In vitro evaluation of antibiotic resistance patterns among Bacillus species
Автор: Harba M., Jawhar M.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 2 т.21, 2025 года.
Бесплатный доступ
Antimicrobial resistance is currently one of the most public health serious concerns because of the overuse and unsuitable use of antibiotics. Therefore, this study was conducted to evaluate the resistance patterns of five Bacillus species including B. subtilis, B.atrophaeus, B. amyloliquefaciens, B. simplex and B. tequilensis in vitro towards antibiotic on NA media. The inhibition of Bacillus growth was detected after 72 hours of incubation. The obtained data indicated that Bacillus resistance to antibiotic was species-related as B. subtilis, (N = 19; % 94.74), B. amyloliquefaciens, (N = 11; % 90.91), and B. atrophaeus (N = 3; % 0), B. simplex (N = 2; % 0) and for B. tequilensis (N = 3; % 66.6) towards Cefazolin. Most of the Bacillus strains were resistant against Cefazolin (79%), Tobramycin (16%), Pefloxacin (11%), and Gentamycin (11%), whereas no resistance was shown against Levofloxacin and Nitrofurantoin. The percentage of zone inhibition of the Bacillus ssp. colonies (in mm diameter) ranged from 0 to 60 mm, compared to the untreated control. The present work has demonstrated species-specific differences in the antimicrobial susceptibility of Bacillus spp. and observation Bacillus resistant strains is of alarm about how this resistance could spread to other bacteria, and then to humans.
Antimicrobial resistance, bacillus species, inhibition zone
Короткий адрес: https://sciup.org/143184711
IDR: 143184711
Текст научной статьи In vitro evaluation of antibiotic resistance patterns among Bacillus species
Antibiotic-resistant bacteria are one of the most important problems affecting public health at the current century, since the natural environment plays a key role in the wider spread of this phenomenon (Bhullar et al., 01 ; Aslam et al., 0 1). Selection of resistant organisms in nature might result from natural creation of antibiotics by soil organisms, from animals or human’s wastes or animal feed or crops runoff (Ben et al., 019). However, although soils are the largest repositories of microbial diversity, the procedures that determine their microbial community dynamics are not completely understood. Increasing our understanding of the antibiotic resistance levels in soils is extremely important to protect public health, since it has been found that the harmless soil bacteria could be the original source of some antibiotic resistance genes identified in hospitals (Forsberg et al., 01 ).
It has been reported that agricultural soils have multidrug resistant bacteria and have the highest level of resistance to three major antibiotic classes (Popowska et al., 01 ). Therefore, more research on soil-associated antibiotic-resistant bacteria, from the ecological reservoir to humans, is urgently needed. Recently, the emergence of antibiotic resistance Bacillus has increased and its likely public health consequences have resulted in many countries (Lee et al., 019; Cha et al., 0 3). To inhibit the spread of Antimicrobial resistance, it is essential to evaluate Bacillus spp. and their antimicrobial resistance profile. Some strains of Bacillus spp. are becoming increasingly resistant to antibiotics, allowing for the acquisition and emergence of new antimicrobial resistance strains (Lee et al., 019).
Although Bacillus species have been used as food microbial supplements, information obtainable on the profiles of antimicrobial susceptibility is fairly limited. Even less evidence is presently accessible on the antimicrobial susceptibility of Bacillus spp. isolated from soils; therefore, the purpose of this study was to identify the antibiotic resistance of local Bacillus spp. isolated from different soil sites in Syria.
MATERIALS AND METHODS
Bacterial strains
In this study, out of 5 5 Bacillus isolates, 38 strains consisting of B. subtilis , B. atrophaeus , B. amyloliquefaciens , B. simplex and B. tequilensis were used (Table 1). They were collected from soil samples of different Syrian regions (Ammouneh et al., 011; Harba et al., 0 0). The strains were grown on nutrient broth (NB) culture and the colonies of prospective Bacillus s sp. were identified according to Wulff et al . ( 00 ) (Table 1).
In vitro susceptibility to antibiotic
Six antibiotics; Cefazolin, Tobramycin , Pefloxacin, Gentamycin, Levofloxacin and Nitrofurantoin were tested in this study. To evaluate susceptibility to each antibiotic , Bacillus spp. strains were grown on NB media . Plates were inoculated ( × 108 CFU/ml) and allowed to dry for 5 minutes, before adding the antibiotic disk and incubated at 5°C for 7 h. Plates without antibiotic were served as controls. Susceptibility to antibiotic was tested using the inhibition zone diameter (Masood and Aslam, 010) which is classified as follows; ≥ 1 mm (susceptible: +), 0-14 mm (intermediate resistant: ++) and >13 mm or no zone of inhibition (resistant: +++).
Data analysis
All experiments were achieved three times with three Petri dishes per replicate, for each bacterium- antibiotic in vitro evaluation. Presence of a clear zone around Bacillus colonies was considered as an index of the antibiotic sensitivity which were analyzed descriptively and presented here using the diameter of colonies.
RESULTS AND DISCUSSION
In the present work the susceptibilities of 38 Bacillus strains comprising five Bacillus spp. ( B. subtilis, B. atrophaeus, B. amyloliquefaciens, B. simplex and B. tequilensis ) against 6 antibiotics were determined on NA media. Susceptibility of Bacillus spp. was concluded and validated by the inhibition zone towards six antibiotics comparing with the control 7 hours post incubation (Fig. 1). The obtained data indicated that Bacillus resistance to antibiotic was species-related as B. subtilis , (N = 19; % 94.74), B. amyloliquefaciens , (N = 11; % 90.91), and B. atrophaeus (N = 3; % 0), B. simplex (N = ; % 0) and for B. tequilensis (N = 3; %
66.6) towards Cefazolin. Most of the Bacillus strains were resistant against Cefazolin (79%), Tobramycin (16%), Pefloxacin (11%), and Gentamycin (11%), whereas no resistance were shown against Levofloxacin and Nitrofurantoin (Table ; Fig. ).
Table 1: Bacillus species used in the study.
|
Bacillus Species |
Number of strains |
Morphology |
|
atrophaeus |
3 |
brown-black, opaque, smooth, circular |
|
amyloliquefaciens |
11 |
creamy white with irregular margins |
|
simplex |
small, creamy, smooth, glossym, circular, sticky texture |
|
|
subtilis |
19 |
fuzzy white, opaque, rough, with jagged edges |
|
tequilensis |
3 |
yellowish, opaque, smooth, circular |
Table 2: Antibiotic sensitivity of Bacillus species
|
No. |
Bacillus spp. |
Nitrofurantoin |
Gentamycin |
T obramvcin |
Levofloxacin |
Cefazolin |
Peflcxacin |
|
1 |
amvlciiquefaciens 82 c |
-4- |
4-4- |
4- |
4- |
||
|
2 |
96 c |
- |
— |
— |
- |
4-4-4- |
- |
|
3 |
96 e |
— |
— |
— |
— |
— |
— |
|
4 |
123.a |
4- |
4-4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
5 |
128.b |
— |
— |
— |
— |
— |
— |
|
6 |
134 c |
4- |
-H- |
4-4- |
4- |
4-4-4- |
4- |
|
7 |
159.d |
4- |
-H- |
4-4- |
4- |
4-4-4- |
4- |
|
8 |
177.C |
4- |
4- |
4-4- |
4- |
4-4-4- |
4- |
|
9 |
185 c |
4- |
4- |
4- |
4- |
4-4-4- |
4- |
|
10 |
19O.d |
4- |
4- |
4- |
4- |
4-4-4- |
4- |
|
11 |
200.d |
4- |
-H- |
4- |
4- |
4-4-4- |
4- |
|
12 |
atrcpheus 15.b |
-1- |
-4-4- |
4-4- |
4- |
4- |
4-4- |
|
13 |
63 e |
4- |
-4-4- |
4-4- |
4- |
4- |
4-4-4- |
|
14 |
199.a |
4- |
4-4- |
4-4- |
4- |
4- |
4- |
|
15 |
simplex 30. a |
-1- |
-4-4- |
4-4- |
4- |
4- |
4-4-4- |
|
16 |
198.b |
4- |
4-4- |
4-4- |
4- |
4- |
4-4-4- |
|
17 |
subtilius 35.a |
-1- |
4- |
4-4- |
4- |
1 I I |
-1- |
|
18 |
41.b |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
19 |
44. a |
4- |
4-4- |
4-4-4- |
4- |
4-4-4- |
4- |
|
20 |
6O.a |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
21 |
73.b |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
22 |
113.C |
— |
— |
— |
— |
— |
— |
|
23 |
116.C |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
24 |
118 c |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
25 |
124.b |
4- |
4-4- |
4-4- |
4- |
4- |
4- |
|
26 |
130.d |
4- |
4-4- |
4-4-4- |
4- |
4-4-4- |
4- |
|
27 |
132.C |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
28 |
132.e |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
29 |
133 |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
30 |
134.d |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
31 |
135.d |
4- |
4-4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
32 |
139-d |
4- |
4-4- |
4-4-4- |
4- |
4-4-4- |
4- |
|
33 |
151.C |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
34 |
168 c |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
35 |
19O.e |
4- |
4-4- |
4-4- |
4- |
4-4-4- |
4- |
|
36 |
tequilensis 69a |
-1- |
-4-4- |
-4-4- |
4- |
-1- |
1 I I |
|
37 |
145.d |
+ |
4-4- |
4-4-4- |
4- |
4-4-4- |
4- |
|
38 |
150.d |
-4- |
4-4- |
4-4- |
4- |
I I I |
-1- |
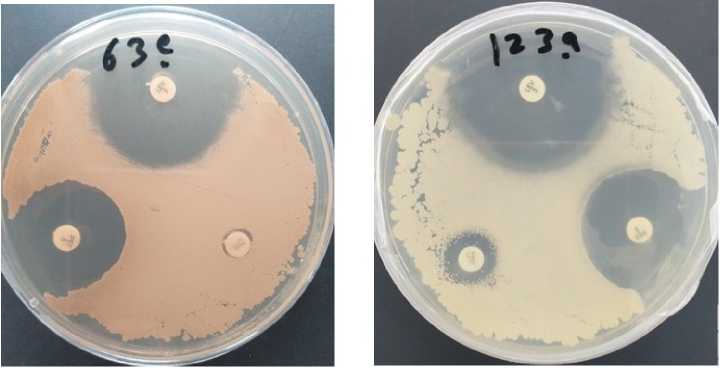
Figure 1. Testing of antimicrobial resistance in vitro . These petri dishes contain Bacillus (creamy yellow and creamy read) cultured on NB media; B. atrophaeus (63e) and B. amyloliquefaciens (1 3a). The white discs each contain antibiotic. Where clear zones appear around the discs, bacterial growth has been prevented by the antibiotic.
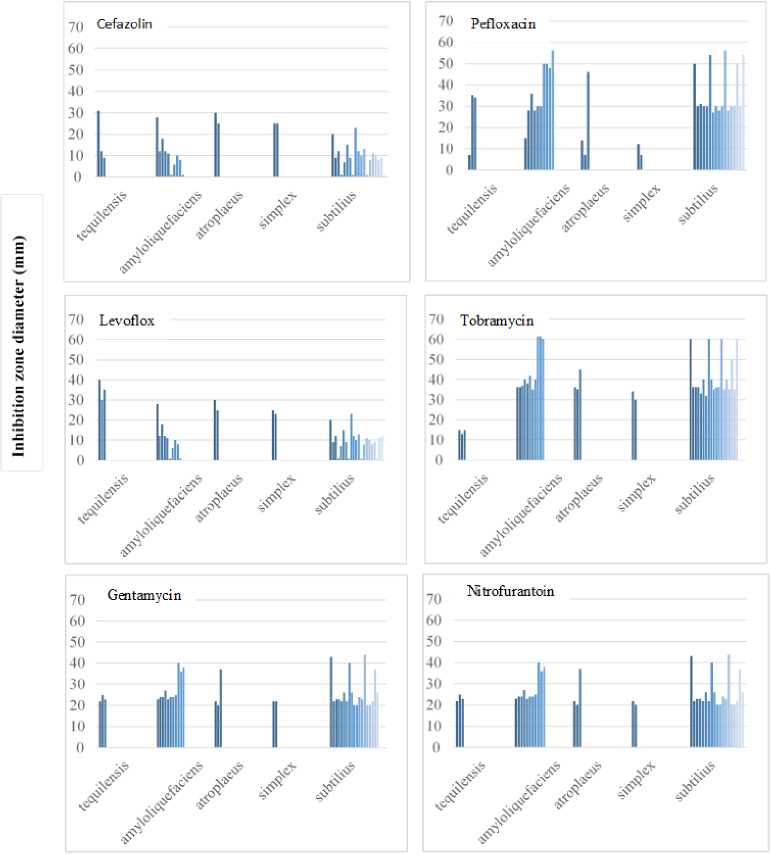
Number of strains
Figure 2. Distribution of inhibition zone diameter in the in vitro six antibiotic resistance test among Bacillus species
The percentage of zone inhibition of the Bacillus ssp. colonies (in mm diameter) ranged from 0 to 60 mm, compared to the untreated control. suggesting the existence of potential risk for Bacillus spp. infections (Fig. 1). These results therefore demonstrated the presence of species-specific variations in the antimicrobial susceptibility of Bacillus spp., parallel to the remarks of other workers (Reva et al., 1995; Lee et al., 019).
Since Bacillus spp. have clinical importance, defining their resistance to antibiotics is necessary for treatment during outbreaks. Our results demonstrated that some Bacillus spp. Such as B. amyloliquefaciens, B. atrophaeus and B. tequilensis were highly resistant towards Cefazolin (Fig. ), and in low resistance levels against Tobramycin, Pefloxacin and Gentamycin, and these are in line with other studies that found such resistance of Bacillus spp. to multi-antibiotic treatments in several sources, including foods (Spyridaki et al., 000 Cha et al., 0 3).
Different mechanisms of bacitracin resistance among bacteria have been reported (Bernard et al., 005; Halawa et al., 0 4) However, little is known about the mechanisms of this resistance in Bacillus spp Podlesek et al ( 000) observed further that deletion of the bcrA or the bcrC gene severely impaired bacitracin resistance. However, nothing is known about other possible mechanisms of resistance to quinolones, namely the elimination of antibiotics through an active efflux system and/or decreased outer membrane permeability.
On the other hand, horizontal gene transfers are the main mechanisms through which antibiotic resistance genes are exchanged among bacteria of diverse origin including environmental, non-pathogenic, human pathogenic, gram- positive and negative via mobile DNA elements such as plasmids and transposons – either with or without mobile integrons (Pruden et al., 006). This might explain the antibiotic resistance among Bacillus species detected in this study.
CONCLUSION
In conclusion, our study demonstrated that Bacillus strains were highly susceptible to Levofloxacin and
Nitrofurantoin. Species-specific variations in the patterns of resistance to Cefazolin Tobramycin, Pefloxacin, Gentamycin were observed. We found also that B. subtilis , (N = 19; % 94.74), B. amyloliquefaciens , (N = 11; % 90.91) and for B. tequilensis (N = 3; % 66.6) were sensitive to Cefazolin, and in low resistance levels against Tobramycin, Pefloxacin and Gentamycin, suggesting the existence of potential risk for Bacillus spp. infections. Therefore, antimicrobial susceptibility testing is required as routine microbiological analyses of soils in Syria.
ACKNOWLEDGMENTS
The authors would like to thank the Director General of AECS and the Head of Molecular biology and Biotechnology Department for their much appreciated help throughout the period of this research. Thanks are also extended to Dr. H. Ammouneh for his assistance in achieving the experiments. We would like also to thank Dr. A. Al-Daoude for critical reading of the manuscript.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.


