Monitoring of EGFR mutations in the circulating tumor DNA from blood plasma of patients with non-small cell lung cancer
Автор: Shamanin Vladimir A., Karpov Igor V., Gervas Polina A., Cherdyntseva Nadezhda V., Simolina Elena I., Kozlov Vadim V., Kovalenko S.P.
Журнал: Сибирский онкологический журнал @siboncoj
Рубрика: Лабораторные и экспериментальные исследования
Статья в выпуске: 5 т.17, 2018 года.
Бесплатный доступ
Activating mutations of egfr are associated with sensitivity of non-small cell lung cancer (NSCLC) to tyrosine kinase inhibitors (TKI). Liquid biopsy using circulating cell-free tumor DNA (cfDNA) is proposed in cases when formalin fixed paraffin embedded (FFPE) tumor tissue is not available and for monitoring of egfr status. In the study we evaluated new qPCR assay for egfr mutations in plasma cfDNA. Sensitivity of the assay was 1 % of the mutant allele for L858R, L861Q, S768I mutations and deletions in exon 19, and 5 % of the mutant allele for G719X or T790M mutations Before surgery, mutation was detected in plasma of 4 out of 7 patients (57 %) with mutant egfr in FFPE tumor tissue. Mutations found in cfDNA completely matched those found in tumor tissue in 2 cases. In one case with G719X and S768I mutations in FFPE tissue, only S768I was found in cfDNA. In another case, T790M was detected in plasma in addition to L858R that was present in tumor tissue. No egfr mutations were detected in plasma DNA from 12 healthy volunteers and 13 cases of NSCLC with wt egfr suggesting 100 % specificity of the assay. Liquid biopsy detected egfr mutations in cfDNA in 8 of 16 cases of NSCLC with mutant egfr being under therapy with TKI. Among them, 7 cases had mutations in liquid biopsy that matched those in tumor tissue and another case had T790M in addition to L858R. In 3 cases increased mutant allele frequency was detected 212 months before clinical progression.
Qpcr, liquid biopsy, egfr, mutation, lung cancer
Короткий адрес: https://sciup.org/140254021
IDR: 140254021 | УДК: 616.24-006.6-076:575.113:577.213 | DOI: 10.21294/1814-4861-2018-17-5-52-59
Текст научной статьи Monitoring of EGFR mutations in the circulating tumor DNA from blood plasma of patients with non-small cell lung cancer
Activating mutations of EGFR are associated with sensitivity of non-small cell lung cancer (NSCLC) to tyrosine kinase inhibitors (TKI). Liquid biopsy using circulating cell-free tumor DNA (cfDNA) is proposed in cases when formalin fixed paraffin embedded (FFPE) tumor tissue is not available and for monitoring of EGFR status. In the study we evaluated new qPCR assay for EGFR mutations in plasma cfDNA. Sensitivity of the assay was 1 % of the mutant allele for L858R, L861Q, S768I mutations and deletions in exon 19, and 5 % of the mutant allele for G719X or T790M mutations Before surgery, mutation was detected in plasma of 4 out of 7 patients (57 %) with mutant EGFR in FFPE tumor tissue. Mutations found in cfDNA completely matched those found in tumor tissue in 2 cases. In one case with G719X and S768I mutations in FFPE tissue, only S768I was found in cfDNA. In another case, T790M was detected in plasma in addition to L858R that was present in tumor tissue. No EGFR mutations were detected in plasma DNA from 12 healthy volunteers and 13 cases of NSCLC with wt EGFR suggesting 100 % specificity of the assay. Liquid biopsy detected EGFR mutations in cfDNA in 8 of 16 cases of NSCLC with mutant EGFR being under therapy with TKI. Among them, 7 cases had mutations in liquid biopsy that matched those in tumor tissue and another case had T790M in addition to L858R. In 3 cases increased mutant allele frequency was detected 212 months before clinical progression.
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer. Activating somatic mutations within the tyrosine kinase (TK) domain in the epidermal growth factor receptor (EGFR) protein are associated with the sensitivity of tumors to EGFR-TK inhibitors [1]. Most of these mutations (~90 %) occur as either in-frame microdeletions that affect codons 746–750 in exon 19 (19del), or a missense mutation (L858R) in exon 21 of EGFR . In addition, rare activating mutations (L861Q, G719S/A/C, S768I and others) make up 10–15 % of cases with mutant EGFR . Besides, T790M mutation occurs in ~50 % of cases of acquired resistance to the first generation EGFR TK inhibitors (TKI). Tumors with T790M mutation are sensitive to therapy with osimertinib, which is the third generation TKI [2]. The European Society for Medical Oncology (ESMO) and American Society of Clinical Oncology (ASCO) recommend an EGFR mutation testing for selection of NSCLC patients who could benefit from anti- EGFR therapy [3, 4].
DNA from formalin fixed paraffin embedded (FFPE) tumor tissue is usually used to test mutations. In cases when tumor tissue is not available, the liquid biopsy using blood plasma that contains circulating cell-free tumor DNA (cfDNA) from tumor cells can be tested for clinically actionable mutations [5]. Major challenges of liquid biopsy are low concentration of cfDNA in plasma, and low frequency of mutant allele in cfDNA due to high background of DNA from normal cells. The assays for liquid biopsy are based on different methods including quantitative PCR (qPCR), digital PCR (dPCR), next generation sequencing (NGS) [6].
NGS can test a broad range of mutations with high sensitivity but the assay is expensive, technically demanding and has turnaround time of 12 weeks. Tests based on qPCR and dPCR detect only hot spot mutations but provide fast turnaround time of several days. dPCR has a high sensitivity and can detect mutations with mutant allele frequency (MAF) below 0,1 % . However, the high analytical sensitivity of dPCR often cannot be fully explored due to not sufficient concentration of cfDNA in plasma. The sensitivity of qPCR can be 1 % MAF. Importantly, equipment and reagents for qPCR are commonly available.
The aim of our study was to evaluate liquid biopsy for EGFR mutations using new test based on qPCR. We used limited multiplex PCR (mPCR) of regions in 1821 exons of EGFR with hot spots for mutations to increase amount of cfDNA targets so that multiple tests for different mutations can be made in clinical setting without necessity for additional blood draw. After mPCR, amplicons of cfDNA were tested for EGFR mutations by qPCR. The developed assay was used for the monitoring of mutations in EGFR gene in cfDNA of patients with NSCLC treated with gefitinib or erlotinib.
Material and methods
Ethics statement
The study was approved by the Ethics Committee of the Cancer Research Institute (Tomsk, Russia). Informed consents were received from all patients.
Clinical samples
Blood samples from 12 healthy volunteers (6 males and 6 females) were collected in the medical center “Status” (Novosibirsk). Paired samples of blood before surgery and formalin-fixed paraffin-embedded (FFPE) tumor tissue after surgical resection from 20 patients with non-small cell lung cancer (NSCLC) were collected at the Novosibirsk Regional Clinical Oncology Center (NRCOC; Novosibirsk, Russia). Blood from 16 patients with EGFR-mutant NSCLC treated with tyrosine kinase inhibitors (TKI) was collected at the NRCOC and Cancer Research Institute (Tomsk, Russia). Demographics of patients and histotypes of tumors are provided in Table 1.
|
demographics of patients with nsclc |
table 1 |
|||
|
Case |
Patient ID |
Sex |
Age |
Tumor histology |
|
Cases with mutant EGFR in FFPE tumor tissue DNA, plasma before surgery |
||||
|
1 |
BK7 |
F |
53 |
NSCLC-NOS |
|
2 |
BK16 |
F |
68 |
NSCLC-NOS |
|
3 |
BK35 |
F |
54 |
AC |
|
4 |
BK40 |
F |
74 |
АC |
|
5 |
BK54 |
М |
60 |
АC |
|
6 |
BK59 |
М |
59 |
АC |
|
7 |
BK74 |
F |
62 |
АC |
|
Cases without EGFR mutation in FFPE tumor tissue DNA, plasma before surgery |
||||
|
8 |
ВК1 |
М |
53 |
NSCLC-NOS |
|
9 |
ВК3 |
М |
47 |
АC |
|
10 |
ВК8 |
F |
59 |
NSCLC-NOS |
|
11 |
ВК9 |
F |
63 |
АC |
|
12 |
ВК10 |
М |
63 |
NSCLC-NOS |
|
13 |
ВК17 |
М |
60 |
АC |
|
14 |
ВК20 |
М |
54 |
SCC |
|
15 |
ВК23 |
М |
64 |
SCC |
|
16 |
ВК24 |
М |
43 |
SCC |
|
17 |
ВК25 |
М |
73 |
АC |
|
18 |
ВК29 |
М |
64 |
АC |
|
19 |
ВК53 |
М |
68 |
SCC |
|
20 |
ВК60 |
М |
56 |
SCC |
|
Cases with mutant EGFR under TKI therapy |
||||
|
21 |
ВК57 |
F |
53 |
АC |
|
22 |
TMB1 |
F |
60 |
АC |
|
23 |
ХАН2 |
М |
80 |
NSCLC-NOS |
|
24 |
МАТ3 |
F |
70 |
NSCLC-NOS |
|
25 |
GLF4 |
F |
73 |
АC |
|
26 |
SLF5 |
F |
62 |
АC |
|
27 |
KVI6 |
М |
na |
NSCLC-NOS |
|
28 |
CAB7 |
F |
64 |
АC |
|
29 |
PIM8 |
F |
52 |
АC |
|
30 |
DVI9 |
М |
60 |
АC |
|
31 |
IEV10 |
F |
69 |
АC |
|
32 |
BVI11 |
М |
60 |
АC |
|
33 |
FVY12 |
М |
67 |
АC |
|
34 |
G013 |
М |
na |
АC |
|
35 |
KMG14 |
F |
77 |
АC |
|
36 |
В16 |
F |
na |
NSCLC-NOS |
Note: NSCLC-NOS – non-small cell lung cancer not otherwise specified, АC – adenocarcinoma, SCC – squamous cell cancer; na – not available.
Blood was collected in 8 ml Vacutainer tubes with EDTA solution and stored at +4°C before preparation of plasma. Plasma was prepared on the same day of the blood draw by centrifugation for 10 min at 2000 g at 4°C and 0.5–1.0 ml aliquots were stored at -70°C.
DNA purification
DNA from FFPE tissue was purified using FFPE DNA kit (Biolink, Russia) and stored at -20°C. cfDNA was purified from 1.0 ml blood plasma using “PME free-circulating DNA Extraction Kit” (Analytik Jena, Germany). cfDNA was dissolved in 50 µl of PCR-grade water and stored at -20°C.
Concentration of cfDNA was estimated by realtime PCR for EGFR exon 21 using control PCR reaction mixture from “Real-time-PCR-EGFR-7RP Kit” (Biolink, Russia) and human placenta DNA (2ngµ/l) as external standard. The average concentration of cfDNA was 9.3 ± 14 ng/ml (range 0.5–81.0 ng/ml) plasma. In a few samples, the concentration of cfDNA was above 250 ng/ml; the samples had pink color due to hemolysis and were excluded from further analysis.
Multiplex PCR of 18-21 exons of EGFR
Multiplex PCR (mPCR) was done in 50µl in reaction mixture with 1x Buffer for Taq-DNA-polymerase; 0.2 mM each of dATP, dGTP, dTTP, dCTP; 1.5 mM MgCl2, 0.5 µM each forward and reverse primers for 18-21 exons of EGFR (Table 2), 2.0 U Taq DNA-polymerase (Biolink), and 20 µl cfDNA from blood plasma. mPCR was performed in iCycler iQ5 (BioRad) using the following protocol: 1 cycle – 95oC 3 min; 8–12 cycles – 95оС 15 sec, 58оС 30 sec, 72оС 20 sec; 1 cycle 72оС 3 min. After mPCR, reaction mixture was diluted 1:25 in PCR-grade water and 5µl was used in qPCR.
Detection of EGFR mutations in cfDNA by realtime PCR
Amplicons after multiplex PCR of EGFR sequences in cfDNA were tested for EGFR L858R, L861Q, G719A/C/S, S768I, T790M mutations and deletions table 2
|
Exon |
Forward primer |
Reverse primer |
Amplicon size, b.p. |
|
18 |
tcccaaccaagctctcttga |
ctgtgccagggaccttacc |
109 |
|
19 |
tggatcccagaaggtgagaaag |
cccacacagcaaagcagaa |
118 |
|
20 |
cctccctccaggaagccta |
gccgaagggcatgagctg |
116 |
|
21 |
ccgcagcatgtcaagatcac |
aatgctggctgacctaaagc |
115 |
|
table 3 |
|||
|
Mutations of EGFR in ffpe tumor dna and plasma dna from nsclc patients |
|||
|
Case ID |
FFPE Tumor DNA(a) |
Plasma cfDNA(a, b) |
|
|
Cases with EGFR mutation in FFPE tumor tissue |
|||
|
BK7 |
G719X; S768I |
S768I |
|
|
BK16 |
19del |
19del |
|
|
BK35 |
19del |
19del; |
|
|
BK40 |
L861Q |
wt |
|
|
BK54 |
L858R |
wt |
|
|
BK59 |
L858R |
L858R; |
|
|
BK74 |
19del |
wt |
|
|
Cases with EGFR mutation, n (%) |
7/7 (100.0) |
4/7 (57.1) |
|
|
Cases without EGFR mutation in FFPE tumor tissue |
|||
|
ВК1 |
wt |
wt |
|
|
ВК3 |
wt |
wt |
|
|
ВК8 |
wt |
wt |
|
|
ВК9 |
wt |
wt |
|
|
ВК10 |
wt |
wt |
|
|
ВК17 |
wt |
wt |
|
|
ВК20 |
wt |
wt |
|
|
ВК23 |
wt |
wt |
|
|
ВК24 |
wt |
wt |
|
|
ВК25 |
wt |
wt |
|
|
ВК29 |
wt |
wt |
|
|
ВК53 |
wt |
wt |
|
|
ВК60 |
wt |
wt |
|
|
Cases with EGFR mutation, n (%) |
0/13 (0,0) |
0/13 (0,0) |
|
Note: (a) – allele-specific real-time PCR for mutations EGFR L858R, L861Q, G719X, T790M, S768I and wild-type blocking PCR for mutations EGFR 19del; (b)– blood draw was done during a week before surgery.
primers for multiplex pcR of sequences in 1821 exons of EGFR
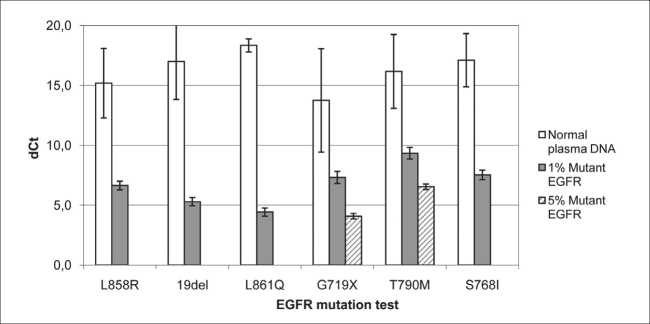
Fig. 1. Sensitivity and specificity of real-time PCR for EGFR mutations. Mutations EGFR L858R, L861Q, G719X, T790M, S768I and deletions in EGFR exon 19 (19del) were detected by “Real-time-PCR-EGFR-7RP Kit” (Biolink, Russia). DNA of positive controls (PC) with 1–5 % mutant allele EGFR L858R, E746_A750delELREA, L861Q, G719S, T790M, S768I and 12 samples of cfDNA from blood plasma of 12 healthy volunteers were tested using six replicates of PC and two replicates of amplicons of each cfDNA. dCt mean and dCt standard variation are shown
in exon 19 using “Real-time-PCR-EGFR-7RP Kit” (Biolink, Russia). In the kit, allele-specific qPCR (as-PCR) is used to detect L858R, L861Q, G719A/C/S, S768I, T790M mutations, and wild-type blocking PCR [8] was used to detect deletions in exon 19.
Results and discussion
Sensitivity and specificity of qPCR test for EGFR mutations
Analytical sensitivity and specificity of the assay was evaluated using human genomic DNA spiked with recombinant plasmid DNA as positive controls (PC) to have 1–5 % mutant allele for tested mutations ( EGFR L858R, E746_A750delELREA, L861Q, G719S, T790M, S768I). As negative controls, we used 12 samples of cfDNA from the blood plasma of healthy volunteers without EGFR mutations.
To have sufficient DNA-copies for mutation test, sequences of 1821 exons of EGFR in cfDNA were amplified by multiplex PCR. Two replicates of amplicons of each cfDNA and 6 replicates of PC were tested for EGFR mutations using ~3000 copies of EGFR DNA in reaction. For each DNA, dCt was calculated as dCt = CtAS – CtC, where CtAS is Ct of the DNA in asPCR for the tested mutation, and CtC is Ct of the DNA in control PCR for constant EGFR sequence.
dCt values were much larger for wt cfDNA in comparison to PC (Fig. 1) indicating that the method differentiated DNA with 1 % of mutant EGFR alleles: L858R, E746_A750delELREA, L861Q, S768I or 5 % of mutant EGFR alleles: G719S or T790M from wt cfDNA with probability p>0.95.
Mutations of EGFR in paired samples of the DNA from FFPE tumor tissue and blood plasma from patients with NSCLC. Clinical sensitivity of liquid biopsy for EGFR mutations was evaluated using FFPE tumor samples from patients with NSCLC and matched blood plasma of the patients that was collected before surgery (Table 3).
Among 7 cases that carried EGFR mutations in the DNA from FFPE tumor tissue, 3 cases had the same mutations in plasma cfDNA. Interestingly, in a case BK7 with G719X and S768I mutations in FFPE tissue, only S768I was found in plasma presumably due to tumor heterogeneity. In the other three cases (BK40, BK54, BK74) mutations present in FFPE tumor tissue were not detected in plasma. Importantly, EGFR mutations were not found in cfDNA of 13 cases without EGFR mutation in FFPE tumor tissue. These data showed that in comparison to FFPE tissue test, the liquid biopsy for EGFR driver mutations had sensitivity of 4/7 (57 %) and specificity of 13/13 (100 %).
Monitoring of EGFR mutations in plasma cfDNA from patients with NSCLC treated with TKI
Changes in MAF of the EGFR in blood plasma of patients treated with TKI can be a molecular marker of sensitivity or development of resistance of tumor to TKI treatment. The accumulation of T790M results in the development of resistance to the first generation TKI that can be treated with the third generation TKI (osimertinib). We started monitoring of EGFR mutations in plasma cfDNA from 16 patients with NSCLC treated with gefitinib or erlotinib. This is ongoing study and interim results are shown in Table 4.
Among 16 cases with NSCLC, 12 carried mutation 19 del and 4 cases carried L858R in FFPE tumor tissue. In 8 cases, EGFR mutations were not detected in plasma cfDNA. Lack of or low concentration of mutant EGFR in the cfDNA of the cases could be due to: 1) tumors being under TKI suppression or 2) metastasis to the brain with the cfDNA level in plasma below the detection limit of our test.
In the other 8 cases, EGFR mutations were detected in cfDNA. The same EGFR mutations that were present in FFPE tumor DNA were detected in cfDNA in 7 cases. In another case (patient TMB1) mutation T790M was detected in plasma cfDNA in addition to EGFR driver mutation L858R. Four table 4
EGFR mutations in cfdna of nsclc patients under treatment with tki
|
Patient ID |
Treatment |
EGFR mutation |
|
|
FFPE DNA |
cfDNA |
||
|
ВК57 |
S,TKI |
L858R |
L858R |
|
TMB1 |
TKI |
L858R |
L858R, T790M |
|
ХАН2 |
ChT, TKI |
19del |
wt |
|
МАТ3 |
S, TKI |
19del |
wt |
|
GLF4 |
TKI |
19del |
wt |
|
SLF5 |
S, ChT, TKI |
19del |
wt |
|
KVI6 |
TKI |
19del |
wt |
|
CAB7 |
TKI |
19del |
wt |
|
PIM8 |
TKI, RT |
19del |
19del |
|
DVI9 |
TKI, G |
19del |
19del |
|
IEV10 |
S, TKI |
19del |
19del |
|
BVI11 |
S, TKI |
19del |
wt |
|
FVY12 |
S, RT, ChT, TKI |
19del |
wt |
|
G013 |
TKI |
L858R |
L858R |
|
KMG14 |
S, ChT, TKI |
19del |
19del |
|
В16 |
TKI |
L858R |
L858R |
Note: ChT – polychemotherapy, G – gamma knife radiosurgery, RT – radiation therapy, S – surgery, TKI – therapy with gefitinib or erlotinib; BM – brain metastasis.
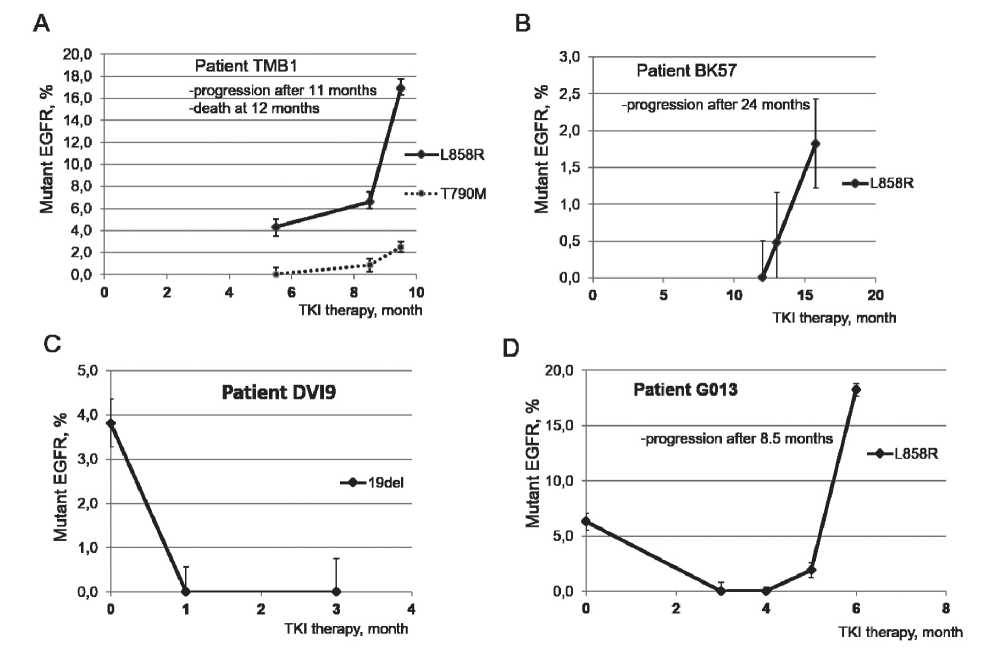
Fig. 2. Mutant allele frequency of the EGFR in plasma cfDNA of patients under TKI therapy. EGFR mutations were detected by qPCR. A. Patient TMB1. B. Patient BK57. C. Patient DVI9. D. Patient G013
representative cases with changes in MAF of EGFR in cfDNA during TKI therapy are shown in Fig. 2. In case TMB1 (Fig. 2A), L858R and T790M mutations were detected 5.5 and 8.5 months after starting TKI therapy, respectively thus suggesting the development of T790M-associated resistance in the tumor. Disease progression was manifested 11 months from the start of TKI therapy, resulting in death at 12 months. Of note, higher MAF of L858R in cfDNA in comparison to T790M suggests heterogeneity of TKI-resistant tumor with subpopulation(s) of resistant tumor cells with and without the T790M mutation. In case BK57, the increased MAF of L858R mutation in cfDNA was detected 12 months after therapy with erlotinib (Fig. 2B), while clinical progression with metastasis to bones developed 24 months after therapy. In case DVI9, tumor with 19 del responded to treatment with gefitinib and the level of 19del became undetectable after the first month of TKI therapy. Later, the patient had metastasis to the brain without accumulation of mutant EGFR in plasma cfDNA (data not shown). In case G013, tumor with the L858R mutation responded to treatment with gefitinib. Mutation in plasma cfDNA dropped to undetectable level 3 and 4 months after starting TKI therapy. However, the L858R mutation in cfDNA was detected 5 months after starting TKI therapy, and the level of the mutation continued to increase 6 months from the start of TKI therapy.
Список литературы Monitoring of EGFR mutations in the circulating tumor DNA from blood plasma of patients with non-small cell lung cancer
- Morgensztern D., Campo M.J., Dahlberg S.E., Doebele R.C., Garon E., Gerber D.E., Goldberg S.B., Hammerman P.S., Heist R.S., Hensing T., Horn L., Ramalingam S.S., Rudin C.M., Salgia R., Sequist L.V., Shaw A.T., Simon G.R., Somaiah N., Spigel D.R., Wrangle J., Johnson D., Herbst R.S., Bunn P., Govindan R. Molecularly targeted therapies in nonsmall-cell lung cancer annual update 2014. J Thorac Oncol. 2015 Jan; 10 (1 Suppl 1): S1-63. DOI: 10.1097/JTO.0000000000000405
- Soria J.C., Ohe Y., Vansteenkiste J., Reungwetwattana T., Chewaskulyong B., Lee K.H., Dechaphunkul A., Imamura F., Nogami N., Kurata T., Okamoto I., Zhou C., Cho B.C., Cheng Y., Cho E.K., Voon P.J., Planchard D., Su W.C., Gray J.E., Lee S.M., Hodge R., Marotti M., Rukazenkov Y., Ramalingam S.S.; FLAURA Investigators. Osimertinib in Untreated EGFRMutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018 Jan 11; 378 (2): 113-125. DOI: 10.1056/NEJMoa1713137
- Kerr K.M., Bubendorf L., Edelman M.J., Marchetti A., Mok T., Novello S., O'Byrne K., Stahel R., Peters S., Felip E.; Panel Members; Panel Members. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann Oncol. 2014 Sep; 25 (9): 1681-90. DOI: 10.1093/annonc/mdu145
- Kalemkerian G.P., Narula N., Kennedy E.B., Biermann W.A., Donington J., Leighl N.B., Lew M., Pantelas J., Ramalingam S.S., Reck M., Saqi A., Simoff M., Singh N., Sundaram B. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol. 2018 Mar 20; 36 (9): 911-919. DOI: 10.1200/JCO.2017.76.7293
- Rolfo C., Mack P.C., Scagliotti G.V., Baas P., Barlesi F., Bivona T.G., Herbst R.S., Mok T.S., Peled N., Pirker R., Raez L.E., Reck M., Riess J.W., Sequist L.V., Shepherd F.A., Sholl L.M., Tan D.S., Wakelee H.A., Wistuba I.I., Wynes M.W., Carbone D.P., Hirsch F.R., Gandara D.R. IASLC Statement Paper: Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC). J Thorac Oncol. 2018 Sep; 13 (9): 1248-1268. DOI: 10.1016/j.jtho.2018.05.030
- Francis G., Stein S. Circulating Cell-Free Tumour DNA in the Management of Cancer. Int J Mol Sci. 2015 Jun 19; 16(6): 14122-42. DOI: 10.3390/ijms160614122
- Wu Y.L., Sequist L.V., Hu C.P., Feng J., Lu S., Huang Y., Li W., Hou M., Schuler M., Mok T., Yamamoto N., O'Byrne K., Hirsh V., Gibson N., Massey D., Kim M., Yang J.C. EGFR mutation detection in circulating cellfree DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer. 2017 Jan 17; 116 (2): 175-185. DOI: 10.1038/bjc.2016.420


