GroEL/GroES mechanism of action and formation of complexes during reaction cycle: a matter of debate
Автор: Kundu Sutrisha, Raychaudhuri Vivek, Saha Sudipa
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 3 т.21, 2025 года.
Бесплатный доступ
The bacterial chaperonin GroEL alongwith its cochaperonin GroES is the paradigmatic molecular chaperone machine for protein folding. Most bacterial proteins require the GroEL chaperonin for proper folding. This review aims to discuss the types of reaction cycles of the GroEL/ES chaperone complex depending upon the concentration of substrate proteins, ATP, and certain ions, through formation of different kinds of complexes. The molecular mechanisms behind formation of these complexes have also been highlighted. The GroEL has been found to undergo the asymmetric and symmetric cycles of protein folding depending on the presence and absence of substrate proteins through formation of different complexes which occur by any of the three mechanisms: active cage model, passive cage model, or iterative annealing model.
Groel, chaperone, protein folding, atp, reaction cycle, substrate
Короткий адрес: https://sciup.org/143184729
IDR: 143184729
Текст обзорной статьи GroEL/GroES mechanism of action and formation of complexes during reaction cycle: a matter of debate
All cellular processes essentially require proteins. Translation of mRNA on ribosomes produces polypeptide chains, which then undergo various post-translational modifications and folding to become active proteins (Anfinsen, 1973). Fundamental insights into the protein folding process was given by Anfinsen's protein cage experiments, which showed that small proteins can refold spontaneously into their active conformations when they are removed from denaturant. The sequence of amino acids itself contains the necessary information required for protein folding but in a highly crowded environment of the cell, they are prone to form non– functional aggregates (Aoki et al. , 2000; Brinker et al. , 2001). Further works demonstrated that, test tube refolding of proteins usually works for single domain, small proteins and those which are not quite akin to the conditions usually encountered inside the cell (Anfinsen, 1973). However, larger protein molecules form aggregates as they fail to reach their functionally active structure. These proteins populate in their kinetically trapped non-native conformations during folding. This exposes their hydrophobic amino acid residues to the solvent and makes them form non-functional aggregates based on a concentration-dependent manner (Aoki et al. , 2000). This aggregation is further exacerbated because of excluded volume effects in the highly crowded environment of the cell, which in turn increases the effective concentrations of macromolecules. However, under in-vivo conditions the protein molecules almost always fold into their functional conformation which is indicative of the fact that cells have evolved various protein folding machineries, called the “Molecular Chaperones” (Anfinsen, 1973), that aid in protein folding by prohibiting improper interactions to occur between the non-native polypeptide chains that ultimately cause aggregation (Aoki et al. , 2000). Most of the essential proteins fold only when assisted by ATP mediated molecular chaperones. Chaperonins are always bimolecular complexes. Different molecular chaperones, like the heat shock proteins (Hsp70, Hsp90) (Chabre, 1990; Chakraborty et al. , 2010), and the chaperonin proteins, assist in protein folding through an ATP-dependent cycle (Chakraborty et al. , 2010).
Chaperonins form a ubiquitous group of molecular chaperones which mediates folding of both nascent polypeptide chains and denatured proteins in an ATP-dependent manner by forming large double-ring (8001000 kDa) complexes (Chiti and Dobson, 2006). They form a unique complex tetradecameric structure that stands out in the proteostasis network. Chaperonins belong to two groups; Group I chaperonin is found in eubacteria (Chiti and Dobson, 2006) and within the chloroplasts and mitochondria (Anfinsen, 1973; Chiti and Dobson, 2006) and Group II chaperonin is found in the cytoplasm of archaebacteria and eukaryotes (Clare et al. , 2009; Clare et al. , 2012). The best characterized among these is the Escherichia coli chaperonin, known as GroEL. The GroEL cannot act alone, in turn it is associated with its cochaperonin or cofactor, GroES (Chang et al. , 2007). It belongs to group I chaperonins. It provides a nano-cage that allows single protein folding. This GroEL/ES chaperonin system is indispensable for the growth of the bacteria at all temperatures (Dobson et al. , 1998) and is directly involved in the folding of most E. coli proteins (Dalton et al. , 2015; Dyachenko et al. , 2013). It also assists folding of many other proteins. GroEL/ES chaperonin thus acts as a passive anti – aggregation cage.
In the in vitro conditions, many denatured proteins fold inside the chaperonin cavity. However, in vivo, GroEL/ES assists in the folding of ∼250 E. coli proteins, which constitue around 10% of the total E. coli cytosolic proteins (Kerner et al., 2005). GroEL/ES is very temperature sensitive in its folding mechanism. Temperature sensitive loss of function mutants have resulted in lots of aggregates, partcilarly for newly synthesized proteins. ∼80 proteins from the prokaryotic system have an obligate dependance on the GroEL machinery for proper folding (Kerner et al., 2005). These proteins usually have complex α/β or α+β structures. TIM barrel proteins have (β/α)8 topology (Kerner et al., 2005) and are mainly dependent on GroEL/ES machinery (Kerner et al , 2005; Azia et al , 2012). They make long range contacts with the hydrophobic amino acids inside the chaperonin cavity and are quickly transformed into kinetically energetically favourable trapped intermediates. These obligate GroEL/ES substrates can easily be identified from their amino acid sequences which, in turn determine their physicochemical properties (Azia et al , 2012; Tartaglia et al , 2010).
Just like living organisms, Darwinian selection is applicable for proteins also. Protein evolution for mutant proteins is based on this selection. However, mutations mostly destabilize the protein structure or cause accumulation of folding intermediates (Zeldovich et al. , 2007). The GroEL/ES system then comes into picture allowing the structural evolution of these mutated proteins by transferring the deleterious effects of mutation to appropriate folding and stability (Williams and Fares, 2010). For the most abundant enzyme Rubisco (ribulose bisphosphate carboxylase/oxygenase), its evolution is attributed to this chaperonin complex (Durão et al , 2015). The large subunit of Rubisco is an obligate substrate for GroEL, which satisfies the notion that compulsory chaperonin dependence slows down the evolutionary process (Liu et al , 2010). Thus, GroEL dependent protein have a slower evolutionary rate. On the other hand, recently it has been revealed that chaperonins also accelerate evolution (Takemoto et al , 2011). Although contrasting, this notion is valid for those proteins that use GroEL/ES system only for a short duration during their evolution. When these proteins get mutated and are positively selected, further secondary mutations make them GroEL independent thereby improving their folding efficiency, compactness and stability. Furthermore, mostly the orthologs of the obligate SPs of GroEL have evolved to become less chaperonin dependent. For example, the TIM barrel protein N-acetylneuraminic acid aldolase (NanA) is GroEL dependent in E. coli but its ortholog in the bacterium Mycoplasma Synoviae has evolved to become GroEL independent because it lacks the chaperonin complex (Georgescauld et al , 2014).
Structure of GroEL, GroES, and GroEL/ES complex
Electron microscopic (EM) images, followed by crystallographic analysis of GroEL, and GroEL/ES complex, have provided a detailed structure of the chaperonin complex. GroEL is a cylindrical double- ring structure, composed of a homo-oligomer having 14
identical subunits of size 57 kDa that are arranged in two heptameric rings. The two rings have a large central solvent filled cavity in between. Each GroEL subunit contains three domains: an apical domain (residues 191–376), an equatorial ATP-binding domain (residues 6–133, 409–523), and an intermediate hinge domain (residues 134–190, 377–408) (Ellis, 2001; Saibil et al. , 2013). The apical domain attaches to GroES and nonnative substrate proteins (SP). Its cavity facing surface is lined on the outer surface with hydrophobic amino acids that facilitate SP binding. These hydrophobic amino acids are exposed mainly on the surfaces of helices H (residues 233–243) and I (residues 255–267). Nonnative SP is bound in the inner channel along the edge. The hydrophobic clusters provide additional sites for a proper binding of the SP on the chaperonin surface. The equatorial domain, being largest of its kind in the GroEL subunit, is involved in both intra- and inter-ring interactions. The N-terminal 5 residues and the C-terminal 25 residues (ending with four Gly-Gly-Met repeats) are located in the central cavity of the equatorial domains, thereby occluding the passage between cavities (Braig et al. , 1994). The inner surface of the central cavity in the upper part of the equatorial domain contains the ATP/ADP binding site. The intermediate domain acts as a bridge between the apical and the equatorial domains of each GroEL subunit and aids in transferring the ATP-mediated conformational changes from the equatorial domain to the apical domain while interacting with ligands, through nucleotide binding and hydrolysis (Saibil et al. , 2013). The two GroEL rings are arranged in a staggered configuration (stacked back-to-back) such that each subunit of a ring contact with two subunits in the other GroEL ring. Thus GroEL, a double ring homo-heptameric structure with ATPase activity, alternates the GroES-sealed foldingactive rings during the reaction cycle which is allosterically regulated by the GroEL ATPase.
GroES is a single dome-shaped heptameric ring made up of 10 kDa subunits. It sits like a cap on one or both the GroEL rings in a nucleotide-dependent manner, forming the cavity where the SP is captured for folding. The GroES subunit has a flexible loop structure made up of 22 amino acid residues that in turn interacts with helices H and I of the apical domain of GroEL rings. Binding of GroEL with GroES involves extensive en bloc domain movements in the GroEL subunit. The chaperonin cage thus formed can assimilate proteins upto 60 kDa. The positive cooperative binding of ATP to each of the seven subunits of the GroEL ring induces binding of GroES by causing 90° clockwise and 60° upwards circular motion in the apical domain, centred around a fulcrum point at Gly192 and Gly375 residues of the intermediate domain (Anfinsen, 1973; Ellis, 2001; Ellis and Minton, 2006). Hydrophilicity is noted in the inner wall of the complex but the hydrophobic sites for binding SP remain buried inside the complex, providing a conducive environment for protein encapsulation (Ellis and Minton 2006). This shows how the group II chaperonins (example: GroEL/ES) have an in-built lid or cap mechanism that facilitates partial encapsulation of large native multidomain proteins that cannot be encapsulated entirely in the folding cavity.
The GroEL/ES chaperonin mediated protein folding cycle
-
• Folding in the absence of substrate protein
Regardless of whether the SP is present or not, the chaperonin complex undergoes ATP-regulated binding and release (Fig. 1). In the apo-state (without any bound SP), there is an equilibrium of the GroEL subunits, between the high affinity for ATP or R-state and the low affinity for ATP, the T-state (Fujiwara et al. , 2010). As a result, there is a complex cooperativity network controlling the protein folding cycle where there is positive cooperativity when ATP binds with GroEL rings but between the two rings, there exists negative cooperativity (Grallert and Buchner, 2001). When the seven ATP hydrolyzes to ADP in the cis-ring, it undergoes a conformational change and the negative inter-ring cooperativity decreases. So, a new ATP binds to the trans-ring. Meanwhile, the cis-ring loses its ADP and GroES and another cis-ring is formed upon subsequent GroES binding to the trans-ring to complete the cycle.
-
• Folding in the presence of substrate protein
The GroEL/ES chaperonin protein folding cycle undergoes different conformational changes in the presence of SPs (Fig. 2). The trans-ring of GroEL without the nucleotide, accepts the non-native SP which binds to the exposed hydrophobic cleft in the apical domains (Anfinsen, 1973). The hydrophobic cleft is exposed between helices H and I and provides a promising opportunity for binding small lipophilic proteins. Subsequently, in an ATP-dependent manner, GroES associates with that same ring (cis-ring). The complex undergoes a conformational change which can be detected by fluorescence resonance energy transfer (FRET). Furthermore, binding of ATP causes an upward clockwise movement in the apical domain, thereby displacing the SP to the dome-shaped GroEL folding cavity (Grason et al., 2008; Grason et al., 2008). Further conformation compaction expands the tightly bound SP, in turn releasing the SP that are weakly bound. This forms an intermediate complex where both GroES and SP are bound and encapsulated simultaneously to GroEL. Folding proceeds in this longest-lived of the GroEL states (Gupta et al., 2014). ATP mediated expansion occurs simultaneously with SP mediated conformational compaction. This prevents SP from being prematurely dissociated into the solution. Although energetically unfavourable, the encapsulation occurs because of the conformational change arising from ATP binding. After ~10s, the seven ATP hydrolyzes to ADP in the cis-ring (Gupta et al., 2014). The ADP still remains bound to GroEL and prevents a second GroES and a new ATP from binding to the opposite trans-ring. The GroES is released from the cis-ring because affinity between the chaperonin and its cofactor reduces due to the loss of γ-phosphate. However, as the non-native SP protein concentration in the vicinity increases, it promotes ADP/ATP exchange, which in turn facilitates new ATP to bind to the trans-ring (Hartl and Hayer-Hartl, 2009; Hartl, 2011). ADP and the folded SP are released from the cis-ring. A new cis- ring is formed that initiates a second ATPase cycle. An incompletely folded SP will be captured by another GroEL molecule to initiate a renewed folding cycle. With rise in temperature from 25 to 37°C, the time required for protein folding by the chaperonin reduces. At 25°C, approximately 6s is essential for protein folding while folding requires just 2s at 37°C. However, at higher temperature, the time increases and the folding also becomes improper.
“Football” and “Bullet” Complexes form during Reaction Cycle
Electron microscopic analysis of GroEL/ES holochaperonin complex tetradecamer isolated from the thermophilic bacterium, Thermus thermophilus revealed the “bullet” and “football” complexes for the first time (Hayer-Hartl et al. , 1999; Hayer-Hartl et al. , 2016). The 1:1 asymmetric “bullet” complex forms when one GroES is bound by only one GroEL. The 1:2 symmetric “football” complex forms when two GroES is bound by one GroEL only (Horovitz and Willison, 2005; Horwich et al. , 2007). When ATP and other nonhydrolyzable ATP analogues are present, both the complexes are formed Horwich, 2011) while when ADP is present alone, only the asymmetric complex forms (Iizuka and Funatsu, 2016). The symmetric complex forms depending on the [ATP/ADP ratio and the potassium ion (K+) concentration (Ishii et al. , 1992). “Bullet” complex is the predominant during protein folding cycle because mostly one GroES binds to one GroEL at a time during the alternate folding ring cycle, promoted by the intricate negative cooperativity of two ring subunits of the GroEL chaperonin. However, the “football” complex was observed using electron microscopy, chemical crosslinking, FRET, and analytical ultracentrifugation (Horovitz and Willison, 2005; Ishii et al. , 1995).
Initially, it was proposed that the football complex is the functional catalytic intermediate of the protein folding cycle but later experiments proved the bullet complex to be the catalytic intermediate. Due to the negative cooperativity between two GroEL rings, it allows only one GroES to bind to one GroEL ring at once, thereby promoting a higher accumulation of the bullet complex compared to the symmetric football complex (Horovitz and Willison, 2005). Similarly, Rye et al. performed an experiment with an ATPase deficient GroEL mutant of D398A (Ala replaces Asp398) (Ishino et al., 2015). D398A can bind ATP but cannot hydrolyze it, so it forms an ‘ATP bullet” (ATP-bound asymmetric GroEL/ES complex). Trans-ring of the “ATP bullet” is unable to bind SP or GroES (Ishino et al., 2015; Kim et al., 2013). Only when the ATP bound to the cis-ring hydrolyzes, can a new SP and ATP bind to the trans-ring. However, further experimentation by Koike Takeshita et al. showed that the mutant GroEL D398A can bind GroES through a conformational change forming a symmetric complex when ATP binds to both rings (Horovitz and Willison, 2005; Kim et al., 2013). This was proved using a singlemolecule assay and it challenged the previous finding. GroES dissociation from GroEL is ATP-dependent and occurs randomly. So, initially, the GroES that associates with GroEL always is not dissociated from the chaperonin before the second GroES dissociates.
The two GroEL rings function sequentially one after the other during protein folding. The “bullet” complex forms first and is the folding active species which eventually forms the “football” complex in the presence of SP. This symmetric complex is the transient intermediate of the reaction cycle. GroES has a high affinity for nucleotide-bound GroEL but the mechanism of GroES release through ADP dissociation is still unknown. Large number of permanently unfolded proteins (eg- reduced lactalbumin) facilitates formation of “football” complexes, as it reduces the negative cooperativity between the GroEL rings (Hayer-Hartl et al. , 2016). This negative cooperativity further persists when the physiological concentration of ATP:ADP is 10:1 (Hayer-Hartl et al. , 2016). Hence, the asymmetric “bullet” complex is the predominant state inside a cell.
Sameshinma et al. later proved through FRET analysis that equal proportions of both complexes were coexisting during the reaction cycle (Kinoshita, 2006). Presence of SP promotes formation of the symmetric complex while ADP prevents accumulation of the complex. Although the SP does not influence the interaction of the first GroES with GroEL, it helps to accelerate binding of the second GroES to GroEL transring by influencing the ATPase kinetics of GroEL (Koike-Takeshita et al. , 2008). As the SP protein concentration in the vicinity increases, it promotes ADP/ATP exchange. The symmetric-asymmetric series reaction is enhanced by raising the concentration of SP (Chang et al. , 2007; Koike-Takeshita et al. , 2008).
In the “football” complex, the cis- and trans-ring are indistinguishable (Chang et al., 2007). So, the concept of negative cooperativity of the two GroEL rings, although appropriate, is incomplete when the SP concentration increases (Chang et al., 2007). Thus, the SP facilitates nucleotide exchange demonstrating the crucial role of the symmetric “football” intermediate in reaction cycle, where both the rings of the symmetric complex aid in folding protein molecules. They lack interring communication and operate parallelly.
Both the football and bullet complexes exist very transiently. Therefore, to study the crystal structures, it was essential to make long-lived complexes. The long-lived football complex was prepared using BeFx in the presence of ATP to produce the (ADP – BeF 3 ) 14 complex, and also the ATPase defective GroEL mutant i.e., GroEL (D52A/ D398A) formed a stable football complex (Taguchi et al. , 1991; Koike-Takeshita et al. , 2014). The crystal structure was solved at 3.7-3.8 Å resolution (Koike-Takeshita et al. , 2014). The football complex has been found to be a copy of two bullet complexes, differing only at the GroEL-ES complex interface and also at the two GroEL rings interface. In the mutant complex, a modified interface was formed between the two GroEL rings through a 7o rotation around the heptamer axis. This hinders the negative cooperativity between the GroEL rings causing a reduced contact surface area, thereby promoting more and more formation of the football complex. Furthermore, the lack of a salt bridge in this football structure contributes to the hindrance of negative cooperativity (Koike-Takeshita et al. , 2014). The mutant GroEL forms the symmetric football complex when both the GroEL rings are occupied with ATP or when the two rings are occupied with ATP and GroES.
The single molecule assay also helped to unravel the protein folding mechanism via the football complex, allowing direct observation and characterization of the different reaction cycle intermediates (Takei et al., 2012). It showed that GroES dissociation is very random and varies whether which GroES: the first one or the second one will dissociate first from the GroEL ring. It rather depends on ATP hydrolysis. The ring where ATP hydrolyzes first, in that ring the GroES dissociates simultaneously. The single molecule assay using GFP demonstrated that both the GroEL rings are simultaneously involved in refolding GFP via symmetric complex. The kinetics of protein folding in both rings being very similar indicated that same reactions can occur parallelly in the two rings as the dissociation of GroES occurs in a very random order (Takei et al., 2012).
Substrate Proteins for GroEL/ES
Being a broad-spectrum chaperone, around 300 E. coli proteins depend on this complex for proper folding. 85% of E. coli proteins ranging from 10 – 150 kDa require the assistance of the chaperonin complex either directly or indirectly for folding (Ewalt et al. , 1997). Mainly the obligate SPs possess the αβ motif, however quite a few unstructured intermediates also associate with GroEL (Tian et al. , 1995). In the case of multidomain proteins, it has been proved that the individual domains first fold followed by subsequent association of the folded monomers which ultimately combine to form an active oligomeric protein. Based on their dependency on the chaperonin complex, the SPs are classified into two types – class I (ATP mediated GroEL/ES assisted folding) and class II (transient binding to GroEL only without the assistance of either ATP or GroES) (Chaudhuri et al. , 2009). Criteria for folding depends upon the nature of the protein, the overall conformation of the protein and the physiological conditions surrounding the protein (Chaudhuri et al. , 2009). Considering RUBISCO, it either requires the entire chaperonin complex or only GroEL for folding based upon the varied physiological conditions. Therefore, RUBISCO is categorized both under class I and class II SPs. Proteins which require the assistance of this chaperonin complex for folding primarily include RUBISCO, aconitase, maltodextrin glucosidase, β galactosidase, methionine biosynthesis enzyme MetE, among others.
The two conditions when GroEL prevents aggregation and promotes folding are – (i) the unfolded polypeptide binds with GroEL which prevents it from associating with other unfolded polypeptides (Fenton and Horwich, 1997) and (ii) the cochaperonin GroES encounters an unfolded polypeptide and forcefully pushes it inside the chaperonin cavity to fold correctly in an isolated environment (Chaudhuri et al., 2009; Fenton and Horwich, 1997). In the first case, GroEL is called holdase since it holds on to the unfolded polypeptide preventing it from other nonproductive interactions while in the second case, GroEL is called foldase as it directly assists in folding by itself (Thirumalai et al., 2003). Further, GroES binding induces such a conformational change in GroEL, that there is almost a doubling in the volume of the folding chamber. This allows even large proteins upto 85 kDa in size to undergo folding inside the chaperonin complex. Without GroES, only small protein molecules can usually fold.
Aconitase, an important enzyme of the Krebs cycle, is 82 kDa in size. If the protein is present in an Hsp60 or Hsp10 deficient environment, it forms insoluble aggregates (Dubaquié et al , 1998). It then requires the assistance of GroEL/ES proper folding. However, due to its large size, GroES alone cannot encapsulate it, so GroES binds to the trans ring of GroEL along with the assistance of ATP which then forms a cavity favourable for accommodating the 82 kDa protein. In E. coli , coexpression of GroEL/ES along with aconitase improves its biological activity through appropriate folding (Chaudhuri et al. , 2001).
β galactosidase, a 116 kDa tetramer falls under the class II SPs as it binds only transiently to GroEL without the involvement of either ATP or GroES (Ayling and Baneyx, 1996). In fact, presence of ATP and GroES reducing the folding efficiency of GroEL for β galactosidase (Chaudhuri et al. 2009). Possibly there can be two proposed mechanisms behind this – 1) when ATP hydrolysis occurs, GroEL undergoes a conformational change that makes it incompetent for binding β galactosidase or – 2) ATP allows folding to occur within 7s but β galactosidase being a large protein requires more time; however, ATP releases the unfolded intermediate by hydrolysis within 15s from the GroEL cavity (Chaudhuri et al. , 2009). They are proposed in vitro mechanisms but the results have not yet been complemented in vivo.
Cnox is a chaperedoxin which acts in association with GroEL to prevent protein aggregation and enhances folding (Goemans et al. , 2018; Goemans et al. , 2018). It acts like a redox quality-control plugin for GroEL. It binds to GroEL via its helix at the C terminus.
When it remains bound to GroEL, it facilitates other proteins binding to GroES to facilitate their folding. It can form different types of disulfide bonds with the bound SPs thereby showing its redox mechanism. It is one of the first chaperedoxins to be discovered, giving an idea that such accessory folding proteins might be present both in prokaryotic and eukaryotic system that transfers substrates for protein refolding inside the chaperonin cavity (Goemans et al. , 2018; Goemans et al. , 2018). Its binding just at the binding site quickly allows protein entry into the GroEL cavity and after GroES binds to GroEL to form the complete chaperonin, Cnox is released. It has recently been identified that the E. coli Cnox can even interact with the mitochondrial Hsp60 of humans (Dupuy et al. , 2023).
Role of denatured protein in the Reaction Cycle
Different groups of scientists performed experiments using different fluorescently labeled variants of the chaperonin to study how denatured proteins affect the formation of symmetric “football” and asymmetric “bullet” complexes for the chaperonin protein folding cycle.
“Football” complex disappears when denatured proteins are absent
As the chaperonin GroEL refolds denatured proteins, the “football” complex disappears gradually. Iizuka et al. used two variants: GroEL-E315C with exposed cysteine residues, and GroES-98C which at the C-terminus of GroES subunits has an extra cysteine residue, labelled using TMR (tetramethylrhodamine) and Cy5 (indicocarbocyanine) respectively, and studied using FRET analysis (Koike-Takeshita et al., 2008). Wild-type variant (wtGroES and wtGroetEL) in presence of the inorganic phosphate analogue beryllium fluoride (BeFx) was used as control, for which the measured FRET efficiency was almost constant for both the symmetric and asymmetric complexes throughout (Langer et al., 1992; Lin et al., 2008). Initially, a value intermediate between the complexes was obtained which gradually decreased and ultimately reached a value nearer to the “bullet” complex in a time-dependent manner (Langer et al., 1992). MDH refolding assay during the reaction cycle showed a result directly correlated with the reduction in FRET efficiency. In the absence of chaperonin complex, the rate of spontaneous folding was much lower for MDH which proved that as denatured MDH is refolding and the amount of denatured MDH decreases, the “football” complex gradually disappears.
“Bullet” complex preferentially forms when denatured proteins are absent
Based on the previous data, Sameshima et al. (2010) further conducted FRET titration experiments using GroEL – E315C labelled with Cy3
(indicocarbocyanine) in the absence of denatured proteins (Lin et al. , 2008). It again showed that in the control setup, irrespective of whether the denatured proteins are present, BeFx led to the formation of both complexes (Langer et al. , 1992; Lin et al. , 2008). On the other hand, when there were no denatured proteins, a saturation ratio for [GroES /[GroEL = 1 was obtained and the “football” complex was not detected, which was further validated using fluorescence correlation spectroscopy (FCS) analysis (Koike-Takeshita et al. , 2008). Thus, only “bullet” complex forms when denatured proteins are absent.
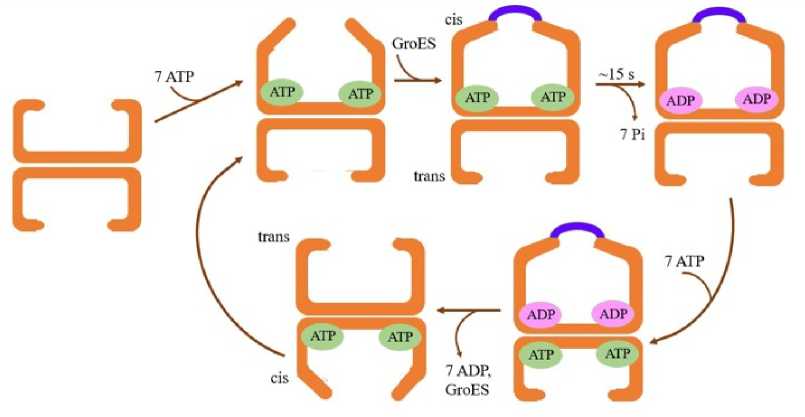
Figure 1. Asymmetric reaction cycle when the substrate protein is absent
SP GroES.
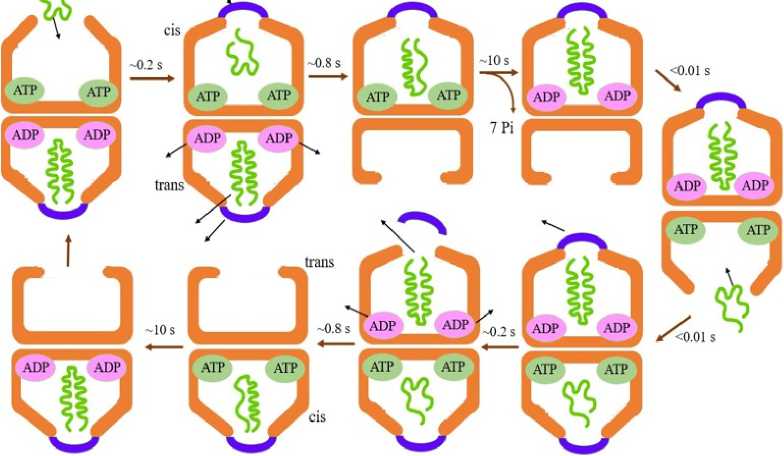
Figure 2. Symmetric reaction cycle when the substrate protein is present
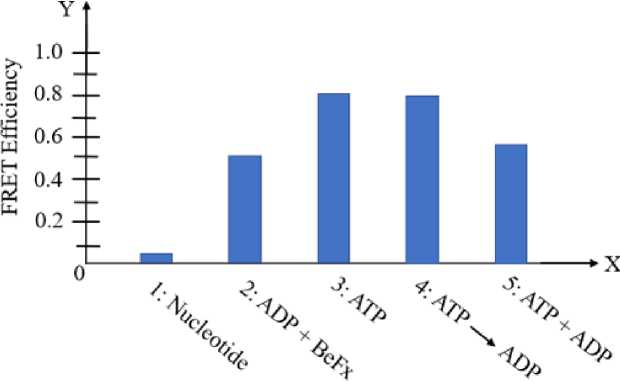
Figure 3. Amount of "football" complex formed was measured with FRET efficiency at different experimental conditions
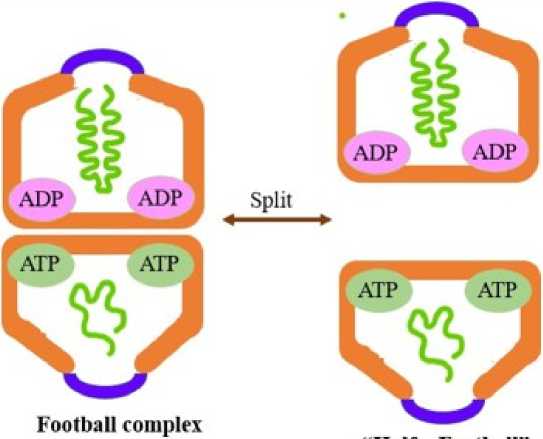
Figure 4. Switch between "football" and "half a football" complex
“Half a Football complex
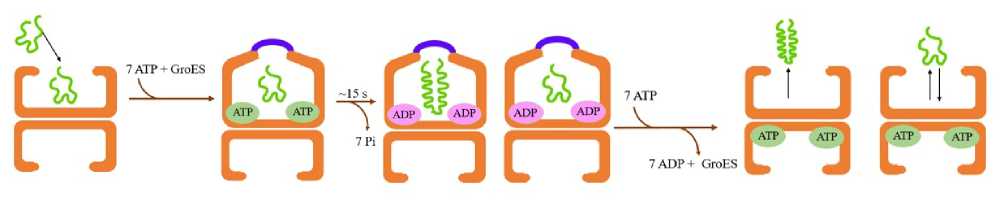
Figure 5. Passive cage model is a representative of the Anfinsen cage
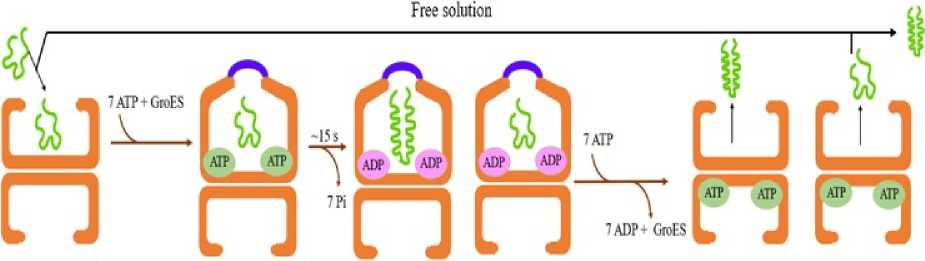
Figure 6. Iterative annealing model shows the mechanism of protein folding mediated by chaperonin and in free solution
Correlation between “football” complex and ATPase activity of GroEL
FRET analysis for the chaperonin and its cofactor at varied concentrations of denatured MDH proved that as the ATPase activity of GroEL increases, concentration of the “football” complex also increases (Fig. 3). It has been observed through increasing FRET efficiency that introducing gradually increasing amounts of dMDH, enhanced GroEL’s ATPase activity, as the amount of “football” complex also increased (Lin et al. , 2006; Llorca et al. , 1996; Llorca et al. , 2008).
“Half a football” complex
Taguchi et al. showed that in native gel electrophoresis, whenever, the chaperonin formed the “football” complex, the complex dissociated at the equatorial plane, dividing the two GroEL rings to form two “half a football” complexes (Fig. 4) (Llorca et al., 1994). It has been inferred that this occurs because of weak inter-ring interaction and also due to reduced contact surface area in the chaperonin as the two GroEL rings lack a salt bridge interaction at the interring interface and are enhanced during electrophoretic conditions. The electrophoretic conditions exaggerate the formation of this complex. This is a very common phenomenon found in group I chaperonins, like the Escherichia coli GroEL (Ec GroEL). In another study, Taguchi et al. (1997) showed that the GroEL/ES chaperonin complex isolated from Thermus thermophilus (Tth GroEL) forms the “half a football” complex under electrophoretic conditions. This was followed by a hybrid complex formation between Tth (GroEL)7: Ec (GroEL)7 in the presence of ATP. The ATPase cycle during protein folding favours formation of this complex. An estimate shows that ~10 % of the GroEL chaperonin exists in this single ring structure under heat shock conditions (Madan et al., 2008). GroES dissociation decreases at high temperatures favouring the notion that the negative cooperativity between the GroEL rings to cause release of GroES decreases with increasing temperature.
Hydrophobic effect on protein stability inside chaperonin cavity
Reduced hydrophobic effect destabilizes protein inside the chaperonin cavity (Korobko et al., 2020). The encapsulated SP undergoes a process very similar to cold denaturation where the relatively ordered water confined in the chaperonin cavity contributes to protein unfolding, thereby making it available for a new proper protein folding cycle. Hydrophobic effect on protein stability was demonstrated using the protein dihydrofolate reductase from the psychropiezophilic organism Moritella profunda (DHFRMp). The protein itself is unstable at room temperature (Xu et al., 2003). To further destabilize it, it was fused to the C terminal end of eGFP (Dave et al., 2016; Sokolovski et al., 2015). Mutations which reduced the hydrophobicity of the protein led to destabilization inside the chaperonin cage and no proper folding was noted. On the other hand, hydrophobic core mutations caused stable encapsulation of the protein inside the GroEL/ES cavity such that the protein could not escape into solution even after incubation of more than 24 hours at room temperature (Korobko et al., 2020). One of the main reasons behind diminishing hydrophobicity is that the volume of the protein and the chaperonin cavity are 58,000 Å3 and 175,000 Å3, respectively (Korobko et al., 2020). So, the SP protein and the cavity wall is at a distance of about 10 Å, which corresponds to three hydration layers, two in the first shell and one in the second shell (Makarov et al., 2002). This arrangement gives a very similar effect to cold denaturation of protein. Such destabilization inside the GroEL/ES corresponds to the iterative annealing model (Thirumalai et al., 2020). Based on this model, the misfolded proteins unfold inside the chaperonin cage, and are further given a second chance to initiate protein folding correctly either inside or outside the cavity. The SP becomes unfolded on binding with GroEL’s apo state and further folding occurs upon binding of ATP and GroES. The reduced hydrophobic effect favours the unfolded state compared to the folded state. Presence of chaperonin then allows proper folding or if the protein is encountered by a proteasome, it undergoes complete degradation because of the hydrophobic effect.
Mechanism of the GroEL/ES chaperonin action
The exact mechanism of action of the GroEL/ES chaperonin during protein folding is a matter of debate. Three different models have been proposed for its mechanism of action based on whether the chaperonin is involved passively in action (passive cage) by preventing aggregation or it additionally promotes the folding process by an active mechanism (active cage and iterative annealing) (Motojima et al. , 2004).
-
• Passive Cage Model
The bacterial chaperonin GroEL/GroES aids in complete folding of various partially folded polypeptide chains post translation. From the onset of this finding two different types of mechanism were proposed. One theory suggested that in each GroEL oligomer, the central cavity acts like a passive sequestration chamber. Inside the chamber, folding of the partially polypeptide chain occurs while protecting it from the external environment to prevent formation of aggregates (Niwa et al., 2016). The other proposition suggested that the chaperonin actively unfolds misfolded polypeptides so as to allow them to fold correctly (Niwa et al., 2016).
Further experiments led to coining the term Anfinsen’s Cage to the GroEL-GroES chaperonin system because GroEL improves the efficiency of folding denatured polypeptides while binding the polypeptide chains into the two GroEL rings (Fig. 5). The probable mechanism demonstrates that ATP hydrolysis catalyzes folding inside the chaperonin cage like as it would occur while folding in free solution in Anfinsen refolding experiment (Rye et al. , 1997). Subsequent in vitro experiments showed that binding of ATP followed by subsequent binding of GroES to the GroEL chains displaces and encloses the polypeptide to the interior of the chaperonin system to prevent it from escaping for a specific duration prior to ATP hydrolysis (Rye et al. , 1997). Further addition of crowding agents to the in vitro system could reduce this release. This suggested that under in vivo conditions of the crowded state of the cell cytoplasm allows any released chain to bind back rapidly to the same GroEL oligomer to minimize the chances of aggregation (Rye et al. , 1997). This is how the protein folding is regulated inside the complex microenvironment of the cell.
The passive cage, also known as the ‘Anfinsen cage’ model represented the idea that protein folding inside the chaperonin cavity occurred with the same kinetics as they would in absence of aggregation in free solution (Rye et al. , 1999). However, this model conflicts in the fact that the chaperonin accelerates protein folding over their spontaneous folding rate even if spontaneous folding occurs rapidly.
Single molecule spectroscopic analysis demonstrated that when aggregation due to spontaneous folding is excluded, protein folding by GroEL/ES gets accelerated. It does not cause accelerated folding. Protein folding occurs with full yield in a single round of GroEL/ES encapsulation (Sameshima et al. , 2008).
-
• Active Cage Model
The active cage model relates the effect of steric confinement of structurally dynamic kinetically trapped folding intermediates to the rate enhancement of folding. These intermediates are produced during hydrophobic collapse of large native protein domains. The accelerated folding is mediated by the volume of chaperonin cavity relative to protein size, net negative charge on the chaperonin cavity wall, and the flexible C – terminal tail containing a Gly – Gly – Met amino acid repeat sequence extending from the equatorial domain of GroEL (Sameshima et al. , 2008). This charged cavity is contributed by the negatively charged amino acids and in turn enhances hydrophobic compaction (Sameshima et al. , 2008; Schmidt et al. , 1994). Molecular dynamics simulations showed that the C – terminal repeat facilitates SP remodeling as it provides a flexible hydrophobic interaction surface for accelerated folding (Sameshima et al. , 2008; Schmidt et al. , 1994). These unstructured C – terminal tails help to capture the SP for efficient encapsulation and compaction by exerting entropic excluded volume effects (Sharma et al. , 2008; Skjærven et al , 2015). Through computer modelling program, it was demonstrated that when the SP is entropically confined in a charged repulsive chaperonin cavity, the conformational freedom of the folding intermediate is restricted through formation of more favourable and stable local and long range contacts (Taguchi, 2015). This helps to accelerate protein folding by atleast one or two orders of magnitude. This model also reported 10-fold higher acceleration fold compared to spontaneous folding (Rye et al. , 1999). It was shown through photoinduced electron transfer coupled to fluorescence correlation spectroscopy (PET – FCS) that inside the chaperonin cavity, chain mobility of SP decreases compared to spontaneous folding and it shows increased chain entropy of the encapsulated SP (Thirumalai and Lorimer, 2001). Accelerated folding occurs with full yield in just a single round of chaperonin encapsulation (Sameshima et al. , 2008). The negatively charged amino acid residues inside the chaperone complex promotes the folding of SPs. Higher the charge
distribution, better is the hydrophobic interaction between the chaperonin and the SP.
-
• Iterative Annealing Model
A third model called the iterative annealing model came up with a new idea about accelerated folding. This suggested that while binding of SP to the chaperonin cage, ATP – driven cycles accelerate protein folding inside the GroEL chamber by periodically unfolding kinetically stabilized misfolded or unfolded states (Fig. 6). Ultimately, a partition is created between rapid protein folding and reformation of the kinetically trapped misfolded state. So, the protein folding can be initiated either inside or outside the cage but inside the cage, the rate of acceleration of protein folding is higher (Xu, 1997).
Evidences have shown that the SP remains inside the GroEL/Es chamber for around 85% of the duration of protein folding while it remains in free solution for a time period of <5%. The greater the time the SP remains in free solution, higher is the chances of aggregate formation and the folding yield decreases. Iterative annealing model thus proved that the binding and subsequent release of SP was not essential for accelerated folding (Xu, 1997), instead annealing required just a single step of productive remodeling (Ye and Lorimer, 2013). Accelerated folding occurs with full yield but requires multiple cycles of active unfolding mediated by the chaperonin binding and release (Sameshima et al , 2008).
CONCLUSION
The GroEL/ES chaperonin complex is a widely studied macromolecular machine (Anfinsen, 1973). Its physiological significance is to provide a folding compartment where a nascent polypeptide or denatured protein molecule is transiently enclosed to form the active state and avoid aberrant interactions and aggregation (Anfinsen, 1973). The surrounding physical environment around the complex enhances the folding of SP. The binding of SP establishes an efficient flip-flop mechanism by which it is enclosed in the chaperonin cage and released upon complete folding. Hence, both the GroEL rings are active during protein folding simultaneously.
The coexistence of the symmetric “football” and the asymmetric “bullet” complexes have been detected under physiological conditions, where the “football” complex is advantageous for protein folding (Yébenes et al , 2011; Yifrach et al , 1996). A direct correlation between the rate of formation of the symmetric complex and protein-folding using MDH has been shown. Association between the two “half a football” complexes forms the symmetric complex, which is very similar to the “two stroke engine” mechanism of chaperonin activity. The role of denatured proteins, and their concentration affect the formation of both the symmetric and asymmetric complexes (Yébenes et al , 2011; Yifrach et al , 1996). Further, under electrophoretic conditions, the “football” complex gets split and exists more prominently as “half a football” complex.
Most of the experiments regarding protein folding by the GroEL/ES chaperonin complex have been performed on model SPs in vitro. Investigations should now be pursued regarding the chaperonin cycling in the presence of in vivo obligate SPs and also elucidating the mechanism of the protein folding cycle inside the cells.
It is also unclear whether this chaperonin acts as a passive cage model or is involved in the active accelerated protein folding process. It is a matter of further investigation. Although there is enough evidence in favour of the accelerated protein folding models (Niwa et al , 2016), yet their mechanisms of action are not well understood (Sameshima et al , 2008; Schmidt et al. , 1994). Advancements over the years are still showing surprising facts about the GroEL/ES chaperonin complex. Future experiments should focus on understanding how the chaperonins mediate protein folding compared to spontaneous folding that occurs in solution and also how few proteins could evolve to become chaperonin independent but their structural homologs could not (Anfinsen, 1973). The understanding of the different sorting signals is also crucial that allow only certain proteins to fold with the assistance of GroEL/ES nanomachine while their homologues fold upon themselves without any aid. Identification of such molecular attributes will further unravel new mysteries about the properties and detailed functioning of the chaperonin machinery.
AUTHOR’S CONTRIBUTIONS
Conceptualization: SK, VR, SS; Data curation: SK, VR; Methodology: SK, VR; Writing – original draft preparation: SK, VR; Writing – review and editing: SK, SS; Supervision: SS.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest and they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.


