Экспрессия C/D МЯКРНК в клеточных линиях лейкоза при хромосомных аномалиях после облучения
Автор: Расторгуева Е.В., Погодина Е.С., Юрова Е.В., Белобородов Е.А., Сугак Д.Е., Тумозов И.А., Саенко Ю.В., Фомин А.Н.
Журнал: Сибирский онкологический журнал @siboncoj
Рубрика: Лабораторные и экспериментальные исследования
Статья в выпуске: 6 т.23, 2024 года.
Бесплатный доступ
Цель исследования - изучить экспрессию C/D мякРНК ((англ. snoRNA) малые ядрышковые РНК) в хромосомах с разным уровнем нарушений и оценить возможность их использования в качестве биомаркеров радиорезистентности при хромосомных аномалиях. Материал и методы. В исследовании сравнивали значения log2FC экспрессии мякРНК C/D семейства в радиочувствительной (HL-60) и радиорезистентной (K562) клеточных линиях, с разным уровнем хромосомных нарушений. Клетки облучались рентгеновским излучением однократно в дозе 4 Гр. Оценивали экспрессию мякРНК C/D через 1, 4 и 24 ч после облучения при помощи секвенирования нового поколения (NGS) MiSeq.
Нкрнк, мякрнк (бокс) семейство, биомаркеры, радиорезистентность, хромосомные нарушения
Короткий адрес: https://sciup.org/140308743
IDR: 140308743 | УДК: 616.15:577.21:576.316 | DOI: 10.21294/1814-4861-2024-23-6-97-106
Текст научной статьи Экспрессия C/D МЯКРНК в клеточных линиях лейкоза при хромосомных аномалиях после облучения
Все чаще в роли регуляторов экспрессии генов выступают некодирующие РНК (нкРНК). Одними из них являются малые ядрышковые РНК (мякРНК), их подавление или экспрессия может привести к различным патогенезам, в том числе онкогенезу [1]. Первые исследования мякРНК начались более 50 лет назад [2]. Это консервативный класс нкРНК, участвующих в модификации (метилирование рибозы и псевдоуридилирование), процессинге и сборке рРНК. мякРНК длиной от 60 до 300 нуклеотидов делятся на два основных бокса в зависимости от структуры и функции: C/D мякРНК, участвующие в 2’-модификации О-метилирования рибосомной РНК; и H/ACA мякРНК, ответственные за псевдоуридилирование рРНК [2, 3].
В данной работе мы рассмотрели экспрессию C/D мякРНК, имеющих длину от 50 до 100 нуклеотидов и характеризующихся своими консервативными мотивами: бокс C/C’ (RUGAUGA) и D/D’. Они взаимодействуют с белками SNU13, NOP56, NOP58 и метил трансферазой фибрилла-рином (FBL) с образованием C/D мякРНП (англ. snoRNP, малый ядрышковый рибонуклеопротеин). C/D мякРНК необходимы для модификации рРНК. Однако многие из них не имеют идентифицированной канонической модификации мишени и называются орфанными мякРНК, некоторые мякРНК также были описаны как управляющие модификацией матричных РНК (мРНК), расширяющих границы потенциальных РНК мишеней [4, 5]. Множество функций C/D мякРНК еще предстоит открыть [5]. Имеются данные, что H/ACA бокс и C/D бокс – эволюционные предки подмножества предшественников микроРНК [6, 7]. В работе M. Ono et al. [6] протестированы предшественники микроРНК, бокса C/D мякРНК (miR-27b, miR-16-1, mir-28, miR-31 и let-7g), они связываются с фибрил-ларином, специфическим белковым компонентом функционального бокса C/D мякРНП комплекса [8]. Недавние исследования выявили аномальную экспрессию и продемонстрировали ключевую роль мякРНК и их генов-хозяев в различных типах гематологических и других злокачественных новообразований [8–13].
мякРНК могут не только регулировать клеточные процессы, но и модулировать радиочувстви- тельность или радиорезистентность клеток [14]. Хорошо известно, что лучевая терапия является одним из основных методов лечения онкологических заболеваний. Было обнаружено, что GAS5 модулирует радиационный ответ путем нацеливания miR-205-5p на путь Wnt/β-catenin и регулирует апоптоз и жизнеспособность клеток после облучения [15]. GAS5 также повышает радиочувствительность в нескольких моделях за счет нацеливания на miR-106b, которая неспособна ингибировать ген IER3, немедленного раннего ответа, супрессора рака шейки матки [16]. В результате радиочувствительность повышала сверхэкспрессию GAS5. В другом исследовании также создавали радиочувствительное состояние клеток рака легких при сверхэкспрессии семейства miRNAslet-7, а снижение экспрессии этого семейства вызывало радиорезистентность [17]. В следующей работе показаны результаты, при которых днкРНК (англ. lncRNA, длинная некодирующая РНК) SNHG12 способствала экспрессии CDK1 для регуляции чувствительности клеток рака шейки матки к радиации при помощи miR-148a, что дает возможность использовать SNHG12 как биомаркер при данном заболевании [18].
В этих и других исследованиях [14, 19–21] подчеркиваются важность некодирующих РНК при онкогенезе и их влияние на чувствительность клеток к лучевой терапии. Есть ряд исследований, показывающих роль микроРНК и длинных некодирующих РНК в ответ на облучение при различных видах рака [22–24]. Но на сегодняшний момент мало данных об участии C/D мякРНК семейства в клеточном ответе на облучение.
Цель исследования – изучить экспрессию C/D мякРНК ((англ. snoRNA) малые ядрышковые РНК) в хромосомах с разным уровнем нарушений и оценить возможность их использования в качестве биомаркеров радиорезистентности при хромосомных аномалиях.
Материал и методы
В работе использовали клеточные линии промиелоцитарного лейкоза человека HL-60 (RRID:CVCL_0002) и хронического миелоидного лейкоза человека К562 (RRID:CVCL_0004) (Институт цитологии РАН, г. Санкт-Петербург). Клетки культивировали в следующих условиях: 37 °С, 5 % CO2, 98 % влажности. Использовали среду RPMI-1640 с L-глутамином (ПанЭко, Россия), с добавлением 50 мкг/мл гентамицина (ПанЭко, Россия) и 10 % эмбриональной бычьей сыворотки (PAA Laboratories GmbH, Австрия).
Клетки облучались в дозе 4 Гр γ-излучением (энергия фотонов 10 МэВ), линейным ускорителем «Elekta Synergy» (Elekta, Швеция) 55 сек, при комнатной температуре. Эксперимент проводили в логарифмической фазе роста клеток. Тотальную РНК выделяли из клеток через 1, 4 и 24 ч после облучения набором Absolutely RNA miRNA Kit (Agilent Technologies, США). Библиотеки кДНК готовили с использованием набора NEBNext Small RNA Library Prep Set (NEB, Великобритания). Их очистку осуществляли в 6 % полиакриламидном геле с помощью электрофореза. Количество кДНК оценивали флуориметром «Qubit» (Invitrogen, США). Затем отбирали эквимолярные количества (2 нМ) образца для пула библиотек. Секвенировали с помощью системы высокопроизводительного секвенирования «MiSeq System» (Illumina, США), применяя набор для одноконцевого чтения 150 п.н. Обработку полученных файлов FASTQ осуществили при помощи сервиса GenXPro omiRas. Величину экспрессии С/D мякРНК выражали через log2 отношения нормализованной экспрессии мякРНК в опыте к аналогичному показателю в контроле (log2fc).
Для определения локализации С/D мякРНК в хромосомах использовали базу данных National Center for Biotechnology Information Search data base (RRID:SCR_006472).
Эксперимент проводили в трех повторах. Для оценки статистической значимости различий применяли критерий Манна‒Уитни, обработка проводилась в программе Origin. Различия между группами считали достоверными при р<0,05.
Эти материалы и методы описаны более подробно в наших предыдущих статьях [25–27], где рассматривались другие классы некодирующих РНК.
Результаты
Количество экспрессирующихся C/D мякРНК через 1, 4 и 24 ч в каждой хромосоме после облучения 4 Гр, в двух клеточных линиях изображено на рис. 1. Через 1 ч мы наблюдаем одинаковое количество C/D мякРНК в 1, 2, 4, 5, 6, 9, 10, 12, 16, 17, 18, 22 и XX хромосоме. В 3, 7, 8, 11, 15, 19 и 20-й хромосоме количество C/D мякРНК выше в клеточной линии HL-60, причем в 15-й хромосоме больше в 2,5 раза. Только в 14-й хромосоме количество экспрессирующихся C/D мякРНК через 1 ч выше в клеточной линии K562.
Че+рез 4 ч после облучения одинаковое количество C/D мякРНК остается в 1, 2, 4, 5, 6, 10, 12, 16, 17, 18, 22 и XX хромосоме, в 9-й их количество стало выше, как и в 3, 7, 8, 11, 15,19 и 20-й хромосоме в HL-60 клеточной линии. В 15-й хромосоме количество C/D мякРНК в 7 раз выше в HL-60 по сравнению с K562. Количество C/D мякРНК больше в К562 только в 14-й хромосоме.
После облучения через 24 ч мы наблюдаем одинаковое количество C/D мякРНК в обеих клеточных линиях в 4, 6, 8, 10, 12, 18, 19 и XX хромосоме. В HL-60 количество C/D мякРНК выше, чем в K562, в 3, 7, 15-й (в 2 раза) хромосоме. В 1, 2, 9,
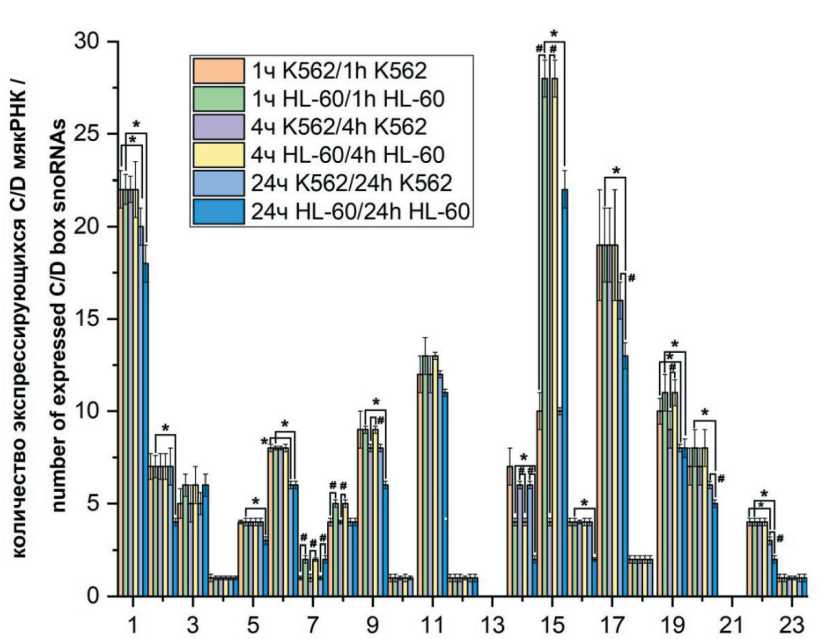
Хромосомы (1-22, XX) /
Chromosomes (1-22, XX)
Рис. 1. Количество экспрессирующихся C/D мякРНК после облучения в каждой хромосоме K562 и HL-60 (* – p<0,05 при сравнении с группой через час после облучения, # – p<0,05 при сравнении между K562 и HL-60 клеточными линиями). Примечания: ось абсцисс – номер хромосом; ось ординат – количество экспрессирующихся C/Dмяк РНК после облучения;
рисунок выполнен авторами
Fig. 1. The number of expressed C/D snoRNAs after irradiation in each chromosome K562 and HL-60 (* – p<0.05 when compared with the group 1hour after irradiation). Notes: Abscissa axisis a chromosome number; ordinate axis is a number of expressed C/D snoRNAs after irradiation; created by the authors
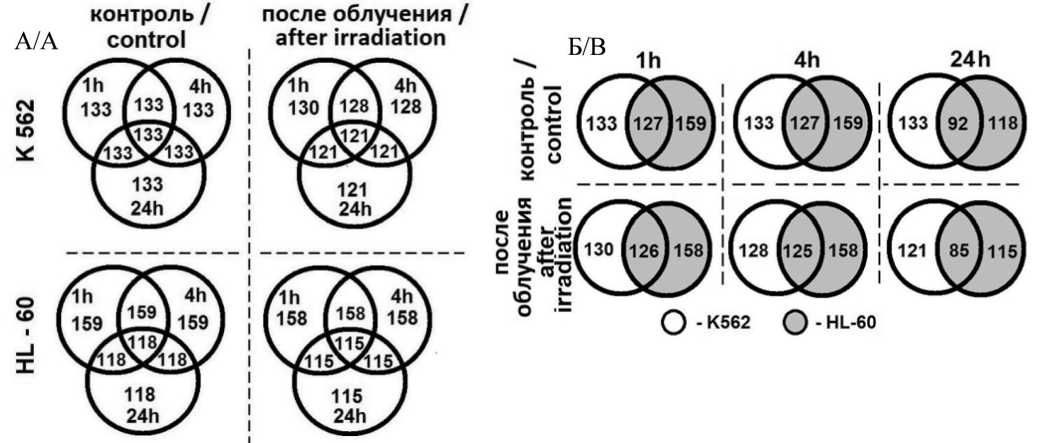
Рис. 2. Общее количество C/D мякРНК в K562 и HL – 60 клеточных линиях (в контроле и после облучения через 1, 4 и 24 ч). Примечания: A – количество по клеточным линиям; Б – сравнение между клеточными линиями; рисунок выполнен авторами Fig. 2. The total number of C/D snoRNA in K562 and HL – 60 cell lines (in control and after irradiation at 1 hour, 4 and 24 hours).
Notes: A – amount by cell lines, B – comparison between cell lines; created by the authors
11, 14, 16, 17, 20, 22-й хромосоме после облучения через 24 ч экспрессирующихся C/D мякРНК стало больше в клеточной линии K562. Статистически значимые отличия получены в основном при сравнении групп после 24 ч облучения с группами через 1 ч облучения внутри клеточных линий, а также в 7, 8, 9, 14, 15, 17, 19, 20, 22-й хромосоме между клеточными линиями в основном через 4 и 24 ч.
На рис. 2 показаны изменения в общем количестве C/D мякРНК после облучения через 1, 4 и 24 ч. У клеточной линии K562 в контроле (рис. 3A) количество C/D мякРНК не меняется в течение всего времени, а после облучения разница с контролем небольшая: в контроле через 24 ч – 133 C/D мякРНК, а через 24 ч после облучения– 121 C/D мякРНК. В клеточной линии HL-60 на этом же рисунке мы наблюдаем уменьшение C/D мякРНК как в контроле (на 41), так и после облучения (на 43) через 24 ч по сравнению с количеством C/D мякРНК через 1 и 4 ч. На рис. 2Б показано общее количество C/D мякРНК в обеих исследуемых клеточных линиях в контроле и после облучения. Как в контроле, так и после облучения через 1 и 4 ч общее количество C/D мякРНК практически не меняется, через 24 ч она снижается на 30 % в контроле и на 33 % после облучения. Данное снижение количества C/D мякРНК происходит за счет клеточной линии HL-60. Несмотря на то, что количество экспрессирующихся C/D мякРНК в данной клеточной линии изначально больше, но с течением времени оно меняется как в контроле через 24 ч, так и после облучения через 24 ч.
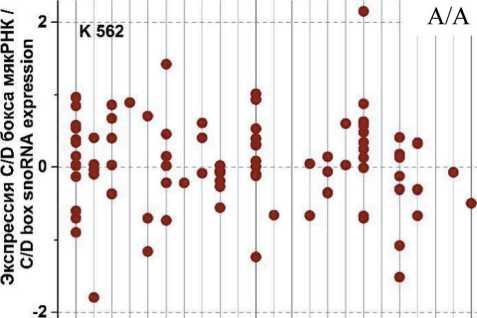
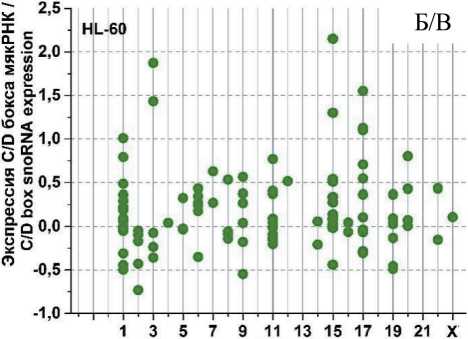
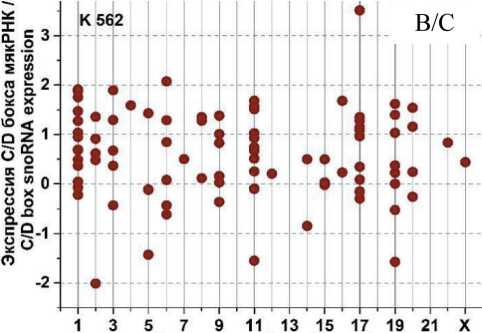
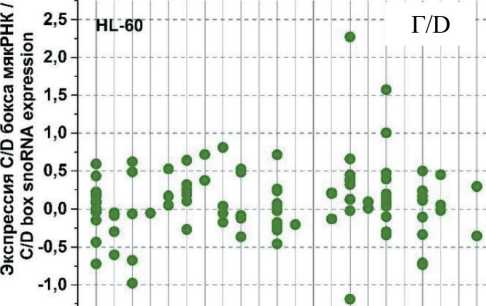
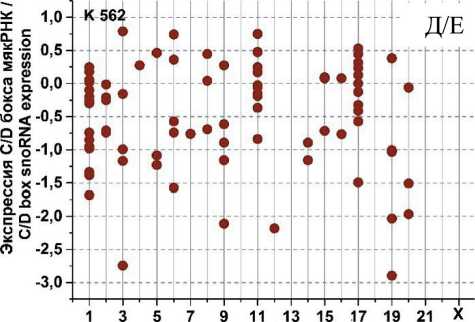
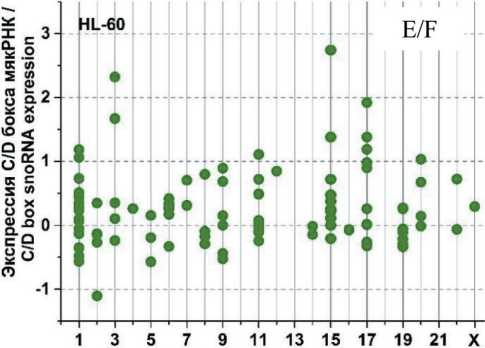
На рис. 3 представлены графики разброса значений log2fc всех экспрессирующихся C/D мякРНК в клеточных линиях HL-60 и К562, распределенных в соответствии с их локализацией в хромосомах, после облучения в дозе 4 Гр через 1, 4 и 24 ч. Через 1 ч (рис. 3А) после облучения в культуре K562 отмечается разброс log2fc в диапазоне от -1,75 до +2. При этом особо выделяется положительная экспрессия C/D мякРНК в 6 (max log2fc=1,4) и 17-й (max log2fc =2) хромосоме и отрицательная в 2, 11 и 19-й хромосоме (min log2fc= -1,75; -1,25 и -1,5, соответственно). Другая картина наблюдается в культуре HL-60 (рис. 3Б). Здесь значения log2fc варьируются в диапазоне от -0,7 до 2. Положительная экспрессия C/D мякРНК наблюдается в хромосомах 3, 15 и 17 (max log2fc = 1,9; 2 и 1,5, соответственно). Однако в половине хромосом фиксируется незначительная отрицательная экспрессия C/D мякРНК.
Через 4 ч после облучения (рис. 3В) в культуре K562 наблюдается разброс log2fc в диапазоне от -2 до +3,5. Положительная экспрессия C/D мякРНК наблюдается в большинстве хромосом, однако фиксируется отрицательный log2fc (в пределах от -1 до -2) для хромосом 2, 5, 11 и 19. Для культуры HL-60 (рис. 3Г) характерна другая картина, диапазон разбросов log2fc от -1,2 до +2,2. Положительная и/или отрицательная экспрессия C/D мякРНК наблюдается во всех хромосомах (большинство значений в диапазоне от -0,5 до +0,7), кроме 10, 13, 16, 18 и 21-й. Отдельно стоит отметить 15-ю хромосому, в которой экспрессия отдельных C/D мякРНК варьирует от минимального до максимального значения для всех групп в целом.
После облучения через 24 ч (рис. 3Д) в обеих культурах наблюдается противоположная картина. Если в культуре K562 можно увидеть в большинстве хромосом экспрессию C/D мякРНК с отрицательным log2fc (min log2fc=-3), то в культуре HL-60 (рис. 3Е) в большинстве хромосом положительная экспрессия и log2fc (max log2fc=+3,2). При этом особо выделяется отрицательная экспрессия C/D мякРНК в хромосомах 3, 9, 12 и 19 в культуре K562 и положительная экспрессия C/D мякРНК в 3, 15 и 17-й в культуре HL-60.
Таким образом, при анализе изменения log2fc всех экспрессирующихся C/D мякРНК после облучения дозой 4 Гр, в течение 1, 4 и 24 ч в культуре
1 3 5 7 9 11 13 15 17 19 21 X
Хромосомы (1-22, X); 1 ч / Chromosomes (1-22, X); 1 h
Хромосомы (1-22, X); 1 ч / Chromosomes (1-22, X); 1 h
Хромосомы (1-22, X); 4 ч / Chromosomes (1-22, X); 4 h
т—'————1——'————'——’—I—'——1——'—Г"
1 3 5 7 9 11 13 15 17 19 21
Хромосомы (1-22, X); 4 ч /
Chromosomes (1-22, X); 4 h
Хромосомы (1-22, Х);24ч / Chromosomes (1-22, X);24h
Хромосомы (1-22, Х);24ч / Chromosomes (1-22, X);24h
Рис. 3. Log2FC дифференциально экспрессирующихся C/D мякРНК через 1, 4 и 24 ч после облучения в дозе 4Гр в K562 и HL-60 клеточных линиях, в каждой хромосоме. Величину экспрессии С/D мякРНК выражали через log2 отношения нормализованной экспрессии мякРНК в опыте к аналогичному показателю в контроле (log2fc). Примечания: ось абсцисс – номер хромосом; ось ординат – экспрессия бокса C/Dмяк РНК после облучения, через 1, 4 и 24 ч; рисунок выполнен авторами
Fig. 3. Log2FC differentially expressed C/D snoRNA 1, 4 and 24 hours after irradiation at a dose of 4Gr in K562 and HL-60 cell lines, in each chromosome. The C/D snoRNA expression was determined by log2 ratio between the normalized snoRNA expression in the experiment and that in the control (log2fc). Notes: abscissa axis is a chromosome number; ordinate axis – expression of C/Dbox snoRNA 1 hour, 4 and 24 hours after irradiation; created by the authors
K562 можно отметить увеличение экспрессии C/D мякРНК в 6 и 17-й хромосоме и уменьшение во 2, 5, 11 и 19-й хромосоме через 1 ч, с последующим сохранением данных показателей через 4 ч и значительным снижением экспрессии C/D мякРНК в большинстве хромосом через 24 ч. В культуре HL-60 в первый час после облучения отмечается увеличение экспрессии C/D мякРНК в 3, 15 и 17-й хромосоме с частичным сохранением показателей в 15 и 17-й хромосоме и снижением экспрессии отдельных C/D мякРНК в 3 и 17-й хромосоме через 4 ч. Однако через 24 ч картина возвращается к первоначальному виду.
На рис. 4 представлена экспрессия отдельных C/D мякРНК, выраженная через log2FC к контролю. Здесь были выбраны хромосомы для сравнения с разным уровнем хромосомных аберраций в исследуемых клеточных линиях. В 3, 6 и 11-й хромосоме наблюдалась экспрессия одинаковых C/D мякРНК в обеих клеточных линиях, но с раз-
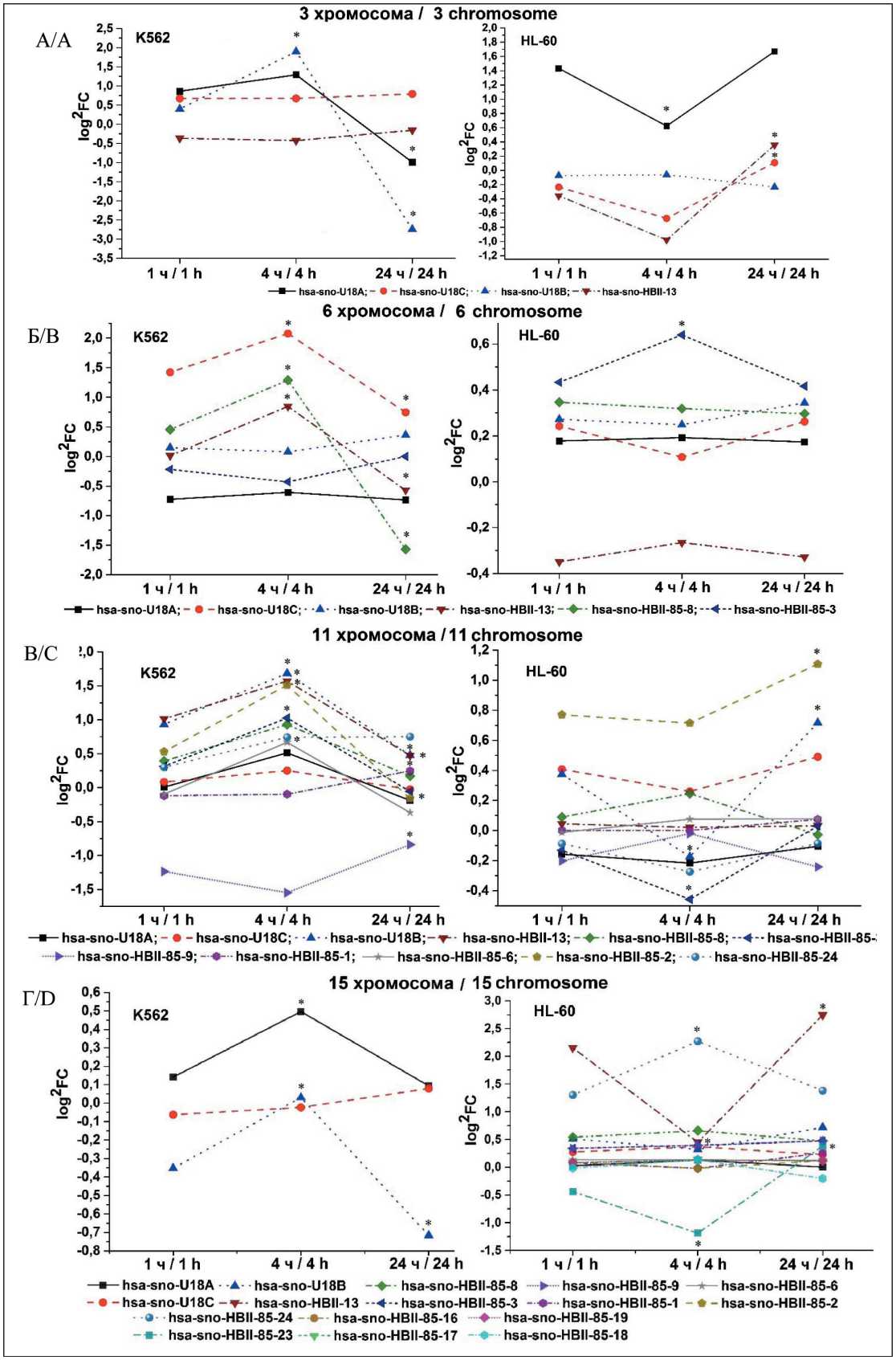
Рис. 4. Log2FC C/D мякРНК в 3, 6, 11 и 15-й хромосоме в K562 и HL-60 клеточных линиях (* – p<0,05 при сравнении с группой через 1 ч после облучения). Примечания: ось абсцисс – время после облучения (1, 4 и 24 ч); ось ординат – значения Log2FC C/D мякРНК после облучения; рисунок выполнен авторами
Fig. 4. Log2FC C/D snoRNA in the 3rd, 6th, 11th and 15th chromosomes in K562 and HL-60 cell lines (* – p<0.05 when compared with control groups without irradiation, # – p<0.05 when comparing expression between K562 and HL-60 cell lines). Notes: abscissa axis – time after irradiation (1, 4 and 24 hours); ordinate axis – Log2FC C/D values of snoRNA after irradiation; created by the authors ными значениями log2FC. Так, в культуре K562 в 3-й хромосоме (рис. 4А) has-sno-snR39B имеет значение +0,9 в первый час с последующим снижением до -1 через 24 ч. В культуре HL-60 для has-sno-snR39B отмечается log2FC в первый час +1,4 с последующим снижением в течение следующих часов до +0,7 и повышением до +1,7 через 24 ч после облучения. мякРНК hsa-sno-HBII-142, как и hsa-sno-E2 в 3-й хромосоме культуры K562, не показывают значительных отклонений от первоначальных значений (для hsa-sno-HBII-142 log2FC в области +0,75, для hsa-sno-E2 – -0,25). Однако в культуре HL-60 другая тенденция. Так, hsa-sno-HBII-142 с первоначального значения log2FC -0,2 через 1 ч после облучения снижается до -0,7 через 4 ч и снова повышается до +0,1 через 24 ч. Для hsa-sno-E2 в культуре HL-60 зависимость аналогична более выраженной экспрессии через 24 ч. В случае с hsa-sno-E3 картина обратная. В культуре K562 в первые 4 ч отмечаются повышение экспрессии мякРНК до log2FC=+2 и резкое снижение в течение следующих 20 ч до log2FC=-2,75. В культуре HL-60 подобного не наблюдается, и значение log2FC сохраняется в пределах от -0,1 до -0,2 в течение всех 24 ч после облучения.
В 6-й хромосоме (рис. 4Б) особое внимание стоит уделить следующим мякРНК: hsa-sno-U52; hsa-sno-U83; hsa-sno-U48 и hsa-sno-U50B). Так, в культуре K562 среди hsa-sno-U52; hsa-sno-U83; hsa-sno-U48 наблюдается похожая тенденция – увеличение экспрессии в первые 4 ч с последующим снижением через 24 ч, особенно hsa-sno-U83, где значение log2FC снижается с +1,25 до -1,75 за 20 ч. В культуре HL-60 подобного не отмечается, все значения log2FC для мякРНК остаются в пределах первоначальных значений (log2FC (hsa-sno-U83)=+0,3; log2FC (hsa-sno-U48)=-0,3) за исключением hsa-sno-U52, где log2FC сначала падает (+0,1), а затем возрастает через 24 ч (+0,3). Также в культуре HL-60 отмечается колебание log2FC для hsa-sno-U50B: увеличение в первые 4 ч до +0,6 с последующим возвращением к первоначальному значению +0,4.
В 11-й хромосоме (рис. 4В) в культуре K562 hsa-sno-U29; hsa-sno-U27; hsa-sno-U97; hsa-sno-U26; hsa-sno-U25; hsa-sno-U22; hsa-sno-U15B демонстрируют похожую закономерность – увеличение в первые 4 ч с последующим снижением через 24 ч от начала облучения. Log2FC мякРНК hsa-sno-U31; hsa-sno-U25; hsa-sno-U14B прослеживается эта же тенденция с увеличением экспрессии через 24 ч относительно 1 ч после облучения. В культуре HL-60 экспрессия hsa-sno-U97; hsa-sno-U30; hsa-sno-U29; hsa-sno-U22; hsa-sno-U14B похожа, но отличается от культуры K562. В первые 4 ч после облучения отмечается снижение экспрессии с последующим увеличением (hsa-sno-U97; hsa-sno-U29; hsa-sno-U22) или возвратом к первоначальным значениям (hsa-sno-U30; hsa-sno-U14B) после облучения через 24 ч. При этом log2FC hsa-sno-U15B; hsa-sno-U25;
hsa-sno-U27; hsa-sno-U28 не претерпевает значительных колебаний после облучения в течение 24 ч, а hsa-sno-U26 и hsa-sno-U31 претерпевают сначала увеличение экспрессии через 4 ч, затем уменьшение через 24 ч.
В 15-й маркерной хромосоме (рис. 4Г) у K562 всего 3 C/D мякРНК: hsa-sno-U18A; hsa-sno-U18B и hsa-sno-U18C. У hsa-sno-U18A и hsa-sno-U18B одинаково повышается экспрессия в первые 4 ч, а затем снижается через 24 ч после облучения, особенно это выражено у hsa-sno-U18B. В HL-60 – 16 C/D мякРНК: hsa-sno-U18A, hsa-sno-U18C, hsa-sno-U18B, hsa-sno-HBII-13, hsa-sno-HBII-85-8, hsa-sno-HBII-85-3, hsa-sno-HBII-85-9, hsa-sno-HBII-85-1, hsa-sno-HBII-85-6, hsa-sno-HBII-85-2, hsa-sno-HBII-85-24, hsa-sno-HBII-85-23, hsa-sno-HBII-85-16. При этом у hsa-sno-HBII-13 log2FC резко падает с +2,25 до +0,5 в первые 4 ч после облучения, а затем возрастает до +2,75 через 20 ч. Log2FC hsa-sno-U18A также уменьшается в первые 4 ч (-1,25) и возрастает в последующие 20 ч (+0,5). Log2FC hsa-sno-HBII-85-24 возрастает через 4 ч после облучения (+2,25) и возвращается к первоначальному значению через 20 ч (+1,25). Экспрессия остальных мякРНК остается в пределах log2FC через 1 ч после облучения.
Обсуждение
В наших исследованиях C/D мякРНК в большем количестве экспрессируется в клеточной линии HL-60, клетках с меньшим количеством геномных аномалий. Клеточная линия HL-60 обладает умеренной радиочувствительностью и небольшими геномными аномалиями [28]. В основном мякРНК бокс C/D образует, по крайней мере, 10 пар оснований полной комплементарности, и их модификация сильно ингибируется несоответствиями внутри этой области [29]. Значит, чем больше хромосомных аномалий, тем меньше консервативных модификаций, так как меньше экспрессирующихся C/D мякРНК, как в случае с K562. К562 является менее радиочувствительной и имеет 15 мутаций и 21-ю маркерную хромосому [30]. После облучения в дозе 4 Гр мы наблюдали, что количество C/D мякРНК в клеточной линии K562 в контроле не меняется в течение 24 ч, а после облучения количество незначительно падает (рис. 2A). В то время как в HL-60 и в контроле, и после облучения через 24 ч число C/D мякРНК уменьшается примерно на 25 %. Можно сделать вывод, что, несмотря на одинаковое воздействие, количество положительно и отрицательно экспрессирующихся C/D мякРНК после облучения разное при рассмотрении значений log2FC в каждой хромосоме в каждой точке эксперимента.
В наших работах мы уже рассматривали влияние хромосомных нарушений на экспрессию всех мякРНК, включающих два основных бокса H/ACA и C/D [25], и влияние хромосомных аномалий на экспрессию мякРНК бокса H/ACA [27] после радиационного воздействия в вышеупомянутых клеточных линиях. В клеточной линии HL-60 в целом наблюдали приблизительно одинаковый разброс положительных и отрицательных значений log2fc, можно отметить небольшое преобладание положительной экспрессии в ходе всего эксперимента, в K562 мы наблюдали переход из большего числа положительно экспрессируемых мякРНК через 4 ч в отрицательные значения log2fc через 24 ч. Похожую картину мы наблюдали и в этой работе, но в ходе эксперимента в клеточной линии HL-60 в основном была отмечена положительная экспрессия С/D мякРНК. Так же, как и здесь, мы наблюдали большее количество экспрессирующихся мякРНК в клеточной линии HL-60 и экспрессию одинаковых мякРНК в нормальных хромосамах. C/D мякРНК в большем количестве (около 70 %) экспрессируются в данных клеточных линиях, поэтому они нами были выбраны для более детального изучения в этом эксперименте. В другой своей работе [26] мы рассматривали влияние хромосомных нарушений на экспрессию микроРНК после облучения в дозе 4 Гр. В целом, как и в данном эксперименте, нами были выявлены похожие закономерности, а именно преобладание нкРНК в клеточной линии HL-60 и связь уменьшения их количества при большем наличии хромосомных аберраций в К562. Исследования не противоречат друг другу, и так же, как и в случае с C/D мякРНК, мы видим как положительные, так и отрицательные значения log2FC в течение всего эксперимента. С/D мякРНК являются предшественниками микроРНК [6], которые на сегодняшний момент больше изучены в роли регуляторов онкогенов.
В настоящее время, несмотря на стремительной рост исследований C/D мякРНК, мало данных по влиянию их экспрессии на радиочувствительность. Но, как и с микроРНК, можно предполагать, что они на разных уровнях экспрессии при раке могут вызывать разные эффекты, такие как повышенная чувствительность к лучевой терапии либо резистент- ность к ней, что затрудняет лечение [31]. Возможно, многие мишени в будущем можно будет объединить с лучевой терапией для изменения радиочувствительности клеток и прогноза заболевания, но при этом надо учитывать хромосомные аномалии в каждом конкретном случае. Эпигенетическая регуляция достаточно сложно устроена, и необходимо накопить данные для окончательных выводов использования C/D мякРНК в качестве биомаркеров.
Список литературы Экспрессия C/D МЯКРНК в клеточных линиях лейкоза при хромосомных аномалиях после облучения
- Coley A.B., DeMeis J.D., Chaudhary N.Y., Borchert G.M. Small Nucleolar Derived RNAs as Regulators of Human Cancer. Biomedicines. 2022; 10(8): 1819. https://doi.org/10.3390/biomedicines10081819.
- Maxwell E.S., Fournier M.J. The Small Nucleolar RNAs. Ann. Rev. Biochem. 1995; 64(1): 897-934. https://doi.org/10.1146/annurev.bi.64.070195.004341.
- Terns M.P., Terns R.M. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002; 10(1-2): 17-39.
- Deschamps-Francoeur G., Couture S., Abou-Elela S., Scott M.S. The snoGloBe interaction predictor reveals a broad spectrum of C/D snoRNA RNA targets. Nucleic Acids Res. 2022; 50(11): 6067-83. https://doi.org/10.1093/nar/gkac475.
- Baldini L., Charpentier B., Labialle S. Emerging Data on the Diversity of Molecular Mechanisms Involving C/D SnoRNAs. Noncoding RNA. 2021; 7(2): 30. https://doi.org/10.3390/ncrna7020030.
- Ono M., Scott M.S., Yamada K., Avolio F., Barton G.J., Lamond A.I. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011; 39(9): 3879-91. https://doi.org/10.1093/nar/gkq1355.
- Scott M.S., Avolio F., Ono M., Lamond A.I., Barton G.J. Human MiRNA Precursors with Box H/ACA SnoRNA Features. PLoS Comput Biol. 2009; 5(9). https://doi.org/10.1371/journal.pcbi.1000507.
- Dong J., Wang H., Zhang Z., Yang L., Qian X., Qian W., Han Y., Huang H., Qian P. Small but strong: Pivotal roles and potential applications of snoRNAs in hematopoietic malignancies. Front Oncol. 2022; 12. https://doi.org/10.3389/fonc.2022.939465.
- Mei Y.P., Liao J.P., Shen J., Yu L., Liu B.L., Liu L., Li R.Y., Ji L., Dorsey S.G., Jiang Z.R., Katz R.L., Wang J.Y., Jiang F. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012; 31(22): 2794-804. https://doi.org/10.1038/onc.2011.449.
- Nachmani D., Bothmer A.H., Grisendi S., Mele A., Bothmer D., Lee J.D., Monteleone E., Cheng K., Zhang Y., Bester A.C., Guzzetti A., Mitchell C.A., Mendez L.M., Pozdnyakova O., Sportoletti P., Martelli M.P., Vulliamy T.J., Safra M., Schwartz S., Luzzatto L., Bluteau O., Soulier J., Darnell R.B., Falini B., Dokal I., Ito K., Clohessy J.G., Pandolfi P.P. Germline NPM1 mutations lead to altered rRNA 2’-O-methylation and cause dyskeratosis congenita. Nat Genet. 2019; 51(10): 1518-29. https://doi.org/10.1038/s41588-019-0502-z.
- Oliveira V., Mahajan N., Bates M.L., Tripathi C., Kim K.Q., Zaher H.S., Maggi L.B. Jr, Tomasson M.H. The snoRNA target of t(4;14) in multiple myeloma regulates ribosome biogenesis. FASEB Bioadv. 2019; 1(7): 404-14. https://doi.org/10.1096/fba.2018-00075.
- Ronchetti D., Todoerti K., Tuana G., Agnelli L., Mosca L., Lionetti M., Fabris S., Colapietro P., Miozzo M., Ferrarini M., Tassone P., Neri A. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012; 2(11). https://doi.org/10.1038/bcj.2012.41.
- Zhou F., Liu Y., Rohde C., Pauli C., Gerloff D., Köhn M., Misiak D., Bäumer N., Cui C., Göllner S., Oellerich T., Serve H., Garcia-Cuellar M.P., Slany R., Maciejewski J.P., Przychodzen B., Seliger B., Klein H.U., Bartenhagen C., Berdel W.E., Dugas M., Taketo M.M., Farouq D., Schwartz S., Regev A., Hébert J., Sauvageau G., Pabst C., Hüttelmaier S., Müller-Tidow C. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol. 2017; 19(7): 844-55. https://doi.org/10.1038/ncb3563.
- May J.M., Bylicky M., Chopra S., Coleman C.N., Aryankalayil M.J. Long and short non-coding RNA and radiation response: a review. Transl Res. 2021; 233: 162-79. https://doi.org/10.1016/j.trsl.2021.02.005.
- Li Y., Ma X., Li J., He S., Zhuang J., Wang G., Ye Y., Xia W. LncRNA Gas5 Regulates Granulosa Cell Apoptosis and Viability Following Radiation by X-Ray via Sponging MiR-205-5p and Wnt/β-Catenin Signaling Pathway in Granulosa Cell Tumor of Ovary. Trop J Pharm Res. 2020; 19(6): 1153-59.
- Gao J., Liu L., Li G., Cai M., Tan C., Han X., Han L. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 2019; 126: 994-1001. https://doi.org/10.1016/j.ijbiomac.2018.12.176.
- Weidhaas J.B., Babar I., Nallur S.M., Trang P., Roush S., Boehm M., Gillespie E., Slack F.J. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007; 67(23): 11111-16. https://doi.org/10.1158/0008-5472.CAN-07-2858.
- Zhang H., Fang C., Feng Z., Xia T., Lu L., Luo M., Chen Y., Liu Y. and Li Y. The Role of LncRNAs in the Regulation of Radiotherapy Sensitivity in Cervical Cancer. Front. Oncol. 2022; 12. https://doi.org/10.3389/fonc.2022.896840.
- Ebahimzadeh K., Shoorei H., Mousavinejad S.A., Anamag F.T., Dinger M.E., Taheri M., Ghafouri-Fard S. Emerging role of non-coding RNAs in response of cancer cells to radiotherapy. Pathol Res Pract. 2021; 218. https://doi.org/10.1016/j.prp.2020.153327.
- Xiao J., He X. Involvement of Non-Coding RNAs in Chemo- and Radioresistance of Nasopharyngeal Carcinoma. Cancer Manag Res. 2021; 13: 8781-94. https://doi.org/10.2147/CMAR.S336265.
- Tian Y., Tang L., Yi P., Pan Q., Han Y., Shi Y., Rao S., Tan S., Xia L., Lin J., Oyang L., Tang Y., Liang J., Luo X., Liao Q., Wang H., Zhou Y. MiRNAs in Radiotherapy Resistance of Nasopharyngeal Carcinoma. J Cancer. 2020; 11(13): 3976-85. https://doi.org/10.7150/jca.42734.
- Masoudi-Khoram N., Abdolmaleki P. Role of non-coding RNAs in response of breast cancer to radiation therapy. Mol Biol Rep. 2022; 49(6): 5199-208. https://doi.org/10.1007/s11033-022-07234-2.
- Li Z., Wang F., Zhu Y., Guo T., Lin M. Long Noncoding RNAs Regulate the Radioresistance of Breast Cancer. Anal Cell Pathol (Amst). 2021. https://doi.org/10.1155/2021/9005073.
- Zhang S., Wang B., Xiao H., Dong J., Li Y., Zhu C., Jin Y., Li H., Cui M., Fan S. LncRNA HOTAIR enhances breast cancer radioresistance through facilitating HSPA1A expression via sequestering miR-449b-5p. Thorac Cancer. 2020; 11(7): 1801-16. https://doi.org/10.1111/1759-7714.13450.
- Rastorgueva E., Liamina D., Panchenko I., Iurova E., Beloborodov E., Pogodina E., Sugak D., Slesarev S., Saenko Y. The effect of chromosome abnormalities on expression of SnoRNA in radioresistant and radiosensitive cell lines after irradiation. Cancer Biomark. 2022; 34(4): 545-53. https://doi.org/10.3233/CBM-210092.
- Liamina D., Sibirnyj W., Khokhlova A., Saenko V., Rastorgueva E., Fomin A., Saenko Y. Radiation-Induced Changes of microRNA Expression Profiles in Radiosensitive and Radioresistant Leukemia Cell Lines with Different Levels of Chromosome Abnormalities. Cancers (Basel). 2017; 9(10): 136. https://doi.org/10.3390/cancers9100136.
- Rastorgueva E.V., Pogodina E.S., Yurova E.V., Beloborodov E.A., Sugak D.E., Saenko Yu.V., Fomin A.N. Ekspressiya H/ACA myakRNK v kletochnykh liniyakh s khromosomnymi narusheniyami posle oblucheniya. Ul'yanovskii mediko-biologicheskii zhurnal. 2022; (4): 149-59. https://doi.org/10.34014/2227-1848-2022-4-149-159.
- Liang J.C., Ning Y., Wang R.Y., Padilla-Nash H.M., Schröck E., Soenksen D., Nagarajan L., Ried T. Spectral karyotypic study of the HL-60 cell line: detection of complex rearrangements involving chromosomes 5, 7, and 16 and delineation of critical region of deletion on 5q31.1. Cancer Genet Cytogenet. 1999; 113(2): 105-9. https://doi.org/10.1016/s0165-4608(99)00030-8.
- Lafontaine D.L., Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998; 23(10): 383-8. https://doi.org/10.1016/s0968-0004(98)01260-2.
- Naumann S., Reutzel D., Speicher M., Decker H.J. Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk Res. 2001; 25(4): 313-22. https://doi.org/10.1016/s0145-2126(00)00125-9.
- Wang Y., Han Y., Jin Y., He Q., Wang Z. The Advances in Epigenetics for Cancer Radiotherapy. Int J Mol Sci. 2022; 23(10): 5654. https://doi.org/10.3390/ijms23105654.
- Brooks W.H., Renaudineau Y. Epigenetics and autoimmune diseases: the X chromosome-nucleolus nexus. Front Genet. 2015; 6: 22. https://doi.org/10.3389/fgene.2015.00022.
- Peitzsch C., Cojoc M., Hein L., Kurth I., Mäbert K., Trautmann F., Klink B., Schröck E., Wirth M.P., Krause M., Stakhovsky E.A., Telegeev G.D., Novotny V., Toma M., Muders M., Baretton G.B., Frame F.M., Maitland N.J., Baumann M., Dubrovska A. An Epigenetic Reprogramming Strategy to Resensitize Radioresistant Prostate Cancer Cells. Cancer Res. 2016; 76(9): 2637-51. https://doi.org/10.1158/0008-5472.CAN-15-2116.


