Клеточные и молекулярные механизмы контроля автофагии: потенциал для повышения стрессоустойчивости и продуктивности культурных растений
Автор: Рабаданова К.К., Тютерева Е.В., Мацкевич В.С., Демидчик В.В., Войцеховская О.В.
Журнал: Сельскохозяйственная биология @agrobiology
Рубрика: Обзоры, проблемы
Статья в выпуске: 5 т.53, 2018 года.
Бесплатный доступ
Сельскохозяйственные растения не способны достичь максимальной продуктивности, если постоянно подвергаются стрессовым воздействиям. При стрессе растения генерируют многокомпонентный метаболический, физиологический и генетический стрессовый ответ, позволяющий им адаптироваться к неблагоприятным условиям. Так, часть содержимого клетки может перевариваться, катаболически высвобождая энергию и вещества для выживания. Этот процесс известен как автофагия (J.H. Hurley с соавт., 2017). Кроме того, часть клеток может отмереть, чтобы позволить другим выжить. Механизм гибели в подобном случае запрограммирован природой и называется программированной клеточной смертью (ПКС) (W.G. van Doorn с соавт., 2011). Оба указанных процесса свойственны всем типам эукариотических клеток и представляют собой эволюционно высококонсервативные программы. Они имеют исключительно важное значение для роста и развития растений, а также для стрессового ответа и выживания в неблагоприятных условиях. Автофагию и ПКС широко изучают на животных и дрожжевых клетках, начиная с 1960-х годов, но на растениях такие исследования проводятся относительно недавно...
Автофагия, калий, программированная клеточная смерть, старение, стресс, транспорт ассимилятов, урожай
Короткий адрес: https://sciup.org/142216595
IDR: 142216595 | УДК: 631.52:581.1:577.2 | DOI: 10.15389/agrobiology.2018.5.881rus
Текст обзорной статьи Клеточные и молекулярные механизмы контроля автофагии: потенциал для повышения стрессоустойчивости и продуктивности культурных растений
Растения преодолевают неблагоприятные внешние влияния (засуху, засоление, резкое изменение температуры и т.д.), не имея возможности их физически избежать. От воздействий неблагоприятных факторов среды в мире ежегодно теряется значительная часть урожая. Остро стоит необхо-
Работа выполнена при поддержке РНФ (грант ¹ 15-14-30008).
димость создания технологий, обеспечивающих повышение устойчивости растений, в первую очередь важнейших сельскохозяйственных культур, к абиотическим и биотическим стрессам. Одной из мишеней направленной селекции стрессоустойчивых сортов сельскохозяйственных растений может стать процесс автофагии, обеспечивающий переживание растением неблагоприятных условий существования на клеточном уровне.
Автофагия — внутриклеточный процесс, в результате которого происходит удаление поврежденных субклеточных структур, обновление органелл, а также рециклирование макромолекул (1-3). В процессе автофагии клеточные компоненты подвергаются деградации в кислых литических компартментах, а высвобождаемые низкомолекулярные вещества и энергия используются на построение новых структур. Автофагия свойственна всем типам эукариотических клеток и представляет собой древнюю, эволюционно высококонсервативную катаболическую программу, однако ее механизмы в животных, дрожжевых и растительных клетках различаются (3). При этом следует отметить, что изучение автофагии у растений значительно отстает от исследований этого процесса у животных и дрожжей.
Процессы программированной клеточной смерти (ПКС) у растений также изучены в значительно меньшей степени, чем у животных. До сих пор не существует достаточно четкой морфологической классификации ПКС в растительной клетке. В отличие от животных, у растений не принято говорить об апоптозе, поскольку особенности клеточной организации растений исключают проявление ряда характерных для этого типа ПКС морфологических признаков (4), хотя встречаются упоминания апоптозоподобного пути и образования апоптотически-подобных тел в растительных клетках (5). По одной из классификаций, у растений существует два основных типа клеточной гибели — вакуолярная и некротическая (4). Известно, что ПКС у растений происходит при обновлении клеток корневого чехлика, элиминации клеток алейронового слоя эндосперма при завершении прорастания семян, обеспечении прорастания пыльцевой трубки к зародышевому мешку, формировании сосудов ксилемы и ситовидных трубок флоэмы. (6-8). В то же время обе программы, автофагия и ПКС, представляют собой важную часть ответа на стресс.
В настоящем обзоре мы обсудим, как протекает автофагия у растений, каковы ее основные функции в растительном организме в отсутствие стресса, а также оценим роль автофагии в стрессовом ответе — ее цито-протекторную функцию и участие в начальных стадиях развития вакуо-лярной программированной клеточной смерти. В связи с выявленной ролью цитоплазматического калия как одного из важнейших регуляторов ответа растений на стресс, в том числе запуска программ автофагии и ПКС, рассмотрены компоненты регуляции количества цитоплазматического калия для определения потенциальных мишеней повышения стрес-соустойчивости растений.
Роль автофагии в физиологических процессах. В растительных клетках, как и в клетках животных и грибов, посредством автофагии удаляются поврежденные (отработавшие и окисленные) либо более не нужные клетке белки. В отличие от протеасомной системы деградации, отвечающей за удаление короткоживущих белков, процесс автофа-гии позволяет клетке избавляться от долгоживущих белков (9). Кроме того, автофагия задействована в деградации целых клеточных органелл. Первоначально было обнаружено, что автофагия индуцируется в ответ на стрессовые факторы, в связи с чем считалось, что ее роль состоит преимуще-882
ственно в адаптации к неблагоприятным условиям (10, 11). Однако, как выяснилось позже, автофагия (базальная, или конститутивная) происходит и в отсутствие стрессовых воздействий и служит одним из ключевых факторов поддержания жизнедеятельности клетки (3, 7, 12, 13).
Конститутивная автофагия необходима для сохранения гомеостаза на клеточном уровне, поскольку белки в клетке неизбежно окисляются в процессе метаболических реакций, а также кислородом воздуха. Растения, мутантные по генам автофагии и неспособные осуществлять этот процесс, даже в благоприятных условиях подвержены раннему старению (7). Кроме того, базальная автофагия обеспечивает пополнение пула аминокислот и других нутриентов, необходимых клетке в качестве строительного материала для осуществления анаболических реакций.
Показано, что автофагия вовлечена в процессы развития растений. При ее участии происходит литическое расщепление крахмала и резервных белков, содержащихся в семенах, во время прорастания последних (14). Образовавшиеся низкомолекулярные соединения (сахара и аминокислоты) транспортируются в клетки формирующихся органов. При созревании семян к ним могут доставляться нутриенты, полученные в результате автофа-гической деградации белков в стареющих листьях (7, 15). Однако значительных нарушений развития у большинства мутантов по генам atg , неспособных осуществлять автофагию, в нормальных условиях не наблюдалось. Это позволяет заключить, что конститутивная автофагия не играет существенной роли в процессах роста и развития растений в отсутствие стресса. Напротив, установлена повышенная чувствительность таких мутантов к недостатку углерода и азота, а также к другим стрессовым условиям (3, 16).
Ночная (nocturnal) автофагия обнаружена сравнительно недавно. Выявлено, что мутанты Arabidopsis и табака по специфическим автофаги-ческим генам (autophagy-related genes, ATG ) не способны за ночь утилизировать крахмал, накопившийся в листьях в процессе дневного фотосинтеза (17). Обработка ингибиторами автофагии приводила к такому же эффекту. В результате тщательных цитологических исследований в клетках мезофилла листьев растений дикого типа были найдены тельца, содержащие крахмал, которые подвергались деградации в вакуолях. Эти тельца не присутствовали в клетках растений, не способных к автофагии вследствие генетических дефектов в atg -генах либо из-за воздействия ингибиторов. Авторы предположили, что ферменты, катализирующие распад крахмала, частично локализованы в лизосомах и при ночном распаде листового крахмала от хлоропластов сначала отпочковываются тельца, которые затем подвергаются деградации по механизму автофагии (17).
Однако наиболее важную роль у растений играет так называемая стресс-индуцируемая автофагия, причем очень часто активация этого типа автофагии связана с продукцией активных форм кислорода (18, 19). Активация автофагии в клетках корня высших растений происходила в ответ на засоление, гипоксию и реаэрацию, водный дефицит, обработку окислителями, генотоксическими агентами и ионизирующей радиацией (18). Установлена ведущая роль автофагии в иммунном ответе растений. Она способствует развитию реакции гиперчувствительности в ответ на атаку некротро-фов или авирулентных биотрофов, но при этом ограничивает ее спонтанное бесконтрольное распространение. Автофагия также усиливает устойчивость растений к биотрофам и некротрофам, основанную на системах салицилатного и жасмонатного сигналинга, участвует в процессах вирус-индуци-рованного замолкания генов (20).
В этом обзоре мы сосредоточимся на роли автофагии в устойчиво- сти растений к абиотическим стрессам, которые в настоящее время наносят максимальный ущерб продуктивности сельскохозяйственных культур по сравнению с другими видами стресса.
Структурные типы автофагии. Изначально считалось, что автофагия представляет собой неспецифический путь деградации клеточных компонентов. Именно неспецифическая массовая деградация одновременно различных структур клетки по механизму автофагии активируется у растений при азотном и углеродном голодании (3). Однако к настоящему времени убедительно показано, что автофагия может быть в высокой степени селективна, и описаны типы автофагии, высокоспецифичные для определенных органелл: митохондрий (митофагия) (21), хлоропластов (хлорофагия) (22), пероксисом (пексофагия) (13, 23), рибосом (24). Селективность достигается при участии белков-рецепторов, специфичных для конкретных органелл (7, 25).
В зависимости от цитологического механизма можно выделить два структурных типа автофагии — микро- и макроавтофагию. При микроав-тофагии доставка цитоплазматических компонентов в кислые литические компартменты (вакуоли у растительных клеток) происходит за счет инвагинации мембран (3, 26). Такой тип задействован, например, при прорастании семян (14, 27). Цитологическим маркером макроавтофагии выступают двумембранные органеллы, называемые автофагосомами. Образование автофагосом начинается с формирования вокруг субклеточных частиц преавтофагосомальной структуры, называемой также фагофором (preau-tophagosomal structure, phagophor assemble site, PAS). В дальнейшем происходит рост этой структуры, что приводит к образованию замкнутой двойной мембраны вокруг компонентов, подлежащих переработке, после чего осуществляется их доставка к месту деградации — в центральную вакуоль растительных и дрожжевых клеток либо в лизосомы животных клеток (7, 22, 26). У растений автофагосомы сначала сливаются с лизосомами, содержащими кислые литические ферменты; при этом их внутренний ком-партмент закисляется и образуются автолизосомы. Затем наружная мембрана автолизосомы сливается с тонопластом, и частично деградированное содержимое автолизосомы, окруженное одной мембраной (автофагическое тело), попадает в вакуоль (7). Часто под термином «автофагия» подразумевают именно этот тип макроавтофагии (рис. 1).
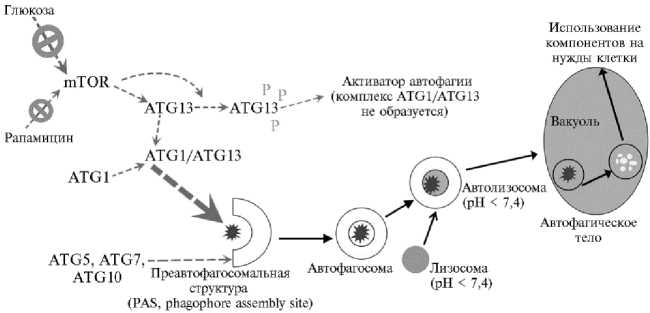
Рис. 1. Основные органеллы и белки, обеспечивающие индукцию и протекание макроавтофагии : mTOR — TOR-киназа; рапамицин, глюкоза — ингибиторы TOR-киназы; ATG1, ATG5, ATG7, ATG10, ATG13 — белки-компоненты основных комплексов автофагии. Прерывистые стрелки — сигнальные процессы, черные стрелки — последовательность событий на уровне субклеточных структур.
Оба структурных типа автофагии были описаны в том числе у растений (10, 11, 14, 28). В животной клетке, помимо них, известен третий тип — шаперон-зависимая автофагия. В ее механизме задействованы белки-шапероны семейства HSP (heat shock proteins), которые связываются с поврежденными белками и доставляют их к лизосомальной мембране (26). У растений и дрожжей сходным образом функционирует путь Cvt (cyto-plasm-to-vacuole targeting), который используется для транспортировки в вакуоль предшественников литических ферментов (3). Таким образом, механизм Cvt представляет собой один из селективных типов автофагии, но относится в большей степени к процессам биосинтеза, а не деградации (29). Кроме того, имеются сообщения, что автофагия задействована в биогенезе центральной вакуоли. В целом в последнее время появляется все больше работ, указывающих на то, что автофагические белки и структурные компоненты, помимо осуществления деградации клеточных компонентов, могут принимать участие в круговороте клеточных мембран, в том числе эндо-и экзоцитозе (30, 31).
Молекулярно-генетические основы и механизмы р азвития авто ф агии у р астений. Гены, кодирующие белки-компоненты автофагического пути ( ATG ), высококонсервативны и представлены у всех групп эукариотических организмов. Первоначально механизм автофагии был открыт на модели дрожжей Saccharomyces cerevisiae , к настоящему моменту у дрожжей описано около 40 ATG -генов (25). Большинство гомологов ATG -генов обнаружили и у растений (3). При этом некоторым одиночным генам автофагии S. cerevisiae у A. thaliana соответствуют целые семейства генов. Несколькими генами представлены, например, гомологи ATG12 , ATG13 , ATG8 , ATG4 и ATG18 (32).
Белки ATG подразделяют на четыре группы, задействованные на разных этапах автофагии: ATG1-киназный комплекс (включает ATG1, ATG13, ATG11, ATG17, ATG29, ATG31, ATG101); фосфатидилинозитол-3-(PI 3 )-киназный комплекс (VPS34, VPS15, ATG6, ATG14, ATG15, ATG38); ATG9-комплекс (ATG9, ATG2, ATG23, ATG27, ATG18); две убиквитин-подобных системы конъюгации, включающие комплекс 1 (ATG12, ATG5, ATG7, ATG10, ATG16) и комплекс 2 (ATG8, ATG4, ATG7, ATG3) (1, 3, 33). Можно выделить пять основных этапов автофагии: индукция, образование преавтофагосомальной структуры, созревание и экспансия автофагосомы, докинг и слияние с тонопастом, деградация автофагического тела (2).
Ключевая структура в индукции автофагии — киназный комплекс ATG1/ATG13 (34, 35). В его образовании участвуют вспомогательные белки ATG17 и ATG11. Их гомологи идентифицированы у растений совсем недавно (33, 36). Образование преавтофагосомальной структуры инициируется связыванием ATG17 с ATG29 и ATG31 (34, 37). Одним из первых к ATG17-ATG-29-ATG31 присоединяется ATG1. Его связывание с трехкомпонентным комплексом опосредуется белком ATG13, который имеет сайты связывания и для ATG1, и для ATG17. Эти взаимодействия способствуют увеличению киназной активности ATG1, обеспечивающей присоединение других белков инициаторного комплекса. Недавно выяснилось, что ATG13 может содействовать формированию димеров белка ATG1, благодаря которому возможна активация этой киназы по принципу положительной обратной связи (34). В результате сложных конформационных взаимодействий между ATG17 и ATG1 возникает инициаторный комплекс ATG1-ATG13-ATG17-ATG29-ATG31 — остов вновь формирующейся автофагосомы (3).
Следующий этап автофагии — рост, или экспансия, автофагосомы.
Для этого необходимо наличие фосфатидилинозитол-3-фосфата (PI3P), который встраивается в мембрану автофагосомы. Количество этого фосфолипида служит еще одним фактором, регулирующим запуск автофагии. Его содержание зависит от активности ферментов-антагонистов — фосфати-дилинозитол-3-киназ (PI3K) и PI3P-фосфатаз. PI3P образуется благодаря работе PI3-киназного комплекса 1 (PI3K 1). В его состав входят следующие белки: VPS34 (vacuolar protein sorting-associated protein 34), который относится к фосфатидилинозитол-3-киназам III класса и выполняет в комплексе роль каталитической субъединицы; VPS15, который служит активирующей субъединицей комплекса и заякоривает его в автофагосомаль-ной мембране; ATG6 (гомолог Beclin-1 млекопитающих) (37). Последний выполняет важную регуляторную функцию: в животных клетках связывание белков Bcl-2 и Bcl-1 служит одним из ключевых этапов инициации автофагии. Однако гомолог Bcl-2 у растений не найден. У дрожжей и млекопитающих известен еще один компонент PI3К-комплекса — ATG14. Сведения об обнаружении этого белка у растительных организмов в литературе и базе данных GenBank (NCBI) до сих пор отсутствуют (33, 38). Исходя из его важной функции, предполагается, что он должен присутствовать в клетках растений (38).
В дальнейшем начинается сборка комплексов конъюгации убикви-тин-подобных белков. Считается, что первым событием становится связывание ATG12 с ATG7. ATG7 обладает E1-подобной активирующей активностью и необходим для сборки обоих комплексов (39, 40). Затем к комплексу ATG7-ATG12 присоединяется ATG10, проявляющий E2-подобную конъюгирующую активность. Эти ферменты осуществляют реакции, необходимые для образования связи между белками ATG12 и ATG5 (20, 40). Для связывания конъюгата ATG12-ATG5 с фагофором требуется еще один белок — ATG16 (40, 41).
Главным белком второго комплекса служит ATG8 — важный регулятор роста и формирования автофагосом. ATG8 — небольшой (14 кД) убиквитин-подобный белок. Он синтезируется в форме предшественника и подвергается значительным посттрансляционным модификациям (42). В процессинге ATG8 задействован редокс-регулируемый фермент — цисте-ин-зависимая протеаза ATG4 (43, 44). Благодаря отрезанию аминокислотной последовательности с C-конца ATG8 становится возможным его связывание с аминогруппой фосфатидилэтаноламина (PE), что обеспечивает заякоривание белка ATG8 в автофагосомальной мембране (45). За активацию ATG8 и присоединение PE ответственны E1-подобный фермент ATG7 и E2-подобный фермент ATG3 (41, 43). В дальнейшем оба комплекса взаимодействуют, и ковалентное связывание белков второго комплекса ATG8 и ATG12 происходит при помощи белка первого комплекса ATG5, обладающего Е3-подобной ATG8-лигазной активностью. ATG12-ATG5 задействован в переносе ATG8 на фагофор (46). Липиды, необходимые для дальнейшего роста автофагосомы, поставляются из эндоплазматического ретикулума с помощью комплекса белков на основе ATG9 (47). Перенос автофагосом и автолизосом в цитозоле осуществляется при посредничестве элементов цитоскелета (48, 49). Слияние автофагосомных мембран с лизосомами и с тонопластом происходит с участием белков SNARE (50).
Регуляция автофагии на молекулярном уровне. К настоящему времени у растений обнаружено два ключевых регулятора (ингибитора) автофагии, реагирующих на концентрации питательных веществ: TOR-киназа (9) и цитозольная изоформа фермента глицеральдегид-3-фосфатдегидрогеназы (ГАФДГ) (51, 52).
TOR-киназа (mTOR, mammalian/mechanistic target of rapamycin) — высококонсервативная серин-треониновая протеинкиназа эукариот, важнейший активатор анаболизма и репрессор катаболизма в клетке (3). TOR-киназа служит регулятором стресс-индуцированной автофагии, связанной в первую очередь с недостаточным снабжением клетки углеродом и азотом. В 2005 году получила подтверждение зависимость процессов автофа-гии от активности TOR-киназы у одноклеточной водоросли Chlamydo-monas reinhardtii (53). В работе Y. Liu с соавт. (54) было доказано, что снижение активности TOR индуцирует автофагию в растительной клетке.
Блокатором TOR-киназы служит рапамицин — антибиотик бактериального происхождения, синтезируемый почвенной бактерией Streptomy-ces hygroscopicus (55). Ранее сообщалось, что, несмотря на регуляцию этого процесса со стороны TOR, растения, в отличие от дрожжей и животных клеток, нечувствительны к рапамицину (54). Затем было установлено, что рапамицин оказывает на TOR-киназу растений ингибирующее действие, однако лишь в бо ́ льших концентрациях по сравнению с животными клетками (9). По данным Y. Xiong с соавт. (9), концентрации, при которых проявлялся эффект рапамицина в растительных клетках, составляют 1001000 нМ, в то время как в животных — 10-50 нМ. Присутствие рапамици-на в указанных концентрациях снижает активность TOR, что морфологически проявляется в замедлении роста корней у Arabidopsis thaliana (9).
Ключевым посредником при индукции автофагии в ответ на стресс служит белок ATG13 (3, 35, 36, 56). В нормальных физиологических условиях TOR-киназа фосфорилирует ATG13. Такая гиперфосфорилированная форма ATG13 имеет низкое сродство к ATG1, и комплекс ATG1/ATG13, инициирующий образование автофагосомы, не формируется. Связывание ATG1/ATG13 становится возможным лишь при снижении активности TOR-киназы (3, 34, 56). Недостаток поступления в клетку питательных веществ становится сигналом, ингибирующим каскад фосфорилирования киназ PI3K/TOR и в результате приводит к снижению активности TOR-киназы (54). То есть стресс, вызванный углеродным или азотным голоданием, инициирует автофагию (см. рис. 1).
Недавно у растений обнаружили еще один ингибитор автофагии — фермент гликолитического пути глицеральдегид-3-фосфатдегидрогеназу (ГАФДГ) (51, 52). Формы Arabidopsis , дефицитные по цитозольной изоформе этого фермента, демонстрировали усиление конститутивной авто-фагии, а также высокую степень окислительного стресса и активации ПКС (52). Продукция клетками АФК в ответ на атаку патогена у таких растений, напротив, снижалась (52). На клетках табака было показано, что ГАФДГ напрямую взаимодействует с компонентом второй системы убик-витин-подобной конъюгации — белком ATG3, подавляя его функцию; ингибирование снимается под воздействием АФК (51). Следовательно, ГАФДГ, как и TOR-киназа, обеспечивает прямую взаимосвязь между метаболическим статусом клетки и индукцией автофагии, но эта связь находится под редокс-контролем.
У дрожжей и млекопитающих важным регулятором автофагии выступают киназы: АМРК (AMP-activated protein kinase) — у млекопитающих, SNF1 (sucrose non-fermenting 1) — у дрожжей (57). Они реагируют на изменение энергетического заряда, который описывается как
([АТФ] + 1/2[АДФ])/([АТФ] + [АДФ] + [АМФ]), и активируют автофагию (прямо или ингибируя TOR-киназы). У растений известно несколько гомологов SNF1/AMPK. Для одного из них (киназы KIN10 у Arabidopsis) недавно показана роль активатора автофагии в усло- виях голодания, гипоксии и водного дефицита (58).
Взаимосвязь автофагии и программированной клеточной смерти. Роль автофагии в развитии ПКС неоднозначна (15, 27, 59). С одной стороны, автофагия может служить способом избежания клеточной гибели, и с этим связана цитопротекторная функция автофагии (60, 61). С другой стороны, активация автофагии в некоторых условиях предшествует запуску программ клеточной гибели, и в этом случае автофагия выступает как один из начальных этапов ПКС (62). Так, в процессе вакуо-лярной клеточной гибели наблюдается уменьшение объема цитоплазмы и увеличение объема, занимаемого вакуолями. Этим событиям сопутствует усиление автофагии и разрыв тонопласта, сопровождающийся выходом гидролаз, что приводит к разрушению протопласта. До момента разрыва тонопласта сохраняется целостность плазматической мембраны, мембран митохондрий и других органелл (4). Весь процесс занимает, как правило, длительное время — до нескольких суток (63). В отличие от вакуолярной гибели, некротическая гибель развивается гораздо более стремительно и характеризуется сжатием протоплазмы, ранним разрушением плазматической мембраны и мембранных органелл, нарушением работы митохондрий и сопутствующим накоплением в цитоплазме активных форм кислорода (АФК) и азота (АФА) (63, 64).
Цитопротекторную роль автофагии демонстрируют исследования нокаутных мутантов, в клетках которых ее развитие невозможно. Инсер-ционные мутанты A. thaliana atg5-1 (46) и atg7-1 (39) при длинном фотопериоде характеризуются нормальным ростом и развитием. Однако при коротком фотопериоде мутанты обеих линий по сравнению с диким типом растут медленнее, имеют меньшую семенную продуктивность и подвержены преждевременному старению. Кроме того, у них повышена чувствительность к стрессу, в особенности к нехватке макроэлементов. В целом эти растения проявляют меньшую жизнеспособность и имеют значительно более низкий относительно дикого типа процент выживаемости, начиная с 10-х сут роста на среде с пониженным содержанием азота. При выдерживании в темноте выживаемость снижается у atg5-1 уже на 2-е сут, а у atg7-1 — на 4-е, в то время как у природных экотипов — соответственно на 6-е и 8-е сут (39, 46). Мутация atg13 в A. thaliana охарактеризована фенотипически в нескольких линиях (35). Они в разной степени подвержены раннему старению в условиях короткого дня. В среде с недостатком азота рост таких проростков замедлен по сравнению с диким типом, а в листьях нарушается синтез хлорофилла. Они также более чувствительны к недостатку углерода в среде. Затемнение в течение 10 ч сказывается на этих линиях не так выраженно, как на растениях с нарушенным формированием комплексов убиквитин-подобной конъюгации. Однако уже после 13 ч выдерживания в темноте обнаруживается заметная разница по устойчивости между растениями atg13 и диким типом, в особенности у двойных мутантов (35). Мутанты atg10 (41) были гиперчувствительны к недостатку углерода и азота, а также демонстрировали спонтанное развитие ПКС.
В клетках корня растений при воздействиях абиотических стрессов, ведущих к развитию ПКС, очень часто наблюдаются симптомы автофагии (18). Можно предположить, что изначально она активируется как цито-протекторный механизм, но после прохождения «точки невозврата» становится необходимым этапом развития ПКС.
Гипотеза калиевой регуляции автофагии и программированной клеточной смерти. С середины 1980-х годов в биологии растений активно развиваются представления о контроле и к 888
rectifyinкоординации физиологических реакций при стрессе посредством цитоплазматического Са2+ и активных форм кислорода (АФК) (65, 66). Известно, что генерация АФК играет значительную роль в регуляции клеточного метаболизма. АФК неизбежно образуются при окислительно-восстановительных реакциях в клетке как в нормальных условиях, так и при воздействии стрессоров (патогенные воздействия, засуха, засоление). Обнаружение повышенного синтеза АФК на ранних этапах стрессового ответа положило начало исследованиям, посвященным функциям этих молекул. Одна из таких функций — регуляция активности ионных каналов (67, 68).
К событиям, сопровождающим реакцию на стресс в растительной клетке, относится быстрый выход K+. В последние годы теория об участии калия в ответе растения на стресс получила развитие (18, 68-70). Калий — наиболее обильный металл и катион в растительной клетке. Его содержание в пересчете на сухую массу составляет 3-10 %, поэтому дефицит этого металла крайне негативно сказывается на продуктивности. Адекватная обеспеченность калием лежит в основе высокой урожайности и устойчивости растений к стрессовым воздействиям. Будучи незаменимым макроэлементом и входя в жизненно важный триплет NPK (азот-фосфор-калий), калий выполняет ключевые функции в жизни растений. В частности, он отвечает за водный баланс и гидроскелет клетки, транспирацию, закрывание устьиц и рост растяжением. Трансмембранные потоки калия формируют диффузионный мембранный потенциал на плазматической мембране, тонопласте и эндомембранах, что служит основой высокой разницы электрохимических потенциалов на этих мембранах. Также калий выступает в роли неспецифического активатора десятков важнейших анаболических ферментов цитоплазмы (71). Возможно, он стабилизирует низкую активность протеаз и нуклеаз, препятствуя незапланированному запуску автофа-гии и ПКС (18, 69).
В высвобождение калия при стрессе и во время некоторых процессов развития вовлечены потенциал-зависимые калиевые каналы, а также несколько неселективных катионных каналов (nonselective cation channels, NSCC) (68, 69). Важно подчеркнуть, что медленный выход K+ происходит и в обычных условиях (70). Более того, он необходим для осуществления важных физиологических процессов, например устьичной регуляции транспирации (72-73). Повышение выхода калия при солевом стрессе было показано достаточно давно (74). Установлено, что выход K+ опосредуется преимущественно деполяризационно активируемыми наружу-выпрямляю-щими К+-каналами (depolarization-activated outwardly-rectifying K+-channels) (75). Детальный механизм этого процесса, а также его влияние на дальнейшие события в клетке еще предстоит выяснить.
Наружу-выпрямляющие K+-каналы, обеспечивающие Гольдманов-ское выпрямление выходящего калиевого тока, активируются при деполяризации плазматической мембраны и относятся к Shaker-типу. Они обычно представляют собой гомо- или гетеротетрамеры (70). В состав каждой субъединицы входят шесть трансмембранных доменов, поровый домен и сенсор напряжения. При сборке тетрамера поровые домены, содержащие специфическую аминокислотную последовательность (GYGD), объединяются таким образом, что внутри поры оказывается четыре калий-связы-вающих сайта, то есть формируют селективный фильтр (76). В плазматической мембране клеток корня A. thaliana экспрессируются два типа нару-жу-выпрямляющих Shaker-каналов — SKOR (stelar K+-outward- rectifier) и GORK (guard cell outward-rectifying K+-channel). При этом каналы типа SKOR представлены в паренхимных клетках и опосредуют ток калия в со- суды ксилемы, в то время как GORK преобладают в эпидермальных клетках и задействованы в выходе калия из корня (70). Оба эти типа непосредственно активируются АФК (77). Открытие канала индуцируется при помощи АФК-чувствительного сайта в структуре молекулы. В случае SKOR роль АФК-сенсора выполняет остаток цистеина (Cys168) в пептидной последовательности домена S3 в составе потенциал-чувствительного комплекса S1-S4 (77). Поскольку структурно GORK и SKOR в значительной степени схожи, предполагается, что у GORK АФК-зависимая активация обеспечивается таким же образом (18).
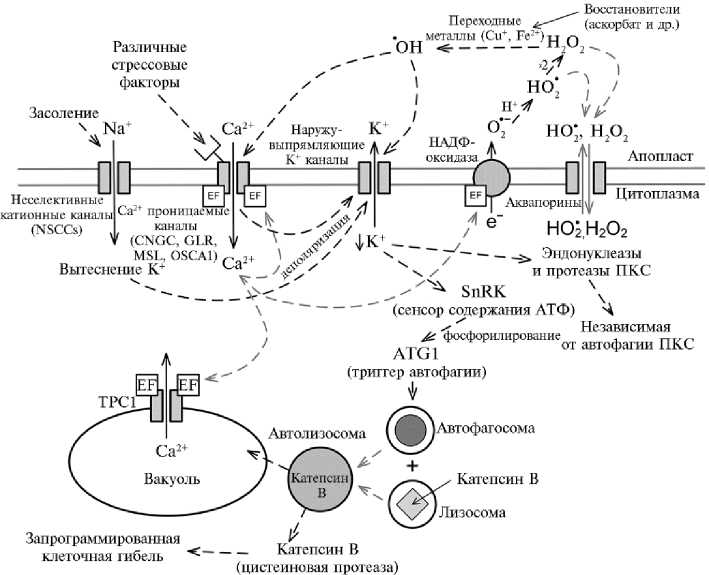
Рис. 2. Схема процессов, лежащих в основе стресс-индуцированной автофагии и программированной клеточной смерти (ПКС). Стрессовые сигналы взаимодействуют со специфическими рецепторами на поверхности клетки, что вызывает деполяризацию плазматической мембраны, повышение цитоплазматической активности кальция, возрастание продукции активных форм кислорода (АФК) вследствие кальций-зависимой активации НАДФН-оксидазы. Деполяризация также приводит к активации наружу-выпрямляющих калиевых каналов, которая дополнительно стимулируется АФК. Резкое падение концентрации цитоплазматического калия приводит к запуску реакций автофагии и ПКС. АФК также продуцируются внутриклеточно и транспортируются в апопласт через аквапорины. Редокс-процессы в апопласте контролируются содержанием восстановленных переходных металлов и аскорбата (по 18, c изменениями). EF — EF-рука (белковый домен); ТРС1 — двухпоровый кальциевый канал. Серыми стрелками обозначены вторичные процессы.
Активность канала GORK стимулируют гидроксильные радикалы ( • OH), которые генерируются Ca2+-зависимыми НАДФН-оксидазами при реакции практически на все виды стресса, включая засоление, засуху, атаку патогенов и др. Снижение концентрации калия в цитоплазме, в свою очередь, стимулирует активность протеолитических ферментов, в том числе каспазоподобных протеаз, играющих важную роль в механизме запуска ПКС у растений (45, 69, 70).
В общем виде механизм развития стрессовой реакции с участием выходящего тока K+ в клетках эпидермиса корня можно представить следующим образом. При связывании стрессового сигнала или патогенного 890
элиситора с рецептором плазматической мембраны, а также в результате поступления Na+ по неселективным каналам происходит активация Ca2+-проницаемых катионных каналов, что приводит к повышению концентрации Ca2+ в цитоплазме. Кальций активирует НАДФH-оксидазу, связываясь с ее цитоплазматическим доменом. Может также наблюдаться Ca2+-зависимая активация эндонуклеаз и протеаз (64). НАДФH-оксидаза генерирует супероксид-анионы (O2 • - ), которые вступают в реакцию с протонами, образуя гидропероксидный радикал (HO2 • ). Дисмутация HO2 • дает пероксид водорода (H2O2), который становится источником кислорода для синтеза гидроксильных радикалов ( • OH) в реакциях Габера-Вайса (78). Пероксид и • OH дополнительно активируют SKOR, GORK и Са2+-прони-цаемые катионные каналы, усиливающие поступление Са2+ в цитоплазму и, в свою очередь, также стимулирующие НАДФН-оксидазу (рис. 2). Действует механизм обратной положительной связи, который может быть остановлен системами выкачивания Са2+ из цитоплазмы (79). Мембранный потенциал восстанавливается при помощи выхода калия и вследствие увеличения активности электрогенной Н+-АТФазной помпы. Если не происходит реполяризации мембраны и восстановления количества клеточного калия, то активируются регулируемые калием протеазы и эндонуклеазы растений и результатом становится инициация молекулярного механизма ПКС (70).
В конце 1990-х годов стало известно, что в клетках млекопитающих снижение концентрации K+ (наряду с притоком Ca2+) играет важную роль в механизме запуска апоптоза (80, 81). Калий — непосредственный ингибитор каспаз, его выход из клетки активирует эти ферменты (82, 83). Скорее всего, похожий механизм существует и в клетках растений (18, 69). Согласно недавно выдвинутой гипотезе, у растений калий (в зависимости от его содержания в цитоплазме) служит переключателем метаболизма, а стресс-индуцированный выход К+ может быть триггером остановки роста клетки, ингибирования биосинтезов, активации катаболизма и в длительной перспективе при сильном стрессе — запуска ПКС (18). Важный шаг в развитии приведенной гипотезы — тестирование потенциальной стимуляции автофагии при потере клетками растений К+. Является ли автофагия К+-зависимым процессом у растений, до сих пор не известно. Показано, что у мутантов A. thaliana gork 1-1 (в отличие от природного экотипа Ws-0) при выдерживании корней в растворе NaCl не происходит резкого накопления автофагосом. Полученные данные свидетельствуют в пользу того, что потеря калия играет непосредственную роль в индукции автофагии (18).
Перспективы исследования автофагии. Согласно современным представлениям, автофагия играет значительную роль в метаболизме клетки, обеспечивая обновление клеточных структур, расщепление поврежденных молекул и получение из них органических соединений, необходимых для извлечения и запасания энергии. То есть это явление направлено на выживание клетки. Особенно важна автофагия при адаптации к различным стрессовым воздействиям. Она во многом определяет выживание растений в неблагоприятных и меняющихся условиях. Несмотря на то, что автофагия изучена достаточно подробно, остается много неразрешенных вопросов. В частности, не до конца понятно, всегда ли резкий выход из клеток ионов калия непосредственно запускает молекулярный механизм автофагии (подобно тому, как он индуцирует программированную смерть клеток). То же касается взаимосвязи автофагии с другими внутриклеточными процессами. Показано, что отсутствие автофагии отрицательно сказывается на перенесении растениями стресса, отражается на накоплении биомассы и семенной продуктивности. Однако влияние ав-тофагии не столь однозначно, поскольку она также задействована в процессах клеточной смерти. Тем не менее, эти процессы представляют неотъемлемую часть развития организма, необходимы для формирования многих структур и прохождения всех этапов жизненного цикла.
В случае подтверждения гипотезы о том, что регуляторным клеточным сигналом для запуска автофагии служит резкое снижение концентрации цитоплазматического калия, его функции представились бы еще более значимыми, особенно в условиях, когда растение подвержено действию неблагоприятных факторов среды. Направленная манипуляция степенью активности клеточных компонентов для поддержания необходимой концентрации калия в цитоплазме при стрессе может использоваться при создании стрессоустойчивых растений, для снижения их гибели и поддержки ростовых процессов в неблагоприятных условиях внешней среды.
Таким образом, автофагия как процесс, имеющий прямое отношение к механизмам стрессоустойчивости, старения и к транспорту ассими-лятов, представляет собой важную потенциальную мишень регуляции, которая до сих пор не была использована при создании новых сортов и в практических приложениях в сельском хозяйстве. Автофагия играет двоякую роль: с одной стороны, она направлена на выживание клетки, с другой — служит частью процесса клеточной гибели. В обоих случаях автофа-гия оказывает непосредственное влияние на развитие растительных организмов. Ввиду этого представляется перспективным дальнейшее изучение регуляции автофагии, в особенности более детальное раскрытие механизмов индукции этого процесса.
Список литературы Клеточные и молекулярные механизмы контроля автофагии: потенциал для повышения стрессоустойчивости и продуктивности культурных растений
- Hurley J.H., Young L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem., 2017, 86: 225-244 ( ) DOI: 10.1146/annurev-biochem-061516-044820
- Klionsky D.J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci., 2005, 118: 7-18 ( ) DOI: 10.1242/jcs.01620
- Reumann S., Voitsekhovskaja O., Lillo C. From signal transduction to autophagy of plant cell organelles: lessons from yeast and mammals and plant-specific features. Protoplasma, 2010, 247(3-4): 233-256 ( ) DOI: 10.1007/s00709-010-0190-0
- van Doorn W.G., Beers E.P., Dang J.L., Franklin-Tong V.E., Gallois P., Hara-Nishimura I., Jones A.M., Kawai-Yamada M., Lam E., Mundy J., Mur L.A.J., Petersen M., Smertenko A., Taliansky M., Van Breusegem F., Wolpert T., Woltering E., Zhivotovsky B., Bozhkov P.V. Morphological classification of plant cell deaths. Cell Death Differ., 2011, 18(8): 1241-1246 ( ) DOI: 10.1038/cdd.2011.36
- Lam E. Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol., 2004, 5(4): 305-315 ( ) DOI: 10.1038/nrm1358
- Самуилов В.Д., Олескин А.В., Лагунова Е.М. Программируемая клеточная смерть. Биохимия, 2000, 8: 1029-1046.
- Liu Y., Bassham D.C. Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol., 2012, 63: 215-237 ( ) DOI: 10.1146/annurev-arplant-042811-105441
- van Doorn W.G., Woltering E.J. Many ways to exit? Cell death categories in plants. Trends Plant Sci., 2005, 10(3): 117-122 ( ) DOI: 10.1016/j.tplants.2005.01.006
- Xiong Y., Sheen J. Rapamycine and glucose-target of rapamycine (TOR) protein signaling in plants. J. Biol. Chem., 2012, 287: 2836-2842 ( ) DOI: 10.1074/jbc.M111.300749
- Aubert S., Gout E., Bligny R., Marty-Mazars D., Barrieu F., Alabouvette J., Marty F., Douce R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J. Cell Biol., 1996, 133(6): 1251-1263 ( ) DOI: 10.1083/jcb.133.6.1251
- Moriyasu Y., Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation Plant Physiol., 1996, 111(4): 1233-1241 ( ) DOI: 10.1104/pp.111.4.1233
- Thompson A.R., Vierstra R.D. Autophagic recycling: lessons from yeast help define the process in plants. Curr. Opin. Plant Biol., 2005, 8: 165-173 ( ) DOI: 10.1016/j.pbi.2005.01.013
- Voitsekhovskaja O.V., Schiermeyer A., Reumann S. Plant peroxisomes are degraded by starvation-induced and constitutive autophagy in tobacco BY-2 suspension-cultured cells. Front. Plant Sci., 2014, 18(5): article 629 ( ) DOI: 10.3389/fpls.2014.00629
- Toyooka K., Okamoto T., Minamikawa T. Cotyledon cells of Vigna mungo seedlings use at least two distinct autophagic machineries for degradation of starch granules and cellular components. J. Cell Biol., 2001, 154: 973-982 ( ) DOI: 10.1083/jcb.200105096
- Guiboileau A., Sormani R., Meyer C., Masclaux-Daubresse C. Senescence and death of plant organs: nutrient recycling and developmental regulation. C. R. Biol., 2010, 333(4): 382-391 ( ) DOI: 10.1016/j.crvi.2010.01.016
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni Ch., Panstruga R., Ohsumi Y., Shirasua K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. The Plant Cell, 2009, 21: 2914-2927 ( ) DOI: 10.1105/tpc.109.068635
- Wang Y., Yu B., Zhao J., Guo J., Li Y., Han S., Huang L., Du Y., Hong Y., Tang D., Liu Y. Autophagy contributes to leaf starch degradation. The Plant Cell, 2013, 25: 1383-1399 ( ) DOI: 10.1105/tpc.112.108993
- Demidchik V., Tyutereva E.V., Voitsekhovskaja O.V. The role of ion disequilibrium in induction of root cell death and autophagy by environmental stresses. Funct. Plant Biol., 2017, 45(1): 28-46 ( ) DOI: 10.1071/FP16380
- Pérez-Pérez M.E., Couso I., Domínguez-González M., Lemaire S.D., Crespo J.L. Redox control of autophagy in photosynthetic organisms. In: Progress in botany. Vol. 79/F. Cánovas, U. Lüttge, R. Matyssek (eds.). Springer, Cham, 2017 ( ) DOI: 10.1007/124_2017_6
- Zhou J., Yu J.Q., Chen Z. The perplexing role of autophagy in plant innate immune responses Mol. Plant Pathol., 2014, 15(6): 637-645 ( ) DOI: 10.1111/mpp.12118
- Minibayeva F., Ponomareva A., Dmitrieva S., Ryabovol V. Oxidative stress-induced autophagy in plants: the role of mitochondria. Plant Physiol. Bioch., 2012, 59: 11-19 ( ) DOI: 10.1016/j.plaphy.2012.02.013
- Ishida H., Wada S. Autophagy of whole and partial chloroplasts in individually darkened leaves: a unique system in plants? Autophagy, 2009, 5: 736-737 ( ) DOI: 10.4161/auto.5.5.8568
- Shibata M., Oikawa K., Yoshimoto K., Kondo M., Mano S., Yamada K., Hayashi M., Sakamoto W., Ohsumi Y., Nishimura M. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. The Plant Cell, 2013, 25: 4967-4983 ( ) DOI: 10.1105/tpc.113.116947
- Niki T., Saito S., Gladish D.K. Granular bodies in root primary meristem cells of Zea mays L. var. Cuscoensis K. (Poaceae) that enter young vacuoles by invagination: a novel ribophagy mechanism. Protoplasma, 2014, 251(5): 1141-1149 ( ) DOI: 10.1007/s00709-014-0622-3
- Reggiori F., Klionsky D.J. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics, 2013, 194(2): 341-361 ( ) DOI: 10.1534/genetics.112.149013
- Ковалева О.В., Шитова М.С., Зборовская И.Б. Аутофагия: клеточная гибель или способ выживания? Клиническая онкогематология, 2015, 2(2): 103-113.
- Bassham D.C. Plant autophagy -more than a starvation response. Curr. Opin. Plant Biol., 2007, 10(6): 587-593 ( ) DOI: 10.1016/j.pbi.2007.06.006
- van der Wilden W., Herman E.M., Chrispeels M.J. Protein bodies of mung bean cotyledons as autophagic organelles. PNAS USA, 1980, 77(1): 428-432.
- Yamasaki A., Noda N.N. Structural biology of the Cvt pathway. J. Mol. Biol., 2017, 429(4): 531-542 ( ) DOI: 10.1016/j.jmb.2017.01.003
- Kim S.H., Kwon C., Lee J.H., Chung T. Genes for plant autophagy: functions and interactions. Mol. Cells, 2012, 34(5): 413-423 ( ) DOI: 10.1007/s10059-012-0098-y
- Yan Q., Wang J., Fu Z.Q., Chen W. Endocytosis of AtRGS1 is regulated by the autophagy pathway after D-glucose stimulation. Front. Plant Sci., 2017, 8: 1229 ( ) DOI: 10.3389/fpls.2017.01229
- Ryabovol V.V., Minibayeva F.V. Molecular mechanisms of autophagy in plants: role of ATG8 proteins in formation and functioning of autophagosomes. Biochemistry (Moscow), 2016, 81(4): 348-363 ( ) DOI: 10.1134/S0006297916040052
- Michaeli S., Galili G., Genschik P., Fernie A.R., Avin-Wittenberg T. Autophagy in plants -what's new on the menu? Trends Plant. Sci., 2016, 21(2): 134-144 ( ) DOI: 10.1016/j.tplants.2015.10.008
- Alers S., Wesselborg S., Stork B. ATG13: Just a companion, or an executor of the autophagic program? Autophagy, 2014, 10(6): 944-956 ( ) DOI: 10.4161/auto.28987
- Suttangkakul A., Li F., Chung T., Vierstra R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. The Plant Cell, 2011, 23: 3761-3779 ( ) DOI: 10.1105/tpc.111.090993
- Li F., Vierstra R.D. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general Autophagy and selective mitophagy. Autophagy, 2014, 10(8): 1466-1467 ( ) DOI: 10.4161/auto.29320
- Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell, 2008, 19(5): 2039-2050 ( ) DOI: 10.1091/mbc.E07-10-1048
- Avin-Wittenberg T., Honig A., Galili G. Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma, 2012, 249(2): 285-299 ( ) DOI: 10.1007/s00709-011-0296-z
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R, Vierstra R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem., 2002, 277(36): 33105-33114 ( ) DOI: 10.1074/jbc.M204630200
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol., 2001, 2: 211-216 ( ) DOI: 10.1038/35056522
- Phillips A.R., Suttangkakul A., Vierstra R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics, 2008, 178(3): 1339-1353 ( ) DOI: 10.1534/genetics.107.086199
- Kellner R., de la Concepcion J.C., Maqbool A., Kamoun S., Dagdas Y.F. ATG8 expansion: a driver of selective autophagy diversification? Trends Plant. Sci., 2017, 22(3): 204-214 ( ) DOI: 10.1016/j.tplants.2016.11.015
- Li F., Vierstra R.D. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci., 2012, 17: 526-537 ( ) DOI: 10.1016/j.tplants.2012.05.006
- Pérez-Pérez M.E., Zaffagnini M., Marchand C.H., Crespo J.L., Lemaire S.D. The yeast Autophagy protease Atg4 is regulated by thioredoxin. Autophagy, 2014, 10(11): 1953-1864 ( ) DOI: 10.4161/auto.34396
- Замятнин А.А. Протеолитические ферменты растений, вовлеченные в процессы регулируемой смерти клеток. Успехи биологической химии, 2015, 55: 145-180.
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol., 2005, 138(4): 2097-2110 ( ) DOI: 10.1104/pp.105.060673
- Le Bars R., Marion J., Satiat-Jeunemaitre B., Bianchi M.W. Folding into an autophagosome: ATG5 sheds light on how plants do it. Autophagy, 2014, 10(10): 1861-1863 ( ) DOI: 10.4161/auto.29962
- Monastyrska I., Rieter E., Klionsky D.J., Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol. Rev. Camb. Philos., 2009, 84(3): 431-448 ( ) DOI: 10.1111/j.1469-185X.2009.00082.x
- Wang Y., Zheng X., Liu Y. Functional links between microtubules, autophagy and leaf starch degradation in plants. Plant Signaling and Behavior, 2016, 11(7): e1201626 ( ) DOI: 10.1080/15592324.2016.1201626
- Moreau K., Renna M., Rubinsztein D.C. Connections between SNAREs and autophagy. Trends Biochem. Sci., 2013, 38(3): 57-63 ( ) DOI: 10.1016/j.tibs.2012.11.004
- Han S., Wang Y., Zheng X., Jia Q., Zhao J., Bai F., Hong Y., Liu Y. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell, 2015, 27: 1316-1331 ( ) DOI: 10.1105/tpc.114.134692
- Henry E., Fung N., Liu J., Drakakaki G., Coaker G. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLOS Genetics, 2015, 11: e1005199 ( ) DOI: 10.1371/journal.pgen.1005199
- Crespo J.L., S. Diaz-Troya S., Florencio F.J. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant. Physiol., 2005, 139: 1736-1749 ( ) DOI: 10.1104/pp.105.070847
- Liu Y., Bassham D.C. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE, 2010, 5(7): e11883 ( ) DOI: 10.1371/journal.pone.0011883
- Yip C.K., Murata K., Walz T., Sabatini D.M., Kang S.A. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell., 2010, 38(5): 768-774 ( ) DOI: 10.1016/j.molcel.2010.05.017
- Chang Y.-Y., Neufeld T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell, 2009, 20(7): 2004-2014 ( ) DOI: 10.1091/mbc.E08-12-1250
- Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic control of autophagy. Cell, 2014, 159(6): 1263-1276 ( ) DOI: 10.1016/j.cell.2014.11.006
- Chen L., Su Z.-Z., Huang L., Xia F.-N., Qi H., Xie L.-J., Xiao S., Chen Q.-F. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci., 2017, 8: article 1201 ( ) DOI: 10.3389/fpls.2017.01201
- Patel S., Caplan J., Dinesh-Kumar S.P. Autophagy in the control of programmed cell death. Curr. Opin. Plant Biol., 2006, 9(4): 391-396 ( ) DOI: 10.1016/j.pbi.2006.05.007
- Liu Y., Schiff M., Czymmek K., Tallócz, Z., Levine B., Dinesh-Kumar S.P. Autophagy regulates programmed Cell death during the plant innate immune response. Cell, 2005, 121(4): 567-577 ( ) DOI: 10.1016/j.cell.2005.03.007
- Shibuya K., Yamada T., Ichimura K. Autophagy regulates progression of programmed cell death during petal senescence in Japanese morning glory. Autophagy, 2009, 5(4): 546-547 ( ) DOI: 10.4161/auto.5.4.8310
- Kabbage M., Kessens R., Bartholomay L.C., William B. The life and death of a plant cell. Annu. Rev. Plant Biol., 2017, 68: 375-404 ( ) DOI: 10.1146/annurev-arplant-043015-111655
- Фомичева А.С., Тужиков А.И., Белошистов Р.Е., Трусова С.В., Галиуллина Р.А., Мочалова Л.В., Чичкова Н.В., Вартапетян А.Б. Программированная клеточная смерть у растений. Успехи биологической химии, 2012, 52: 97-126.
- Collazo C., Chacуn O., Borras O. Programmed cell death in plants resembles apoptosis of animals. Biotecnologia Aplicada, 2006, 23: 1-10.
- Trewavas A., Knight M. Mechanical signalling, calcium and plant form. Plant Mol. Biol., 1994, 26(5): 1329-1341 ( ) DOI: 10.1007/BF00016478
- Demidchik V., Maathuis F.J.M. Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol., 2007, 175(3): 387-405 ( ) DOI: 10.1111/j.1469-8137.2007.02128.x
- Demidchik V. Reactive oxygen species and oxidative stress in plants. In: Plant stress physiology. 2nd edition/S. Shabala (ed.). Wallingford, CABI, 2012: 24-58 ( ) DOI: 10.1079/9781780647296.0064
- Demidchik V., Shabala S.N., Coutts K.B., Tester M.A., Davies J. Free oxygen radicals regulate plasma membrane Ca2+-and K+-permeable channels in plant root cells. J. Cell Sci., 2003, 116: 81-88 ( ) DOI: 10.1242/jcs.00201
- Demidchik V., Cuin T.A., Svistunenko D., Smith S.J., Miller A.J., Shabala S., Sokolik A., Yurin V. Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci., 2010, 123: 1468-1479 ( ) DOI: 10.1242/jcs.064352
- Demidchik V. Mechanisms and physiological roles of K+ efflux from root cells. J. Plant Physiol., 2014, 171(9): 696-707 ( ) DOI: 10.1016/j.jplph.2014.01.015
- Maathuis F.J.M, Amtmann A. K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Annals of Botany, 1999, 84(2): 123-133 ( ) DOI: 10.1006/anbo.1999.0912
- Hos E., Vavasseur A., Mouline K., Dreyer I., Gaymard F., Porée F., Boucherez J., Lebaudy A., Bouchez D., Very A.A., Simonneau T., Thibaud J.B., Sentenac H. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. PNAS, 2003, 100(9): 5549-5554 ( ) DOI: 10.1073/pnas.0733970100
- Li J., Zhang H., Lei H., Jin M., Yue G., Su Y. Functional identification of a GORK potassium channel from the ancient desert shrub Ammopiptanthus mongolicus (Maxim.) Cheng f. Plant. Cell. Rep., 2016, 35(4): 803-815 ( ) DOI: 10.1007/s00299-015-1922-6
- Nassery H. The effects of salt and osmotic stress on the retention of potassium by excised barley and bean roots. New Phytol., 1975, 75(1): 63-67 ( ) DOI: 10.1111/j.1469-8137.1975.tb01371.x
- Shabala S., Demidchik V., Shabala L., Cuin T.A., Smith S.J., Miller A.J., Davies J.M., Newman I.A. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol., 2006, 141: 1653-1665 ( ) DOI: 10.1104/pp.106.082388
- MacKinnon R. Potassium channels and the atomic basis of selective ion conduction (Nobel lecture). Angew. Chem. Int. Edit., 2004, 43(33): 4264-4277 ( ) DOI: 10.1002/anie.200400662
- Garcia-Mata C., Wang J., Gajdanowicz P., Gonzalez W., Hills A., Donald N., Riedelsberger J., Amtmann A., Dreyer I., Blatt M.R. A minimal cysteine motif required to activate the SKOR K+ channel of Arabidopsis by the reactive oxygen species H2O2. J. Biol. Chem., 2010, 285(38): 29286-29294 ( ) DOI: 10.1074/jbc.M110.141176
- Halliwell B., Gutteridge J.M.C. Free radicals in biology and medicine. Oxford University Press, USA, 2015 (doi: 10.1093/acprof:oso/9780198717478.001.0001).
- Demidchik V., Shabala S. Mechanisms of cytosolic calcium elevation in plants: the role of ion channels, calcium extrusion systems and NADPH oxidase-mediated ‘ROS-Ca2+ Hub’. Funct. Plant Biol., 2017, 45(1): 9-27 ( ) DOI: 10.1071/FP16420
- Bortner C.D., Hughes F.M. Jr., Cidlowski J.A. A primary role for K+ and Na+ efflux in the activation of apoptosis. J. Biol. Chem., 1997, 272(51): 32436-32442 ( ) DOI: 10.1074/jbc.272.51.32436
- Yu S.P., Yeh C.H., Sensi S.L., Gwag B.J., Canzoniero L.M., Farhangrazi Z.S., Ying H.S., Tian M., Dugan L.L., Choi D.W. Mediation of neuronal apoptosis by enhancement of outward potassium current. Science, 1997, 278(5335): 114-117 ( ) DOI: 10.1126/science.278.5335.114
- Park I.-S., Ja-Eun K. Potassium efflux during apoptosis. J. Biochem. Mol. Biol., 2002, 35(1): 41-46 ( ) DOI: 10.5483/BMBRep.2002.35.1.041
- Remillard C.V., Yuan J.X. Activation of K+ channels: an essential pathway in programmed cell death. Am. J. Physiol. -Lung C., 2004, 286(1): 49-67 ( ) DOI: 10.1152/ajplung.00041.2003


