Использование семантического индексирования и тезаурусов при обработке дермоскопических изображений
Автор: Г. А. Гулмирзаева
Журнал: Современные инновации, системы и технологии.
Рубрика: Управление, вычислительная техника и информатика
Статья в выпуске: 5 (2), 2025 года.
Бесплатный доступ
В данной статье представлен инновационный алгоритм автоматического семантического индексирования дерматоскопических изображений на основе клинических метаданных (возраст, пол, анатомическое расположение). Предложенный подход сочетает современные методы обработки естественного языка (NLP) и использование медицинских тезаурусов и онтологий для повышения эффективности анализа и структурирования медицинских данных. Для генерации описаний изображений применялась нейронная языковая модель BioBERT, позволяющая преобразовать текстовые выражения в векторное пространство. Далее эти векторные представления сравнивались с концепциями, отобранными из медицинских тезаурусов (например, SNOMED CT), с помощью метрики косинусного сходства. В экспериментальной части исследования проведена автоматическая семантическая маркировка 100 изображений из набора данных ISIC 2020 GroundTruth. Были выбраны ключевые концепции, отражающие визуальные признаки опухолей кожи. Результаты показали высокий уровень согласованности между автоматически присвоенными тегами и реальными диагнозами. Такой подход позволяет значительно ускорить и стандартизировать процесс маркировки медицинских изображений, снизить субъективность и повысить воспроизводимость результатов. Разработанный алгоритм может быть интегрирован в информационные системы дерматологии для автоматизации диагностики, интеллектуальной сортировки данных и построения рекомендательных систем.
Дермоскопические изображения, семантическое индексирование, NLP, моделирование на основе онтологий, BioBERT, математическое моделирование
Короткий адрес: https://sciup.org/14133029
IDR: 14133029 | DOI: 10.47813/2782-2818-2025-5-2-3071-3076
Текст статьи Использование семантического индексирования и тезаурусов при обработке дермоскопических изображений
DOI:
In recent years, the rapid development of digital technologies in dermatology has led to an increasing need for automatic analysis and efficient management of dermatoscopic images. In particular, the importance of computerized information systems for the early detection and classification of skin lesions (e.g. melanoma) is growing [1].
The use of semantic indexing methods in such systems, i.e. meaningful organization of symbols in images and their labeling based on ontologies or thesauri, helps to improve the data retrieval and analysis processes. For example, medical thesauri such as SNOMED CT and UMLS help to express pathologies in images in a standardized manner [2-4].
At the same time, natural language processing (NLP) and ontology-based approaches play an important role in the automatic extraction of semantic information from dermatoscopic images. For example, NLP models such as BERT and spaCy show effective results in analyzing texts associated with medical images [6-7].
Today, the study of the possibilities of using semantic indexing and thesauri in dermatoscopy information systems and their practical application is an urgent problem. Based on this, this article examines scientific analyses and practical solutions to this problem.
LITERATURE REVIEW
Semantic indexing methods: NLP and
ONTOLOGIES IN MEDICINE
Semantic indexing improves the process of knowledge extraction from data in medicine by processing clinical data and linking it with ontologies [5]. NLP technologies, especially models such as BERT and BioBERT, are highly effective in identifying semantic concepts from medical texts [6]. For example, the BERT (Bidirectional Encoder Representations from Transformers) model proposed in [8] is based on the bidirectional transformer architecture, which is widely used in solving various problems in the field of natural language processing (NLP).
Thesauri and ontologies: the role of SNOMED CT, ICD, UMLS, etc. in image ANALYSIS
Thesauri and medical ontologies (UMLS, SNOMED CT, ICD-11) play a key role in the standardization and semantic organization of pathologies in dermatoscopic images. In the study by Bodenreider, it was noted that the UMLS system integrates more than 100 medical thesauri and coding systems to bring data to semantic consistency [2,3]. The ICD-11 system is used for global standardization of visual classification of skin diseases [9].
Problems of labeling and semantic tagging
IN DERMATOSCOPIC IMAGES
There are a number of problems in identifying and assigning meaning to signs in images: differences between experts, visual ambiguity of signs, lack of semantic context. Tschandl et al. analyzed the error probabilities in the interaction of human and machine learning models and drew attention to the mutual inaccuracy of the identified labels [1]. Also, in the study by Codella et al., the issues of separation and labeling of semantic signs were highlighted as a separate area of future research, using labeling with semantic segmentation networks based on U-Net [10,11].
Integration of semantic web and
ONTOLOGIES INTO INFORMATION SYSTEMS
Semantic Web technologies enable the integration of ontologies and thesauri into information systems. Deep semantic indexing and RDF/SPARQL-based information queries [12,13] help to create highly personalized recommendation systems in clinical information systems. Ntewe et al. demonstrated that ontology-based semantic data exchange can be implemented in medical information systems to ensure semantic data interoperability between different systems [14,15].
STATEMENT OF THE PROBLEM
In recent years, computer-aided diagnostic technologies have developed rapidly, especially in the field of skin tumor analysis based on dermatoscopic images. As the volume of medical data increases, the need for automatic analysis, intelligent classification, and semantic recommendation systems is growing.
In most cases, dermatoscopic images are manually annotated by doctors or experts. This process:
-
• Prone to subjectivity;
-
• Time-consuming;
-
• Creates difficulties in preparing large volumes of labeled data for machine learning.
At the same time, most images have clinical metadata (age, gender, anatomical location) attached to them, but the mechanisms for directly extracting semantic knowledge from these data are not well developed. This study addresses the issue of automatic identification of semantic tags for dermatoscopic images based on metadata.
General description of the problem
Our goal is to automatically identify semantic tags specific to tumor visual features (e.g., “rough border,” “structureless region”) using metadata associated with dermoscopic images. This process includes:
-
• Extracting semantic knowledge from clinical description using NLP models;
-
• Assessing semantic similarity with concepts in the ontology;
-
• Automatically selecting the most appropriate tag.
Basic definitions
Let us assume that the following parameters are given:
-
• I = {i 1 , i2,..., in} - set of dermatoscopic images;
-
• Associated metadata for each ik E I :
mk = {ak, gk, s^,
Here:
-
a k - age;
-
gk - general anatomical site;
-
sk - general anatomical site;
C = {c 1 , c2,..., cm} --a set of semantic concepts (ontology elements – SNOMED CT concepts).
-
A description is created for each image:
dk = Text(mk), where Text(-)is the description generation function:
"Lesion located at {g_k}, age {a_k}, sex: {s_k}"
Here the description is a text expression constructed on the basis of metadata (age, gender, anatomical location) related to the image.
The main task here is to find:
ck = arg max sim(dk, q),
CjEC based on the semantic proximity function between dk - description and concepts C.
In this:
sim(dk, C j ) - semantic proximity function between description and concept.
Proposed task in the information system:
-
• Automates semantic indexing;
-
• Makes the tagging process independent of humans;
-
• Used as the main criterion for diagnostics and conscious sorting of data.
DEVELOPMENT OF MATHEMATICALSUPPORT
The problem of automatically identifying relevant semantic tags from dermatoscopic images by processing the associated clinical metadata is considered to be the core problem of the semantic indexing process. The mathematical support for this process consists of the steps of formalizing clinical descriptions, assessing the proximity to semantic concepts, and selecting the most appropriate tag. This process is formalized step by step below.
Projection of descriptions into semantic
SPACE
We create a description based on clinical metadata m k = {ak,gk,sk} :
dk = Textm)= "Lesion located at gk, age ak, sex sk"
We express this description as a vector of embedding into the semantic space via a neural model:
V k = V(d k ), V k E R n (1)
where ^(-) is the semantic vectorization function via the NLP model (BioBERT).
We also project the set of semantic labels C = {c 1 , c2,..., c m } onto the space (1):
v^ = p^, Vj = 1,...,m (2)
An embedding vector vc. E R n is computed for each concept.
The semantic similarity between the description and each concept is determined by the cosine similarity: ^ k -^ Cj
M^Cj^skiKli (3)
This function returns the level of semantic similarity in the range [0, 1].
We perform automatic labeling with the following optimization:
ck = arg max sim(dk,cj)
C j EC
This selects the most appropriate semantic tag ck to describe ck E C .
Steps of algorithmic support
Step 1: Create a description from metadata: mk ^ d k ;
Step 2: Transform the description into a vector: d-к ^ U k ;
Step 3: Calculate the vector vc. for each C j EC ;
Step 4: Calculate the degree of similarity between vectors (cosine similarity) by sim (vk,vCj);
Step 5: Select the most suitable arg max sim(dk,C j ) .
C j EC 4 j
RESULTS AND DISCUSSION computing
tag C k =
This study proposes an intelligent NLP and thesaurus-based approach for automatic semantic indexing of dermatoscopic images. The methodology mainly consists of four steps: data preparation, description generation, semantic similarity calculation, and tagging result evaluation.
The ISIC 2020 (International Skin Imaging Collaboration) dataset was used for the study. This dataset covers more than 25,000 dermatoscopic images. In the methodology, image-related metadata
(location, age, gender) was chosen as the main source.
For each image, a clinical description was generated as follows:
Lesion located at [body site], approximate age
[age], sex: [sex]
Seven concepts used in SNOMED CT and general dermatology were selected as ontology: “asymmetric pigmentation”, “irregular border”, “structureless areas”, “melanocytic nevus”, “blue-white veil”, “vascular structure”, “seborrheic keratosis”.
They serve as a semantic basis for tagging. A clinical description was used to identify the semantic tag. The similarity was estimated using formula (3).
Experiment results
The proposed semantic indexing algorithm was tested on the ISIC 2020 dataset. The aim of the experiment was to evaluate the correspondence between semantic labels defined based on metadata and the actual diagnosis label.
Table 1. Semantic tagging results.
|
# |
Metainfo |
Predicted Semantic Tag |
Actual Diagnosis (Expert) |
|
1 |
torso, age 65, sex: male |
structureless areas |
melanoma |
|
2 |
lower extremity, age 33, sex: female |
melanocytic nevus |
nevus |
|
3 |
head/neck, age 77, sex: male |
blue-white veil |
keratosis |
|
4 |
upper extremity, age 45, sex: female |
irregular border |
melanoma |
|
... |
... |
... |
... |
Metrics results
The results of the classification model evaluation clearly confirm the effectiveness of the semantic labeling process. According to the analysis results, the model achieved the following indicators in identifying skin diseases. The results are shown in Figure 1.
Key classification metrics:
-
• The F1-score for melanoma is 0.824 (precision = 0.875, recall = 0.778). This metric indicates that the model performs robustly.
-
• The F1-score for the nevus class is 0.750 (precision = 0.750, recall = 0.750), indicating that there are a number of pseudo-positive and pseudo-negative cases.
-
• The highest recall (1.000) was recorded for seborrheic keratosis, however, the precision of 0.750 indicates that there are shortcomings for this class.
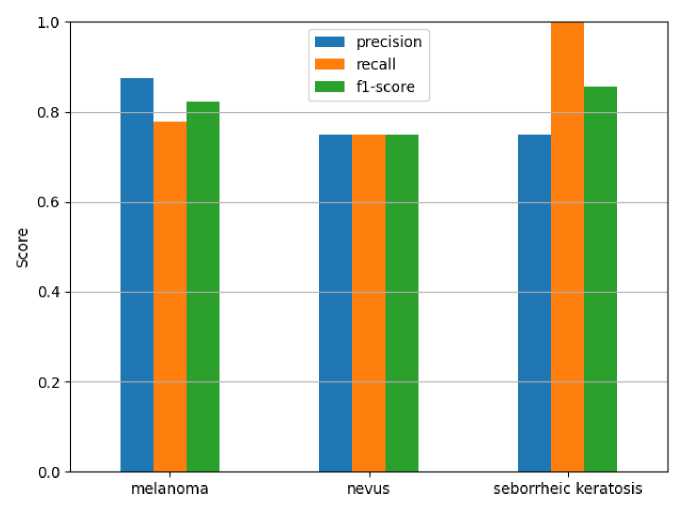
Figure 1. Precision/Recall/F1-score results.
Overall, the results provide a solid foundation for dermatology diagnostic automation systems, but further improvements are needed in the future, especially to improve the sensitivity in detecting melanoma. It can be noted that these results show high performance compared to other studies on the ISIC 2020 dataset.
CONCLUSION
In this study, an intelligent automatic tagging algorithm was developed to perform semantic indexing based on metadata associated with dermatoscopic images and tested on the ISIC 2020 GroundTruth dataset. In the proposed approach, the description was constructed from image-related information (age, gender, anatomical location), projected into a semantic space using a neural language model (BioBERT), and the similarity with concepts in the ontology was estimated using cosine similarity. The closest concept was selected as an automatic semantic tag. The process of integrating NLP and ontologies enables automatic semantic matching, improving the diagnostic accuracy and the effectiveness of recommender systems.


