Image Classifiers in Endoscopy for Detection of Malignancy in Gastro Intestinal Tract
Автор: K V Mahendra Prashanth, Vani V
Журнал: International Journal of Image, Graphics and Signal Processing(IJIGSP) @ijigsp
Статья в выпуске: 6 vol.9, 2017 года.
Бесплатный доступ
Wireless Capsule Endoscopy (WCE) is one of the methods for examination of gastrointestinal (GI) disorders such as obscure GI bleeding, Crohns disease, polyps etc. WCE has been recognized as a less expensive and painless procedure for the diagnosis of GI tract. This paper examines the various image classifiers designed and developed for the purpose of endoscopy focusing specifically on WCE. It is revealed that designing a suitable image classifier is an important prerequisite for accurate and precise diagnosis of malignancy in WCE. The assessment on various image classifiers used for the diagnosis of pathologies in different parts of GI tract shows that classifiers have reduced the diagnosis time for medical experts and also provided reasonably accurate diagnosis of malignancy. However, correlating classifiers and related pathologies is still observed to be challenging. In view of the fact that early detection may decrease the mortality rate significantly, inclination towards computer aided diagnosis are expected to increase in future. There is a need for advanced research in the development of a robust computer aided diagnosis system, capable of diagnosis of various pathologies in GI tract with higher degree of accuracy and reliability. Further, the study depicts that a direct comparison of results of classifier such as accuracy, prediction, sensitivity, specificity and precision to evaluate its performance is challenging due to diversity of image databases. More research is needed to identify and reduce the uncertainties in the application of image classifier to improve the diagnosis accuracy.
Image classification, Wireless Capsule Endoscopy (WCE), Machine Learning, Support Vector Machine (SVM)
Короткий адрес: https://sciup.org/15014197
IDR: 15014197
Текст научной статьи Image Classifiers in Endoscopy for Detection of Malignancy in Gastro Intestinal Tract
Published Online June 2017 in MECS DOI: 10.5815/ijigsp.2017.06.06
Wireless Capsule Endoscopy is one of the standard imaging techniques today for examination of gastrointestinal tract(GI tract), especially the small intestine; which is not easily reachable by normal upper endoscopy and colonoscopy methods [1].By conventional endoscopic techniques, only the proximal duodenum and distal ileum of the small intestine could be examined. The discovery of capsule endoscope led to the new technology by means of which even a small bowel became visible to the gastroenterologist [2]. The swallow able wireless capsule endoscopy takes images of the GI tract which are later downloaded for analysis in a computer [3]. Conventional method of diagnosis involves visual analysis of images by an expert physician. Computer-aided diagnosis includes extraction of features of the images and automatic classification of images indicating the presence or absence of malignancy; thereby, easing the task of objective interpretation by a physician in endoscopy.
A wireless capsule provided continuous video stream of the inner mucosa lumen tubular structure. Typically, in WCE the image examination for each patient consists of 8hours of video which is around 55,000 frames. The procedure involving examination and reviewing the video from capsule endoscopy has become cumbersome; as it requires concentration for long durations. Computerized image analysis algorithms can reduce the time required to review for image examination by experts and can augment the decision making processes [4]. Thus, automatic CAD methods are required which can help in analysis and diagnosis. Significant research has been observed from the last decade and the number of publications in area of design and development of computer aided diagnostic systems for WCE has substantially increased to 35% starting from year 2004 [5].
The objective of this paper is to provide a comprehensive overview of the latest advancements and techniques used for improving classification accuracy in WCE. Further, the issues and limitations in existing image classification techniques; which are being used in WCE are discussed. Possibilities to improve the image classifiers, providing a faster, more accurate and precise diagnosis of malignancy in GI tract are being deliberated. Literature has been reviewed on keying the keywords: capsule endoscopy, histogram, classifier learning, computer aided diagnosis, medical image data, image classifier in various prevalent publications such as Springer, IEEE, SAGE, Elsevier, PubMed, ACM, Hindawi, ScienceDirect which have been published during 2000-2015.
Section 2 discusses the pathologies under investigation in GI tract. Development of better image classifiers is described in Section 3. Section 4 provides a comprehensive study of the general classification of Image Classifiers and Image Classifiers employed for capsule endoscopy. Section 5 describes the evaluation of classifier performance. Advantages and challenges in classifiers are discussed in Section 6. Section 7 depicts the overall conclusion of the study.
-
II. Pathologies Under Investigation in GI Tract Using WCE
It is observed that a variety of pathologies; that are targeted by different Computer Aided Diagnostic systems for detection and classification in GI tract do exist. Some of the commonly made diagnoses are polyps, tumor, cancer, motility assessment, ulcer detection, Barretts oesophagus, Celiac disease, Chrons disease, GI bleeding detection and distinction between normal and abnormal lesions. The investigation of intestinal motility allows a medical expert to detect the presence of different intestinal dysfunctions. Gastrointestinal ulcers are mucosal erosions; exceeding a certain size. They may arise in several parts of the GI tract, such as oesophagus, the stomach, or the small intestine. Ulcers may lead to complications like GI bleeding or perforation of the mucosal wall [5]. Barretts oesophagus is a specific disorder of the oesophagus. Celiac disease is a complex autoimmune disorder which affects the small bowel after introduction of gluten containing food in certain individuals and can affect all age groups. Crohns disease is also an autoimmune disorder. It may affect all parts of GI tract; characterized through a patchy inflammation of the GI tract.
However, the most frequently affected parts of the GI tract are ileum and colon. Studies [6–12] have shown that WCE is much superior for diagnosis of GI tract not only because it is well accepted by patients as it is less painful compared to other endoscopic/colonoscopy procedure, but also provides better accuracy and yield. A survey of 14 studies [7] including 396 patients with Obscure GI bleeding showed a higher yield for clinically significant lesions with WCE (56%) than with Push Enteroscopy (26%) or small-bowel follow through (6%). These studies indicate that WCE is more suitable for finding small intestinal pathology than with push enteroscopy.
-
III. Need of Developing Better Image Classification Techniques in WCE
It is observed that Lower adenoma detection rate is due to failure to perceive a lesion as abnormal due to misinterpretation of adenoma as a hyperplastic polyp [13]. As discussed in [14], the reliable understanding of the WCE procedure requires experienced reader of adequate number of studies. Experimentally it is proved that Clinical analysis of WCE images is scoring and requires training. As observed in [15], WCE produces high quality image of small intestinal mucosa at a rate of 2 per sec. But it is not always possible to visually recognize the subtle differences between the images especially in low contrast areas. As indicated in [16], the optimal review time is 15 images per second and it takes over 1 h to read a full 8hours procedure and hence it is time consuming process and requires long reading time. As depicted in [17], the visualization and imaging techniques employed for diagnosis of small bowel diseases are not precisely accurate. There is a need for better solutions that provides reasonable visual imaging for detection of abnormalities of small bowel, easy to use by medical experts and relatively inexpensive. These observations from the literature have shown that there is a need of developing better image classification techniques in WCE.
-
IV. Review of Classifiers Used in Medical Endoscopy
The common steps involved in image processing of medical endoscopy are shown in Fig 1. Once the image is acquired, the initial step is pre-processed which is required to enhance the quality of degraded images. Region of Interest (ROI) is detected on choosing an appropriate feature extraction technique. A postprocessing may be required in certain cases such as removal of invalid feature combinations in high-level features. The pathologies are detected in most of the cases through classification of image, using a previously trained classifier. But there also exists certain systems wherein pathologies are detected through features eg. using feature thresholds [5].
Some of the systems are being targeted at Content Based Image Retrieval (CBIR) or Content Based Video Retrieval (CBVR) [5]. As the image classifier is an automated system; the output is generated without having any intervention of a medical expert. In case of CBIR/CBVR system; a number of similar images/videos (on demand) will be diagnosed by an expert for analysis. This enables CBIR/CBVR systems to act as an interactive tool and cannot be used for offline image processing.
It is observed that Classifier based approach provides second opinion to medical expert, while CBIR/CBVR based systems can be considered as digital valuator to help the medical expert to compare cases with unknown pathologies
-
A. Classification of Image Classifiers
Image Classification is a critical task in diagnosis of WCE image. In computer aided medical diagnosis, it very difficult to obtain set of rules for disease classification by classical engineering methods. Alternatively, machine learning methods are applied to obtain classifiers from sets of data pre-classified by medical experts. Machine learning investigates the study and development of algorithms that can learn from and predict from the data [18,19].
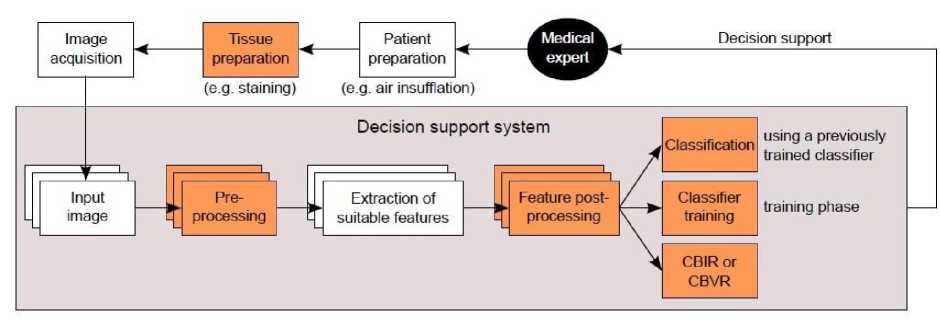
Fig.1. Block diagram of a decision support system. Colored boxes represent optional steps. Layers indicate multiple frames from endoscopy that may be simultaneously processed to represent the inter-frame relationships [5]
Common paradigms of machine learning are:
-
1. Supervised learning: wherein each data point set is associated with a training label. It includes Artificial Neural Network (ANN) [20], Support Vector Machine (SVM) [21] and Linear Discriminant Analysis (LDA).
-
2. Unsupervised learning: wherein statistical number of expected classes such as K NN [18, 21] are used. As seen in [22,23,20,21] Supervised learning can be used to solve classification and regression while data clustering can be performed through unsupervised learning [18].
Classifier performance can be improved through two other methods namely bagging and boosting. These methods help to control the trade-off between precision and accuracy through changes in thresholds and weights [18]. It is revealed that various challenges are involved while selecting best suited classifier for endoscopy.
-
B. Image Classifiers in Capsule Endoscopy
The major classifiers used in many computer aided diagnosis of endoscopy images are K-NN Nearest Neighbor classifier, Support Vector Machine (SVM) and Decision trees [22]. K-nearest neighbor learning (K-NN) [22] is the most basic lazy learning method. As compared to eager learners, lazy learners do not explicitly induce a representation of the target function. Training phase consists of simply storing the training data [23]. Support vector machines (SVMs) are powerful kernel based learning methods which belongs to class of eager learners. SVMs introduce linear and non-linear decision function in feature space [21]. Decision Tree (DT) belongs to the class of eager learning which builds typical hypotheses based on set of if-then-rules or decision trees. With this type of hypothesis, the learned rules or trees concepts can be easily expressed and understood by humans [19].
Kodogiannis et. al. in their work on Neuro-fuzzy Classification System for WCE; calculated a total of 54 features(9 statistical measures for 6 image planes). The nine statistical measures estimated were standard deviation, variance, skew, kurtosis, entropy, energy, inverse difference moment, contrast, and covariance. The textual descriptors were calculated for both RGB (Red, Green, Blue) and HSV (Hue, Saturation, Value) planes. New texture features from texture spectra in chromatic and achromatic domains were also extracted. The results achieved a good accuracy of 95.71% [20].
-
C. Classifiers for detection of Polyp
The findings of Berner et.al.[24] indicates that VO-DOVA (Valley Orientation-Depth Of Valley Accumulation) (accuracy: 97.53%) is able to determine polyp better than EF-DOVA ( Ellipse Fitting-Depth Of Valley Accumulation ) (accuracy: 74.61%). Figueiredo et al. proposed another method based on geometric and mean curvature of capsule endoscopy image assuming that polyps show up as protrusions. However the algorithm suffers from strong dependence on the measure of protrusion of the polyp to identify potential candidates. Hence, the polyp may be missed if it is not protruding enough from the surrounding mucosal folds [25]. Karargyris and Bourbakis proposed an algorithm for WCE images mainly based on Log Gabor filters and Susan edge detector with SVM as the binary classifier [26,27]. The experimental result shows that Log Gabor filter is able to extract the dominating texture segments while Susan edge detector is able to provide details of the boundaries of the polyp. However the experiment demonstrates that it achieves lower specificity of 67.5%.
Further, Kodogioannis and Boulougoura have proposed a texture based approach; wherein new texturebased features were computed from the achromatic and spectra of the ROI (Region of Interest) that may contain a polyp. Neuro fuzzy classification scheme was also used. The results of the algorithm implemented shows that textural information is of primary importance for the discrimination between non-polyps and polyps [20]. As observed, the authors [24–26,28–30] detected polyps using shape features. While Zhoa et.al. [31] focused on detection of polyps through extraction of color and texture features with SVM classifier for polyp detection whereas Isabel et.al. [30] used curvature information to obtain polyp segments. Hwang et.al.[32] focused on detection of abnormalities such as polyp, blood and ulcer through color and texture features. Then bag-of-visual- words [33] were applied for image classification.
Multimodal embedded technique for detection and classification of polyp in wireless capsule endoscopic images was developed and tested by Romain, et. al [28]. The developed multimodal wireless capsule used 3dimensional and 2-dimensional data to identify the presence of polyps and provide cancerous information of the polyps. A Support Vector Machine (SVM) classifier is used for final classification [29]. The boosting-based polyp classification achieved specificity of 95%, sensitivity of 91% and false detection rate of 4.8% on a database composed of 300 hundred positive examples and 1200 negative ones.
-
D. Classifiers for detection of bleeding
The main effort in computer aided diagnosis for WCE has also been in detection of another kind of malignancy i.e. bleeding in GI tract [34, 58]. Bleeding is usually detected by performing an analysis of color (usually in Hue-Saturation-Intensity space). The method as depicted in [35] uses chromaticity moment with a NN classifier, where as in the paper [36] the adaptive color histogram together with a SVM classifier is used; while the algorithm [37] uses color spec trum transformation with threshold value to detect bleeding regions in WCE frames. Given M2A newest software provides a new feature for detection of luminal blood called Suspected Blood Indicator (SBI); achieving a sensitivity of 72% and specificity of 85%. The performance of SBI performance was improved through Bayesian In-formation Criterion (BIC) and Expectation Maximization (EM) clustering algorithm [9]; which was used to train probability models for the bleeding pixels and non-bleeding pixels with a sensitivity of 92% and specificity of 98% [38]. While Barbara Penna et al., have proposed blood detection algorithm based on color modeling, edge masking and Reed-Xiaoli RX detection [39,40]. The bleeding detection algorithm was performed on different values of RX detection threshold. The threshold was selected so as to obtain False Alarm Rate lower than 10% and missed detection rate close to 10%. K-NN classifier used for classifying bleeding and non-bleeding images into two classes using HUE based statistical features yielded an accuracy of 98% [41].
-
E. Classifiers for detection of Chrons disease and ulcer
The small bowel is the most affected site of Chrons disease [17]. Hence detection of Chrons disease and ulcer has become a vital part of research. In the paper [42], wavelets based LBP algorithm were being used to detect tumors and bag-of-words approach; based on LBP (Local Binary Patterns) and SIFT features are exploited in [29] to detect ulcers. In [43], pixel brightness and image texture descriptors together with nonlinear classifier are used to detect celiac disease. In the paper [44], MPEG-7 descriptors for color, texture and edge are developed and tested for Chrons disease. Valdeavilla et. al [45] detected suspected pathologies such as chime blocked, suspected blood indicator and ulcer through methods such as standard LBP and Sobel LBP as recognition features and
SVM as classifiers with resulting accuracy of 98.10% [45].
C.S.Lima et.al. showed that color textural information using Radial basis function as classifier is adequate to classify lesions with sensitivity of 930.4%, specificity of 950.1% and accuracy of 940.25% [45] . An innovative CR-ULBP (Color Rotation Uniform Rotation Variant LBP) scheme forWCE image analysis was proposed to boost the chromatic attributes of ulcer regions in WCE image on dataset consisting of 10 of the most normal and 10 of the hardest ulcer images. It could be noted that the majority of physicians could hardly recognize the ulcer regions i.e (15.7% 5.3) mean sensitivity while CR-ULBP scheme achieved 70% correct positive predictions [46]. A set of weak classifiers was constructed by using weighted least square regression and AdaBoost learning to fuse the ensemble of weak classifiers to a strong classifier for detection of ulcer in WCE [47]. This method provided an accuracy of 82% and sensitivity of 70%.
-
F. Classifiers for detection of diverse organs
Another line of computer aided diagnosis is focused on identification of diverse organs of the intestinal tract like esophagus, stomach, duodenum, jejunumileum and cecum. In the reference [48], the textural features captured by Gabor filters are exploited in the problem of duodenum discrimination. The locations of the esogastric junction, pylorus, and ileo-cecal valve are estimated with an algorithm based on MPEG-7 visual descriptors in [49], while the color change pattern is exploited to detect different organs in [50].
-
G. Classifiers for detection of non-informative frames
Detection of non-informative frames has been one area of prime research. In this effort, Gabor filters have been used to detect bubble like shape of intestinal juices [51], while color histograms together with a SVM classifier were being used to detect intestinal content [52]. A three-stage cascade to detect informative frames has been proposed in [51]. Color information (histogram and color moments with a SVM classifier) was used to characterize turbid in the first stage. In the second stage, texture segmentation (Gauss Laguerre Transform segmentation) was applied to characterize bubbles, and, finally, a threshold on the segmented regions is applied in order to detect informative frames.
-
H. Classifiers for detection of intestinal motility
It is clinically relevant to assist the physician to study intestinal motility by identification of intestinal lumen in endoscopic images.. Zabulis et al [53] proposed a system based on a Mean Shift Segmentation algorithm variant to locate lumen regions in WCE frames. Giovanni Gallo et. al suggested an intestinal lumen detection method, based on boosting. The experiments were conducted on nearly 5000 images (1500 positive and 3500 negative, rescaled to 24 *24 pixels) by using customized set of Haar-like features and Adaboost ; combined with a cascade of strong classifier resulting in an accuracy of 92; 4% , a recall of 90.5% and precision of 71%[53]. Further,
Fernando Vilarino et.al. [54] proposed to use ROC curves to evaluate several classifier models. A bagging ensemble of 25 decision trees reveals that decision tree classifier is very good for accuracy while the choice of bagging over AdaBoost were more suitable for datasets with substantial amount of noise.
These observations on various studies reveal that different methodologies are being employed to assist medical experts for detection of pathologies in GI tract. Further it is also observed that significant research has been undertaken over the last decade. However, the correlation between image classifier and data to be classified is yet to be discovered. Table 1 summarizes the image classifiers used for endoscopy mentioned in above literature.
Table 1. Summary Of Image Classifiers In Capsule Endoscopy
|
Sl.No. |
Author |
Publ ication |
Proposed Algorithm |
Results |
Training sets used |
|
1 |
Kodogiannis et. al[20] |
2005 |
Neuro-fuzzy Classification |
Accuracy: 95.71% |
35 normal;35 abnormal images |
|
2 |
Bernal et al.[24] |
2012 |
Region descriptor based depth of valleys (SA-DOVA) Accuracy: 74.61% (EF-DOVA) |
97.53%(VODOVA) |
300 images |
|
3 |
Figueiredo et al[25] |
2011 |
Mean and Geometric curvature |
80% polyp detection |
Full video from 5 patients |
|
4 |
Karargyris and Bourbakis[26] |
2009 |
Susan edge detector and Log Gabor filters |
Sensitivity:97% Specificity:94% |
10 polyps and 40 non-polyp images |
|
5 |
Zhoa et.al.[31] |
2011 |
Color and textures with SVM classifier |
Accuracy : 97% |
2 videos valuated by domain specialist |
|
6 |
Karargyris et. al[27] |
2011 |
Gabor filters based segmentation |
Sensitivity: 75%;Specificity :73.3% |
20 frames with ulcer |
|
7 |
Isabel et.al.[30] |
2010 |
Curvature information |
NS |
NS |
|
8 |
Baopu Li[42] |
2009 |
Wavelet based LBP and SVM |
Sensitivity: 91.33%(RGB) Accuracy: 93.67%(RGB) and 96.67%(HIS) |
150 normal;150 abnormal images |
|
9 |
Ciaccio et. al. [42] |
2010 |
Pixel brightness and image texture descriptors together with nonlinear classifier |
s ensitivity :92.7%; Specificity:93.5% |
200 consecutive image frames |
|
10 |
Kumar et.al.[44] |
2012 |
MPEG-7 descriptors |
Precision 90%;recall 90% |
Images:212(normal),213(mild);7 4(moderate),34(severe) |
|
11 |
Romain et. al [28] |
2013 |
Boosting-based polyp |
Sensitivity:91% Specificity:95% |
300 positive examples and 1200 negative ones |
|
12 |
Liedlgruber et. al.[36] |
2012 |
Adaptive color histogram together with a SVM classifier |
Accuracy: 95% |
NS |
|
13 |
Jung et. al[37] |
2008 |
Color spectrum transformation with threshold value |
Sensitivity :92.86% Specificity :89.49% |
1000 images: abnormal patient 1000:normal patient |
|
14 |
Karkanis, et. al. [38] |
2006 |
Color wavelet covariance (CWC) |
Sensitivity:90% Specificity:97% |
60 video corresponding to adenoma |
|
15 |
Barbara Penna et al[40] |
2010 |
Reed-Xiaoli RX detection |
False Alarm Rate lower than 10% and missed detection rate close to 10% |
11 sequences;8 bleeding cases;3:normal |
|
16 |
Vilari et.al[51] |
2006 |
Gabor filters |
Accuracy:95% |
200 valid frames,10 videos |
|
17 |
Vilarino et. al.[52] |
2010 |
Color histograms together with a SVM classifier |
Sensitivity:70% |
Training:8 videos(leaveout-one) strategy |
|
18 |
Valdeavilla et.. al[55] |
2010 |
Standard LBP and sobel LBP and SVM as classifier |
Accuracy : 98.10% |
136 normal images; 214abnormal images |
|
19 |
Zabulis et al [53] |
2008 |
Mean Shift Segmentation algorithm |
NS |
NS |
|
20 |
Giovanni Gallo et. al [46] |
2011 |
Customized set of Adaboost and Haar-like features |
Accuracy: 92% |
1500 positive images; 3500 negative images |
|
21 |
Charisis et. al[39] |
2013 |
CR-ULBP (Color Rotation Uniform Rotation Variant LBP) |
70% positive predictions |
100 ulcer images; 100 normal images |
|
22 |
Zhoa et.al. [31] |
2011 |
Color and textures with SVM classifier |
Accuracy : 97% |
2 videos valuated by domain specialist |
|
23 |
T.Ghosh[41] |
2014 |
Statistical features with K-NN |
Accuracy:98%; Sensitivity:97% Specificity:99% |
100 bleeding;100 non bleeding images |
|
24 |
That Mon Htwe[47] |
2010 |
AdaBoost |
Accuracy:82% Sensitivity:70% Specificity:94% |
800 positive samples; 800 negative samples |
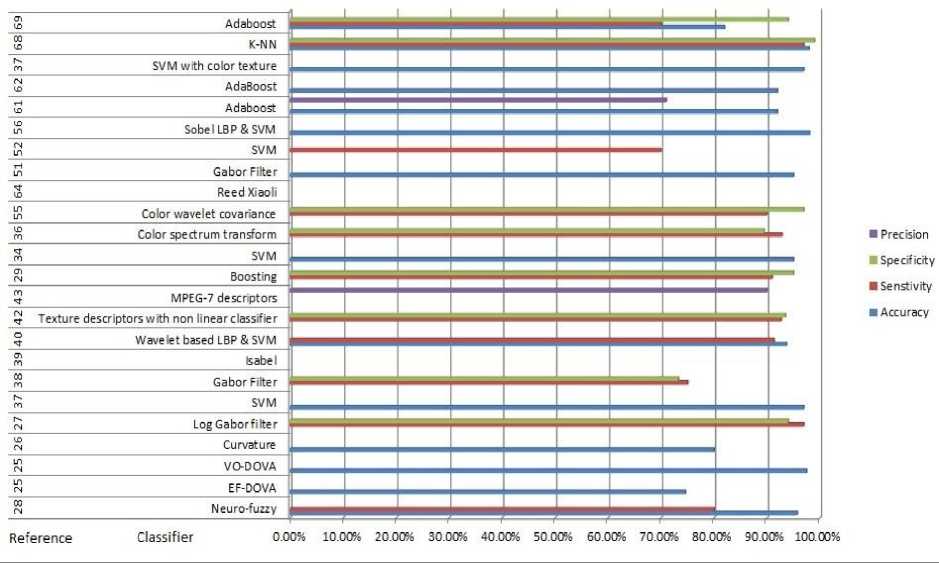
Fig.2. Graphical details of Table 1, depicting methodology/algorithm and references with respective accuracy, sensitivity, specificity and precision in %

Fig.3. Graphical details of Table 1, depicting the number of samples/training sets employed for different classifiers
-
V. Evaluation of Classifier Performance
Results obtained during evaluation of classifier plays a significant role in the classification procedure. The classifier performance could be evaluated through the following parameters: precision, accuracy, sensitivity and specificity.
accuracy = £ TrPo + £ TrNe / £ TrPo + ^ TrNe + £ FaPo + £ FaNe specificity = ^ TrNe) / (^ TrNe + ^ FaPo)
sensitivity =£ TrPo) / (£ TrPo -У FaNe)
precision = ^ TrPo) / (^ TrPo + ^ FaPo)
where TrPo = true positive, TrNe = true negative, FaPo = false positive and FaNe = false negative. The classifier performance for CAD systems for Wireless Capsule Endoscopy has been compared as shown in Table 1.
Table 1 and Fig 2 provides summary of the reviewed studies on various image classifiers used in Wireless Capsule Endoscopy. Fig.3 shows the impact of number of training sets on the classifier. The observation reveals that all algorithms mentioned in the Table 1 could achieve higher accuracies with sufficient training sets. It is observed that SVM could produce very high accuracy; even with a very small number of training sets. Though the number of training sets used in Adaboost outnumbers the number of training sets used in SVM and K-NN, the accuracy is still less comparatively. This observation reveals that SVM and K-NN serves as a very stable classifier. The study on boosting based classification employed in [53,28,47] demonstrates that as compared to other mentioned classifiers; it does not require any predefined routines extraction of high-level features. This technique has great generalization performance and low complexity.
-
VI. Assessment of Classifier: Advantages and Challenges
Identification of malignancies using WCE requires various computer aided diagnosis methodologies such as geometrical features, colour space techniques, texture identification along with strong image classifiers. The literature shows that machine classifiers score fairly well with accuracy of more than 80%. As observed from the Table 1, SVM classifier provided higher sensitivity of 90% while Adaboost provided lower sensitivity(less than 80%).
A texture based image classification reveals that Feed forward neural network yields higher classification accuracy as compared to Naïve Bays classifier, K NN and Cascaded NN [59]. Another study [60] shows that multi classifier scheme helps to boost the performance of the different classifiers. The brain style computation model
A-NN has been combined with the best known classification model SVM with Adaboost to create a strong classifier out of the set of weak classifiers yielding a higher precision. However the processing time increases due to increase in complexity.
Based on the literature; the following salient points were being observed
-
• The classifiers might reduce the image analysis time and provides better accuracy and sensitivity.
-
• One of the biggest challenges in assessment of classifier is that the performance parameters such as accuracy, sensitivity, specificity and precision cannot be directly measured since it depends not only on type of classifiers chosen, but also on the selection of training and testing data.
-
• Higher the number of training and testing data; better is the classifier performance. However the segregation of training and testing data remains a challenge.
-
• The training data should be a good indicator of performance on future data. The number of training and testing data can be selected based on hold-out approach wherein 1/3 data are taken for testing and remaining 2/3 data are taken for training. In case of limited image database, Leave-One-Patient-Out cross-validation (LOPO) [56] can be used wherein one image out of image database is taken as a testing image and the remaining images are used as training images.
-
• Another important challenge is the number of times the experiment has to be repeated to get better performance. Michael Drozdal [57] repeated sequential image analysis on WCE 50 times (Once for each video in the training set). It was observed, the accuracy was high and variance was less when videos such as (#10, #21 and #38) were used as training set. While the accuracy was low and variance was high, when videos #36, #37 or #39 were chosen as training set.
-
• The variation in accuracy could be justified through two possible reasons:
Firstly, some videos were found to be very homogenous and hence do not offer much enough information to learn a good classifier. Secondly, certain videos contain lot of valuable information like strange turbid colour and thereby offer good information to learn a good classifier.
-
• Selection of data for classifiers remains a challenge as it requires validation and explanation for correctness, repeatability and consistency.
-
• WCE requires a well-defined database with ground truth by expert Gastroenterologists; which would help to establish a benchmark and test the different methodologies for detection of malignancies.
-
VII. Conclusion
The paper presented a comprehensive literature assessment on image classifiers for endoscopy images. This comparative study of unsupervised and supervised classifiers used in the field of endoscopy examined to validate the classification capabilities for various types of malignancies in GI tract using WCE. It is observed that the image classifiers have reduced the diagnosis time to a large extent for analysis. The classifiers provided more accurate and precise diagnosis of malignancy. The bagging and boosting rules might help to control the trade-off between precision and accuracy. Furthermore, the study reveals that SVM is a very strong and stable classifier providing a high degree of accuracy. To facilitate higher accuracy of classification, detailed learning with CNN is one of the future directions of research.
However, a correlation of classifiers and related pathologies is difficult due to issues like limited heterogeneous image databases and missing information for assessment of accuracies among systems. It is evident from the study that classifier is more reliable and accurate when the training data used in the process represents well the future data. Further, the data suggests that the number of times the experiments were conducted also matters. In view of the fact that early detection may decrease the mortality rate significantly, computer aided diagnosis are being preferred.
As a step towards exploration of new avenues of classifiers such as CNN, the biggest challenge is unavailability of a large data set for WCE; validated and tested by experts which can be used a benchmark for an effective comparative experimental assessment.
There is a need for advanced research in the development of a robust computer aided diagnosis system capable of diagnosis of various pathologies in GI tract, with higher degree of accuracy and reliability. These features would definitely enable an automatic image analysis and diagnosis system embedded within the WCE empowering the Gastroenterologists to make real time decisions.
Acknowledgment
The authors would like to thank Dr. Ravindra B.S, Senior Consultant Gastroenterologist, Dr. Nagabhushan J.S, Senior Consultant in Gastrointestinal and Colorectal Surgery and team from BGS Global Hospitals, Kengeri, Bangalore for sharing their experiences regarding WCE. We also thank the journal editors and reviewers for their suggestions and reviews.
Список литературы Image Classifiers in Endoscopy for Detection of Malignancy in Gastro Intestinal Tract
- G. Iddan, G. Meron, A. Glukhovsky, P. Swain, Nature 405, 417 (2000)
- J.D. Mellinger, Surgical Innovation 10(1), 3 (2003)
- A. Moglia, A. Menciassi, P. Dario, A. Cuschieri, The Lancet 370(9582), 114 (2007)
- V.S. Prasath, R. Delhibabu, in Computational Intelligence in Medical Informatics (Springer, 2015), pp. 73–80
- M. Liedlgruber, A. Uhl, Department of Computer Sciences, University of Salzburg, Austria,http://www. cosy. sbg. ac. at/research/tr. html, Tech. Rep 1, 2011 (2011)
- S.L. Triester, J.A. Leighton, G.I. Leontiadis, D.E. Fleischer, A.K. Hara, R.I. Heigh, A.D. Shiff, V.K. Sharma, The American journal of gastroenterology 100(11), 2407 (2005)
- L. Fisher, M.L. Krinsky, M.A. Anderson, V. Appalaneni, S. Banerjee, T. Ben-Menachem, B.D. Cash, G.A. Decker, R.D. Fanelli, C. Friis, et al., Gastrointestinal endoscopy 72(3), 471 (2010)
- B.S. Lewis, P. Swain, Gastrointestinal endoscopy 56(3), 349 (2002)
- M. Mylonaki, A. Fritscher-Ravens, P. Swain, Gut 52(8), 1122 (2003)
- A. Mata, J. Llach, J. Bordas, F. Feu, M. Pellis´e, G. Fern´andez-Esparrach, A. Gin´es, J. Piqu´e, Gastroenterologia y hepatologia 26(10), 619 (2003)
- J. Saurin, M. Delvaux, J. Gaudin, I. Fassler, J. Villarejo, K. Vahedi, A. Bitoun, J. Canard, J. Souquet, T. Ponchon, et al., Endoscopy 35(7), 576 (2003)
- M. Mu˜noz-Navas, (2008)
- J. Church, Diseases of the Colon & Rectum 51(5), 520 (2008)
- T. Kav, Y. Bayraktar, World J Gastroenterol 15(16), 1934 (2009)
- E. Rondonotti, F. Villa, C.J. Mulder, M. Jacobs, R. de Franchis, World Journal of Gastroenterology 13(46), 6140 (2007)
- J.A. Leighton, V.K. Sharma, K. Srivathsan, R.I. Heigh, T.L. McWane, J.K. Post, S.R.Robinson, J.L. Bazzell, D.E. Fleischer, Gastrointestinal endoscopy 59(4), 567 (2004)
- Z. Fireman, E. Mahajna, E. Broide, M. Shapiro, L. Fich, A. Sternberg, Y. Kopelman, E. Scapa, Gut 52(3), 390 (2003)
- R. Kumar, T. Dassopoulos, H. Girgis, G. Hager. System and method for automated disease assessment in capsule endoscopy (2010). US Patent App. 13/382,855
- J.R. Quinlan, C4. 5: programs for machine learning (Elsevier, 2014)
- V. Kodogiannis, M. Boulougoura, in Neural Networks, 2005. IJCNN’05. Proceedings. 2005 IEEE International Joint Conference on, vol. 4 (IEEE, 2005), vol. 4, pp. 2423–2428
- B. Sch¨olkopf, C.J. Burges, Advances in kernel methods: support vector learning (MIT press, 1999)
- J. Mennicke, C. M¨unzenmayer, T. Wittenberg, U. Schmid, in 4th European Conference of the International Federation for Medical and Biological Engineering (Springer, 2009), pp. 629–632
- T.M. Cover, P.E. Hart, Information Theory, IEEE Transactions on 13(1), 21 (1967)
- J. Bernal, J. S´anchez, F. Vilarino, Pattern Recognition 45(9), 3166 (2012)
- P.N. Figueiredo, I.N. Figueiredo, S. Prasath, R. Tsai, Diagnostic and Therapeutic Endoscopy 2011 (2011)
- A. Karargyris, N. Bourbakis, in Life Science Systems and Applications Workshop, 2009. LiSSA 2009. IEEE/NIH (IEEE, 2009), pp. 143–147
- A. Karargyris, N. Bourbakis, Biomedical Engineering, IEEE Transactions on 58(10), 2777 (2011)
- O. Romain, A. Histace, J. Silva, J. Ayoub, B. Granado, A. Pinna, X. Dray, P.Marteau, in Bioinformatics and Bioengineering (BIBE), 2013 IEEE 13th International Conference on (IEEE, 2013), pp. 1–6
- C. Cortes, V. Vapnik, Machine learning 20(3), 273 (1995)
- I.N. Figueiredo, S. Prasath, Y.H.R. Tsai, P.N. Figueiredo, ICES REPORT 10, 36 (2010)
- Q.Zhao, M.Q.H. Meng, in Intelligent Control and Automation (WCICA), 2011 9th World Congress on (IEEE, 2011), pp. 948–952
- S. Hwang, in Advances in Visual Computing (Springer, 2011), pp. 320–327
- D.A.R. Vigo, F.S. Khan, J. Van de Weijer, T. Gevers, in Pattern Recognition (ICPR), 2010 20th International Conference on (IEEE, 2010), pp. 1549–1553
- M.W. Mackiewicz, M. Fisher, C. Jamieson, in Medical Imaging (International Society for Optics and Photonics, 2008), pp. 69,140R–69,140R
- B. Li, M.Q.H. Meng, Computers in biology and medicine 39(2), 141 (2009)
- M. Liedlgruber, A. Uhl, Biomedical Engineering, IEEE Reviews in 4, 73 (2011)
- Y.S. Jung, Y.H. Kim, D.H. Lee, J.H. Kim, in BioMedical Engineering and Informatics, 2008. BMEI 2008. International Conference on, vol. 1 (IEEE, 2008), vol. 1, pp. 859–862
- S. Hwang, J. Oh, J. Cox, S.J. Tang, H.F. Tibbals, in Medical Imaging (International Society for Optics and Photonics, 2006), pp. 61,441P–61,441P
- V.S. Charisis, C. Katsimerou, L.J. Hadjileontiadis, C.N. Liatsos, G.D. Sergiadis, in Computer-Based Medical Systems (CBMS), 2013 IEEE 26th International Symposium on (IEEE, 2013), pp. 203–208
- I.S. Reed, X. Yu, Acoustics, Speech and Signal Processing, IEEE Transactions on 38(10), 1760 (1990)
- T. Ghosh, S. Bashar, S. Fattah, C. Shahnaz, K. Wahid, in Computer and Information Technology (ICCIT), 2014 17th International Conference on (IEEE, 2014), pp. 354–357
- B. Li, M.Q. Meng, in Intelligent Robots and Systems, 2009. IROS 2009. IEEE/RSJ International Conference on (IEEE, 2009), pp. 498–503
- E.J. Ciaccio, C.A. Tennyson, S.K. Lewis, S. Krishnareddy, G. Bhagat, P.H. Green, computer methods and programs in biomedicine 100(1), 39 (2010)
- R. Kumar, Q. Zhao, S. Seshamani, G. Mullin, G. Hager, T. Dassopoulos, Biomedical Engineering, IEEE Transactions on 59(2), 355 (2012)
- E.B. Valdeavilla, S.G. Miaou, (AIT 2010, 2010)
- G. Gallo, A. Torrisi, in The Third International Conferences on Pervasive Patterns and Applications (2011), pp. 25–30
- Htwe, L. Shen Weijia, J. Poh Chee Khun, O. Lim Joo Hwee, K.Y. Ho, in Asia PacificSigna l and Information Processing Association Annual Summit and Conference (APSIPA) (APSIPA, 2010), pp. 653–656
- L. Igual, J. Vitria, F. Vilarino, S. Segu´ı, C. Malagelada, F. Azpiroz, P. Radeva, in 4th European Conference of the International Federation for Medical and Biological Engineering (Springer, 2009), pp. 1536–1539
- J.S. Cunha, M. Coimbra, P. Campos, J.M. Soares, Medical Imaging, IEEE Transactions on 27(1), 19 (2008)
- J. Lee, J. Oh, S.K. Shah, X. Yuan, S.J. Tang, in Proceedings of the 2007 ACM symposium on Applied computing (ACM, 2007), pp. 1041–1045
- F. Vilarino, P. Spyridonos, O. Pujol, J. Vitria, P. Radeva, F. De Iorio, in Pattern Recognition, 2006. ICPR 2006. 18th International Conference on, vol. 4 (IEEE, 2006), vol. 4, pp. 719–722
- F. Vilarino, P. Spyridonos, F. DeIorio, J. Vitria, F. Azpiroz, P. Radeva, Medical Imaging, IEEE Transactions on 29(2), 246 (2010)
- X. Zabulis, A.A. Argyros, D.P. Tsakiris, in Intelligent Robots and Systems, 2008. IROS 2008. IEEE/RSJ International Conference on (IEEE, 2008), pp. 3921–3926
- F. Vilari˜no, L.I. Kuncheva, P. Radeva, Pattern Recognition Letters 27(8), 875 (2006)
- S.A. Karkanis, D.K. Iakovidis, D.E. Maroulis, D.A. Karras, M. Tzivras, Information Technology in Biomedicine, IEEE Transactions on 7(3), 141 (2003)
- M. H¨afner, M. Liedlgruber, A. Uhl, A. V´ecsei, F. Wrba, Medical image analysis 16(1), 75 (2012)
- M. Drozdzal, L. Igual, J. Vitria, C. Malagelada, F. Azpiroz, P. Radeva, in Computer Vision and Pattern Recognition Workshops (CVPRW), 2010 IEEE Computer Society Conference on (IEEE, 2010), pp. 117–124
- A Novel Algorithm for Color Similarity Measurement and the Application for Bleeding Detection in WCE
- Choudhary, Dolly, et al. "Performance analysis of texture image classification using wavelet feature." International Journal of Image, Graphics and Signal Processing 5.1 (2013): 58.
- Hai, Tran Son, and Nguyen Thanh Thuy. "Image classification using support vector machine and artificial neural network." International Journal of Information Technology and Computer Science (IJITCS) 4.5 (2012): 32.


