Mutation of the mitochondrial transcription termination factor gene affects the expression of organellar genes and nuclear genes encoding alternative respiratory pathway proteins in arabidopsis
Автор: Katyshev Alexander I., Fedoseeva Irina V., Katysheva Natalya B.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 1 т.16, 2020 года.
Бесплатный доступ
Characterization of mterf13 mutant Arabidopsis plants showed the possible participation of the mTERF13 in the modulation of genetic processes in mitochondria and chloroplasts of Arabidopsis. Moreover, according to data obtained, this gene can encode a potential mitochondrial transcription factor. Mutation of the studied gene through retrograde regulation can also lead to a change in the expression of nuclear genes encoding components of the alternative respiratory pathway.
Mitochondrial transcription termination factors, organellar gene expression, mitochondrial alternative respiratory pathway
Короткий адрес: https://sciup.org/143171158
IDR: 143171158
Текст научной статьи Mutation of the mitochondrial transcription termination factor gene affects the expression of organellar genes and nuclear genes encoding alternative respiratory pathway proteins in arabidopsis
Mitochondria, which are energy stations of eukaryotic cells, are one of the main sources of generation of reactive oxygen species (ROS) in plant cells (Møller, 2001). The formation of ROS in plant mitochondria is primarily associated with impaired functioning of the main and alternative respiratory pathways (Møller, 2001; Vanlerberghe, 2013). The protein components of the main and alternative mitochondrial respiratory complexes are predominantly encoded by the nucleus, only certain subunits of the respiratory complexes are encoded by the mitochondrial genome (Gualberto et al. , 2014). Expression of a relatively small number of mitochondrial genes in plant mitochondria undergoes complex regulation at various levels (Millar et al. , 2004; Holec et al. , 2006). Transcription of mitochondrial genes is carried out by 2 phage-type polymerases — RpoTm and RpoTmp, the last of which has double localization — in mitochondria and in chloroplasts (Hedtke et al. , 2000; Kühn et al. , 2009). It has been shown that, at least in the case of RpoTmp, the implementation of its function requires the presence of some unknown protein cofactors. (Kuhn et al, 2007). Several potential candidates have been proposed as potential mitochondrial transcription factors, including homologs of the bacterial sigma factor, individual PPR proteins, homologs of mitochondrial transcription factors (mtTFA and mtTFB) found in animals, and others (reviewed by Liere et al. , 2011). Reliable data confirming the participation of these factors in the expression of plant mitochondrial genes has not yet been obtained.
In recent years, data have been obtained showing an important role in the implementation of genetic processes in plant organelles of proteins homologous to the mitochondrial transcription termination factor (mTERF), firstly identified in metazoans (reviewed by Klein and Leister, 2015). In Arabidopsis, the mTERF gene family consists of 35 representatives, encoding chloroplast, mitochondrial, or double-localized proteins (Kleine et al., 2012). Mutations of Arabidopsis mTERF genes may lead to perturbations of plastid and mitochondrial gene expression homeostasis, which in turn, may result in the activation of acclimation and tolerance responses, presumably via retrograde signaling. (Kim et al., 2012; Robles et al., 2015; Leister et al., 2017; Xu et al., 2017; Robles et al., 2018). The aim of this work was to study the role of the mTERF13 gene of Arabidopsis in modulating the expression of organellar genes and nuclear genes encoding the components of the mitochondrial alternative respiration pathways.
MATERIALS AND METHODS
Wild-type Arabidopsis thaliana (L.) Heynh plants (Col-0) and the T-DNA insertion SALK_076739 (N658768) line were obtained from the ABRC seed stock center. Seeds were surface sterilized and plated on containing 0.5× Murashige and Skoog salts (Sigma-Aldrich, USA) and 0.8% (w/v) Phytagel (Sigma-Aldrich). Seeds were stratified at 4°C for 3 d. Plants were grown in KBW720 (Binder, Germany) growth chambers at 24/22 ºС day/night, 16-h photoperiod and irradiance of 150 μmol m -2 s -1. Plant for RNA isolation were harvested after 14 d.
GeneJET Plant RNA Purification Kit (ThermoFisher Scientific Baltic, Lithuania) was used for RNA isolation according to the manufacturer’s instructions. The RNA was then subjected to two consecutive treatments with DNase (ThermoFisher Scientific Baltic, Lithuania). One microgram of RNA was reverse-transcribed with RevertAid Reverse Transcriptase (ThermoFisher Scientific Baltic, Lithuania) according to the manufacturer’s protocol using random hexamers (Evrogen, Russia). qRT-PCR analysis was performed using qPCRmix-HS SYBR (Evrogen, Russia) and the CFX96 Real-Time PCR Detection System (Bio-Rad, USA) according to the manufacturer’sinstructions. The primer sequences are provided in the Table 1. All reactions were performed in duplicates with the three independently isolated RNA samples. The expression of each gene was normalized to the expression of gapdh (At1g13440) and pp2a (At1g13320) genes.
RESULTS AND DISCUSSION
In Arabidopsis, the mTERF gene family consists of 35 representatives, which are divided by Klein (Kleine, 2012) into several clusters according to co-expression patterns, together with data pertaining to the subcellular localizations of the proteins encoded by the genes coexpressed with each mTERF defined by the Gene
Ontology (GO) Annotations. To date, the role of two mTERF genes from designated by Kleine as a “mitochondrial” cluster, mTERF15 and mTERF18/SHOT1, is best characterized in arabidopsis organelles. The mTERF15 is not, apparently, a transcription factor, but takes part in the maturation of the transcript of the mitochondrial nad2 gene. Disruption of splicing of intron 3 of the nad2 gene in the mTERF15 mutant leads to a decrease in the activity of the respiratory complex I in mitochondria of arabidopsis (Hsu Y.-W. et al. , 2014).
SHOT1 / mTERF18 is more favorable canditate for the role of mitochondrial transcription modulating factor, since expression of mitochondrial genes is significantly altered in mterf18/shot1 mutant Arabidopsis plants (Kim et al. , 2013). At the same time, a mutation of this gene led to the opposite change in the expression of chloroplast genes (Kim M. et al. , 2012). This effect can be determined by experimentally not shown double localization of mTERF18, the possibility of which allows bioinformatic methods of analysis (Kleine, 2012) or can be mediated by complex mechanisms of retrograde/anterograde signaling (Kleine and Leister, 2016). Moreover, a change in the expression of a number of nuclear stress genes and an alternative oxidase Aox1a gene was observed in SHOT1/mTERF18 mutant plants as a result of retrograde regulation. A perturbation in organellar gene expression homeostasis and altered expression of stress response genes resulted in enhanced thermotolerance of the mterf18/shot1 mutant plants (Kim et al. , 2012).
In this work we studied the effect of a mutation of another mTERF gene from the “mitochondrial” cluster (Kleine, 2012) – mTERF13 - on the organellar gene expression as well as the expression of nuclear genes encoding the components of the main and alternative mitochondrial respiration pathways. Seeds of T-DNA insertion SALK_076739 line were obtained from the ABRC seed stock center.
The phenotypic characteristics of mutant plants under standard growing conditions did not reveal significant differences in the growth rate of mutant and wild-type plants.
An analysis of the transcript amount of a number of mitochondrial and chloroplast genes in 2-week-old mutant plants showed that the mutation of the mTERF13 gene similar to mutation of the mTERF18 gene leads to a decrease in the expression of chloroplast genes (Fig.1B). But in contrast to the mterf18/shot1 mutants in the mterf13 plants the expression of the mitochondrial genes was reduced or unchanged (Fig.1A).
Observed by Kim et al. in mterf18/shot1 mutant plants an increase of abundances of the most of the investigated mitochondrial transcripts can be differently explained and suggests various possible functions of the mTERF18 in the realization of genetic processes in mitochondria of Arabidopsis. In case of the mterf13 mutant line, the observed decrease of certain mitochondrial gene expression indicates primarily the possible role of this protein as a mitochondrial transcription factor. This assumption will be further verified using other molecular biological approaches.
Table 1. Primers used for quantitative RT-PCR analysis.
|
Gene |
Forward primer, 5’->3’ |
Reverse primer, 5’->3’ |
|
cox1 |
GACTCTCTATTTCATTTTCGGTGCCA |
TGATCGCCGGGTCGTGCT |
|
matR |
TACGATCCCGAGTTTCCAGACACAT |
TTCCCCACTCTTCTTTGATCCGTCT |
|
ccmC |
CAACCTGGGAGCATTAGCCGA |
AGATACGGGTTGAGAAGGGGAAGTTA |
|
nad6 |
ATGATACTTTCTGTTTTGTCGAGCCCT |
CTGGGATGGGAAACAAAACGGA |
|
cob |
CGATGTCCACCCCGCCTCA |
GGCTACACCTCCCGCTTTGTCA |
|
orf153a |
ACAGACAGTTCCTTTATCACACCTTCACA |
GGGAAGATGTCGAGATAGCAGGAGAA |
|
atpI |
TCAAGGGGAGTTAGCAGCACCAA |
GGGTAAAAGAATTGGGGTTGGTTGA |
|
accD |
CCTTGGGGTTATGGATTTTCGGTT |
ACTTCCTTCTTGCATTCGTGCTCCT |
|
rrn16 |
GGGTAGCTATATTTCTGGGAGCGA |
GTTGCATTACTTATAGCTTCCTTCTTCGT |
|
clpP |
TCGGAGGAGCAATTACCAAACGT |
AATTCTCCCGTTTGTGCCTCATAA |
|
aox1a |
CGGCTGGACCACGTTTGTTCT |
CCAATCGTCGGAGCTCTAGTCCATA |
|
aox1d |
GACATCTCATTAGCCACTTGCCCA |
TTCCCACTTACCGGAGATGACGT |
|
ndb2 |
CTCCAAGGCTCCAATACACATATCTCTC |
ATCTCTCCTCGTTACTTGTGGTCATTATTT |
|
gapdh |
GGCGAGAGTTTGTGTGTGGTTGA |
AAGCAGGGAAACATTAAGAGAAAGCAA |
|
pp2a |
AAAAGGTAAAGAAGACAGCAACGAATT |
TAGCAGAGAGAAACAATGGTCACGTA |
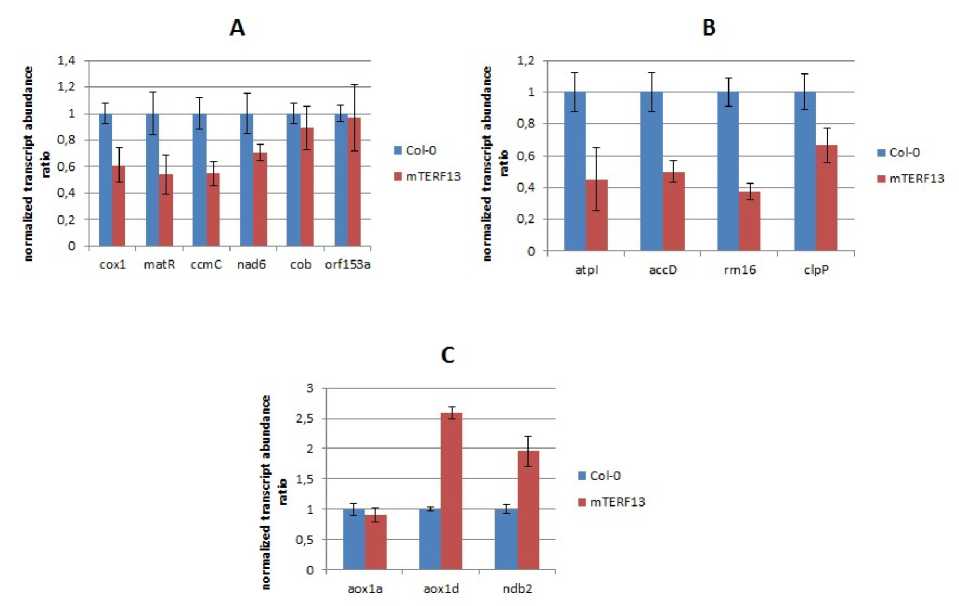
Figure 1. Differential gene expression of organellar and nuclear genes encoding alternative respiration pathway proteins in mterf13 mutant Arabidopsis plants . (A) Transcript abundance of mitochondrial genes from mterf13 mutant plants (mTERF13) and wild type (Col-0) plants. (B) Transcript abundance of chloroplast genes from mterf13 mutant (mTERF13) plants and wild type (Col-0). (C) Transcript abundance of nuclear alternative respiration patway genes from mterf13 mutant (mTERF13) plants and wild type (Col-0) plants. Error bars are SD based on three biological replicates.
As a perturbation in organellar gene expression homeostasis may result in change of an expression of nuclear genes encoding alternative mitochondrial pathway proteins (Kim et al. , 2013) we also tested the transcript abundances of Arabidopsis alternative oxidase and rotenone-insensitive NADH dehydrogenase genes in mterf13 mutant plants. The increase in the expression of aox1d and ndb2 genes in mutant plants was observed compared to control plants (Fig.1C). The amount of the aox1a gene transcript did not significantly differ in the control and mutant plants. These data together with data on changes in the expression of organellar genes, suggests that, by analogy with other characterized mterf mutants (Kim et al. , 2013; Robles et al. , 2015 and 2018), the mterf13 plants that are phenotypically not different from control plants when grown under control conditions, may demonstrate altered resistance to abiotic stress factors, what will be interesting to test in the further experiments.
CONCLUSION
Thus, сharacterization of mterf13 mutant plants showed the possible participation of the mTERF13 in the modulation of genetic processes in mitochondria and chloroplasts of Arabidopsis. Moreover, according to data obtained, this gene can encode a potential mitochondrial transcription factor. Mutation of the studied gene through retrograde regulation can also lead to a change in the expression of nuclear genes encoding components of the alternative respiratory pathway.
ACKNOWLEGMENTS
The work was financially supported by RFBR grant No.17-04-01515. We thank the ABRC for providing seed stocks.
Список литературы Mutation of the mitochondrial transcription termination factor gene affects the expression of organellar genes and nuclear genes encoding alternative respiratory pathway proteins in arabidopsis
- Gualberto JM, Mileshina D, Wallet C, Niazi AK, Weber-Lotfi F, Dietrich A. (2014) The plant mitochondrial genome: dynamics and maintenance. Biochimie., 100, 107-120
- Hedtke, B., Borner, T., Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep., 1, 435-440
- Holec S, Lange H, Kühn K, Alioua M, Börner T, Gagliardi D. (2006) Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell Biol. 26, 2869-2876
- Hsu Y.-W., Wang H.-J., Hsieh M.-H., Hsieh H.-L., Jauh G.-Y. (2014) Arabidopsis mTERF15 Is Required for Mitochondrial nad2 Intron 3 Splicing and Functional Complex I Activity. PLoS ONE, 9, e112360
- Kim M., Lee U., Small I., des Francs-Small C.C., Vierling E. (2012) Mutations in an Arabidopsis Mitochondrial Transcription Termination Factor-Related Protein Enhance Thermotolerance in the Absence of the Major Molecular Chaperone HSP101W. Plant Cell, 24, 3349-3365
- Kleine T. (2012) Arabidopsis thaliana mTERF proteins: evolution and functional classification. Front. Plant Sci., 15, 233
- Kleine T., Leister D. (2015) Emerging functions of mammalian and plant mTERFs. Biochim. Biophys. Acta, 1847, 786-797
- Kleine T., Leister D. (2016). Retrograde signaling: organelles go networking. Biochim. Biophys. Acta, 1857, 1313-1325
- Kühn K., Richter U., Meyer E.H., Delannoy E., de Longevialle A.F., O'Toole N., Börner T., Millar A.H., Small I.D., Whelan J. (2009) Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell, 21, 2762-2679
- Leister D., Wang L., Kleine T. (2017) Organellar Gene Expression and Acclimation of Plants to Environmental Stress. Front. Plant. Sci., 8, 387
- Liere K., Weihe A., Börner T. (2011) The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol., 168, 1345-1360
- Millar H., Day D.A.., Whelan J., Somerville C.R., Meyerowitz E.M. (2004) Mitochondrial Biogenesis and Function in Arabidopsis. The Arabidopsis Book, In Somerville C.R., Meyerowitz E.M., ed., pp.1-36
- Møller I.M. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol,. 52: 561-591
- Robles P., Micol J.L. Quesada V. (2015) Mutations in the plant-conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana. Physiol. Plant., 154, 297-313
- Robles P, Navarro-Cartagena S, Ferrández-Ayela A, Núñez-Delegido E, Quesada V. (2018) The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress. Int. J. Mol. Sci., 19, E2388
- Vanlerberghe G.C. (2013) Alternative Oxidase: A Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis during Abiotic and Biotic Stress in Plants. Int. J. Mol. Sci., 14, 6805-6847
- Xu D, Leister D, Kleine T. (2017) Arabidopsis thaliana mTERF10 and mTERF11, but Not mTERF12, Are Involved in the Response to Salt Stress. Front. Plant Sci., 8, 1213.


