data are based on the expression of multiple tissues in the plant where we get a global gene expression (Fig. 5). Oryza sativa indica expresses least level of protease with respect to the other species taken for the study and thus the pathogen interaction can be observed to be more among the plant species under study. The root and callus of rice has a significant level of expression where the pathogenic interaction can be prevented by the plant immune system. Unlike A. thaliana, rice has a better level of gene expression in endosperm. On the other hand, the jute leaf has a high level of expression in leaf and root, but in case of corn, the leaf has the least protease gene expression. The highly expressed gene in Arabidopsis is At1g09730 which encodes as cysteine proteinases superfamily protein. This gene is expressed in most of the tissues in A. thaliana, except in endosperm and embryo. The least expression of proteases was predicted in endosperm. The parts of flowers also have a very less amount of expression in among plant species. Genes like CCACVL1_19617, CCACVL1_23281, CCACVL1_29348, CCACVL1_30294 and CCACVL1_16581 encodes for peptidase C19 are mostly expressed in various tissues of Jute. Other genes like UBP15, UBP27, ULP1B, and ULP2A are also seen to be expressed in most of the tissues, which belongs to the ubiquitin family and thus helps in the plant-pathogenic interaction via 26S/ubiquitin proteasome pathway. It can be observed that in Arabidopsis thaliana, most of the genes are expressed in tissue specific region, where ubiquitin-specific protease expression level is elevated in particular regions like leaf, flower, root, stem, and other specific parts of the plant for their defense system against pathogens.
DISCUSSION
The pathway analysis have shown a series of events which helps in the expression of proteases in response to biotic stress (Erika I and NagelMK, 2014). As pathogen invades the cell, the events of pathogen recognition are initiated. In Oryza sativa indica and Zea mays, Chitin acts as the initial component as it acts as the invader for the cell which in turn acts as a signal for the cell to initiate the recognition events. The generation of ROS helps in the induction of defense genes through signaling pathways of Jasmonic Acid, Salicylic acid or Ethylene (Yaeno et al., 2008; Thines et al., 2007; Binder et al., 2007). The genes which are identified belongs to Ubiquitin-specific proteases, cysteine proteases, SUMO proteases, ubiquitin-like proteases and genes from ubiquitin family, has a certain expression level in plant tissues across the species and this expression can be obtained from the EST databases (Davis et al., 2015). Since Biotic stress signaling pathway is a complex process, hence the expression is observed to be different in each plant tissue.
In Arabidopsis thaliana the digital expression was taken from its TAIR. Most of the identified genes were expressed in root, flower (including young flower) and leaf. It has been analyzed that in these parts of the plant has the maximum level of expression of protease which can cleave most of the peptide bonds in the cell for the regulation and defense mechanism. Thus it can be interpreted that in these area, pathogen defense interactions are in maximum amount compared to the other parts of the plants. So from the result we have inferred that the root, leaf and flower are more exposed to pathogen.
Since the database of the plant species like Oryza sativa, Zea mays and Chorchorus capsularis do not have a curated and proper set of EST database available, so the expression is checked from NCBI EST database for performing cross-species comparisons. Digital expression analysis also has some drawbacks which is the availability of large unbiased cDNA libraries from the tissues of interest, for which the resolution can be low (Fei et al., 2004). Hence, during EST database information analysis, the cDNA library is often taken for the whole plant analysis or rather multiple tissues taken at a time in large scale. In case of Corn, ubiquitinspecific protease genes play a huge role in the expression, and so it can be interpreted as the deubiquitination to be the key role. Thus with ubiquitin specific protease gene helps the plant in the recognition of the pathogens followed by the hypersensitive response (HR) and transcriptional reprogramming in the infected region and thus can help in the restriction of the pathogenic spread (Zhou et al., 2017).
In case of Corn, few genes are expressed in the endosperm, which help the plant growth and development as risk of pathogenic activity can destroy the whole seed if the damage persists. Since expression is not in specific region in corn, hence the risk of pathogenic entry can be random. The global gene expression may not always get expressed in huge amount, which can allow the pathogen to enter the plant via various routes like tassel, roots, leaves or flowers.
On the other side, Jute is long, resilient and tender plant which has a very high economical value and thus the defense mechanism must be studied. It has been observed that protease expression is higher in leaves of Jute plant. Thus the plant has a vital defense region where the defense is required for the effective function of the plant. Like Arabidopsis, protease also gets expressed in the root part, which helps in the defense against the soil-borne diseases. The stem of the Jute is seen to have a moderate level of defense where genes of ubiquitin-protease family is getting expressed and it is highly essential because of its extreme economic value for fibers and protecting them from the air-borne diseases and biotic stresses (Devoto et al ., 2002).
In case of Rice, the expression of protease genes was less compared to the other three sample plants taken for the study. The expression is mostly occurring globally. The expression was observed in root, embryo, leaf and seedling mostly. Rice is one of the most important crops and consumed by more than half a population of India. Hence the defense system is quite important in rice where the expression in the seedling helps in the developmental growth of the plant. Diseases like rice blast caused by fungal infection in leaf can cause the plant to produce spot like symptoms and speed the damage in whole plant gradually. Thus the expression of genes in the leaf part is important and effective against the fungal pathogens. Other disease like sheath blight, which is also a fungal pathogen can invade the plant via roots as it is a soil-borne disease. Hence the expression in roots can be potent with respect to the immunity of the respective plant.
From this it can be concluded that the most common gene which act against the biotic stress in plants are UBPs which belongs to DUBs and the plants have their own defense system against pathogen to sustain in the environment (Lu et al., 2011). There are various risks of disease caused due to pathogen invasion which can be quite harmful for economically important plants. So the elucidation of digital expression in specific parts of these four plants can focus in understanding and dealing the agronomical challenges and can be an effective approach in improving the crop production by working on the specific gene regulation in the specific parts and also focusing on those parts where the gene expressions are least, so that plant can have a better immune system against most of the biotic stresses.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.
Список литературы Genome wide identification and in-silico transcriptional and expression analysis specific protease against biotic stress in plant species
- Binder, B., Walker, J., Gagne, J..... Bleeker, A. A. (2007). The Arabidopsis EIN3 Binding F-Box Proteins EBF1 and EBF2 have distict but overlapping roles in ethelene signaling . The Plant Cell,19: 509-523.
- Colwell, R.R. (2000). Viable but nonculturable bacteria: A survival strategy. J. Infect. Chemother 6: 121-125.
- Dangl J, J. J. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826833.
- Davis, M. I., & Simeonov, A. (2015). Ubiquitin specific proteases as druggable targets. Drug Target Rev., 60-64.
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, ... J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin ligase complex in Arabidopsis. The Plant Journal,32: 457-466. Diaz, I. (2018). Plant defense genes aginst biotic stresses. International Journal Of Molecular Sciences. 19(8), 2446
- Dielen, A.-S., Badaoui, S., Candresse, T., & GermanRetana, S. (2010). The ubiquitin/26S proteasome system in plant pathogen interaction: a never ending hide and seek game. Molecular Plant Pathology, 293-308.
- Doherty, F.J., Dawson S., & Mayer, R.J. (2002). The ubiquitin proteasome pathway of intracellular proteolysis. Essays Biochem, 51-63.
- Fei Zhangjun; Xuemei Tang; Rob M. Alba; Joseph A. White; Catherine M. Ronning; Gregory B. Martin. (2004). Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. The Plant Journal 40, 47-59.
- Figaj D.; Ambroziak P.; Przepiora T. and Skorko-Glonek J.. (2019). The role of protease in virulence of plant pathogenic bacteria. International Journal of Molecular Sciences, 20: 672.
- Grudkowska, M., & Zagdanska, B. (2004). Multifunctional role of plant cystine proteinase. Acta Biochemica Polonica 51: 609-624.
- Hou S, Jamieson P. and He Ping. (2018). The cloak, dagger and sheild: protease in plant-pathogen interactions. Biochemical Journal 475: 2491-2509.
- Isono E., Nagel. M.-K. (2014). Deubiquitylating enzymes and their emerging role in plant biology. Front Plant Sci 5: 56.
- Lu, D., Lin, W., Gao, X., Wu, S., Cheng, C., Avila, J. ... & Shan, L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science, 332(6036): 1439-1442.
- Moon J.; Parry G.; Mark E. (2004). The Ubiquitin-Proteasome pathway and plant development. The Plant Cell, 16: 3181-3195.
- Pickart. C.M.; (2003). Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503-533.
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D; Huang, X.; Bateman, A.; FInn, R.D. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acid Res., 46: D624-D632.
- Reyes-Turcu, F. E., Ventii, K. H., & Wilkinson, K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual review of biochemistry, 78: 363-397.
- Thines, B., Katsir, L., Melotto, M.....J. (2007). JAZ repressor proteins are targets of the SCF complex during jasmonate signalling. Nature, 661-665.
- Van der Hoorn; R.A.L. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu.Rev. Plant Biol., 191-223.
- Vorholt, J.A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10: 828-840.
- Yaeno, Iba, T. a., & K. (2008). BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiology 148: 1032-1041.
- Zhou, H., Zhao, J., Cai, J., & Patil, S. B. (2017). Ubiquitin-specific protease function in plant development and stress responses. Plant Mol Bio 94: 565-576.


