Natural antiviral flavonoids: striking successful repurposing against COVID-19 through in-silico docking
Автор: Velayutham Gurunathan, Chidambaram Sathishkumar, Hariharan Govindasamy, Karumalaiyan Palanisamy, Periyanayagam Arockia Doss
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 3 т.21, 2025 года.
Бесплатный доступ
Repurpose of recognized compounds and medications as anti-COVID 2019 (anti-CoVID-19) agents, in anti-SARS-CoV-2 operation, via biological re-assessment of their activities, is a recent and durable development for pandemic COVID-19 novel drugs in 2020. Even so, nearly all of the recorded inhibitors of the various phases of SARS-CoV-2 progression lacks severe forces towards main fateful SARS-CoV-2 enzymes (like the papain protease "PLpro", main protease "Mpro", and RNA-based RNA polymerase "RdRp"). The main objective of this article is to estimate and classify natural antiviral flavonoids as inhibitor medicines like Quercetin, Quercetagetin, Volkensiflavone, Ternatin, Meliternatin, Formononetin, Afromosin, Chrysosplenol B, Chrysosplenol C and Axillarin for COVID-19 main protease and compared with antiviral medication Remdesivir. In-silico docking studies devours natural flavonoid derivative Volkensiflavone was of exceptional inhibition ability (Binding energy-8.9, -9.0 kcal/mol) of 5N5O and 6LU7 enzyme, relative to the other compounds and Remdesivir antiviral medication (Binding energy -7.4 and -7.7 Kcal/mol). The need for the most time is the prompt discovery and commitment of appropriate medication to tackle and convince the global COVID-19 crisis. Besides, timely in vivo experiments takes place to approve inhibition efficacy of anti-SARS-CoV-2 compounds might save people are warranted. 115
Antiviral, adme, covid-19, remdesivir, volkensiflavone, molecular docking
Короткий адрес: https://sciup.org/143184726
IDR: 143184726
Текст научной статьи Natural antiviral flavonoids: striking successful repurposing against COVID-19 through in-silico docking
The COV D-19 global epidemic has driven the planet to pursue possible therapies to fight these destructive infectious diseases. According to the study, drugs produced by repurposing current medications are focussed on the destruction of virus pathogenicity (Manisha et al ., 2020). A modern coronavirus (2019-nCoV), generally alluded to as SARS-CoV-2 (severe acute respiratory syndrome Coronavirus 2), later appeared in Wuhan City in December 2019 (Hui et al ., 2020). Coronavirus is a non-segmented, enveloped virus of the Coronaviridae family, which has positive senses of genomic RNA. This specific epidemic is suspected to be linked to a crowded maritime sector in Wuhan, China, and closed since 23 January 2020 (Rupal et al ., 2020). The virus infects by overall 10,896,029 cases reported and 521,862 fatalities by 3 July 2020 and dispersed to 188 countries as per the CoV D-19 Console from the Johns Hopkins University Centre for Systems Science and Engineering (JHU) (Abo-Zeid et al ., 2020).
The dissemination of this enigmatic bread-packed virus, prompting highly effective control steps, continues to propagate to the human respiratory system (i.e. detected by a characteristic pulmonary infection) the virus-characteristic illness, coronavirus illness (COV D-19) in 2019 (i.e., key signs and symptoms localized and located). The epidemic of China's 2019 nCoV virus (starting with the Wuhan epidemic) takes feast through our world and devises world-wide epidemic of devastation and catastrophic catastrophe (Hui et al ., 2020; Li et al ., 2020). Since then, the intense besides finest exertions of global pharmacological firms, medication development testing centres/organizations, foreign health agencies, pharmaceutical/medical colleges and other relevant organizations have concentrated on prompt quest on behalf of successful effective drugs and rehabilitations capable of killing SARS-CoV-2, repetition of SARS-CoV-2, and combating the utmost stark and lethal possessions of COV D-19. No new antiviral drugs were formally and globally licensed so far in mid-2020 (Jiang et al ., 2020) for effective management of COV D-19.
Given the present non-appearance of proven successful anti-SARS-CoV-2 treatments and unadorned circumstance under the evolving global condition, several investigators have proposed the usage of some established active, potent drugs (Dong et al ., 2020), for instance, arbidol (anti-influenza) (Wang et al ., 2020), remdesivir (antiviral) (Choy et al ., 2020; Elfiky et al ., 2020; Elfiky et al ., 2020a; Shannon et al ., 2020; Wang et al., 2020; Yin et al ., 2020), hydroxychloroquine (antimalarial and anti-rheumatic disorientation) (Derwand et al ., 2020: Liu et al ., 2020), favipiravir (anti-influenza) (Cai et al ., 2020; Du et al ., 2020; Shiraki et al ., 2020), and ivermectin (anti-parasite) (Caly et al ., 2020), towards combat the original COV D-19. The prospects of several therapeutic and pharmacological chemists uses in-silico based molecular modelling studies to provide foundation for the production of innovative persuasive compounds active beside SARS-CoV-2, utmost of that placid in recently reported literatures (Amin et al ., 2020; Goyal & Goyal, 2020).
Environmental and economic characteristics can significantly promote the efflux of secondary metabolites such as tropical plant bioactive compounds. Additionally, secondary plant-concealed metabolites are deemed prodigiously in tropical regions and are progressed in remedies (Guerriero et al., 2018; Yang et al., 2018). nnumerable medicinal plant natural products were already evaluated for antiviral action (Zakaryan et al., 2017; Thayil & Thyagarajan, 2016; Jo et al., 2020). The natural flavonoid analogue Quercetin was extracted from the medicinal plant Chenopodium quinoa (Quenopodiaceae) and displayed remarkable antiviral activity against potato virus X (PVX) with 80% inhibition at <1µg/ml-1 (French et al., 1992). Antiviral flavonoid Quercetagetin showed significant antiviral effect against several retroviruses comprising H V and Rauscher murine leukaemia (RLV), along with cellular DNA polymerases and isolated from the many spp of Compositae flowers (Cody et al., 1986). The natural antiviral flavonoid analogue Volkensiflavone displayed remarkable antiviral activity against influenza B virus and isolated from the therapeutic plant Rhus succedania L. (Anacardaceae) (Beladi et al., 1977; Lin et al., 1997). Natural antiviral 3-methoxyflavone derivative Ternatin and Meliternatin showed significant antiviral effect against several viral pathogens including VSV type 2, HSV-1, poliovirus type 2, HSV-2, and adenovirus type 2; extracted from medicinal plant Evodia madagascariensis Baker (Rutaceae) (Simoes et al., 1990). Natural isoflavonoids Formononetin and Afromosin were displayed significant antiviral inhibitory effect against Epstein-Barr virus and obtained from the therapeutic plant Wisteria brachybotrys Sieb (Leguminosae) (Konoshima et al., 1989). Natural antiviral flavonoid analogues Chrysosplenol B, Chrysosplenol C and Axillarin, were exhibited remarkable antiviral effect against rhinovirus and isolated from the medicinal plant Chrysospleniumtosaense (Saxifragaceae) (Tsuchiya et al., 1985). Fig. 1 represents the natural antiviral analogues. This overall prevailing theme in the quest for vital compounds with structural stability to repurpose effectively as potential replacement drugs targeting COV D-19 directed us to screen natural antiviral flavonoids Quercetin, Quercetagetin, Volkensiflavone, Ternatin, Meliternatin, Formononetin, Afromosin, Chrysosplenol B, Chrysosplenol C and Axillarin as possible SARS inhibitor candidates (PDB D: 5N5O and 6LU7), as well as Remdesivir, an antiviral medication. The findings of this report will give more researchers the prospect of discovering the correct COV D-19 medicines.
MATERIALS AND METHODS
In-silico Docking
Molecular docking tests were being used for binding mode examination, an association of compounds Quercetin, Quercetagetin, Volkensiflavone, Ternatin, Meliternatin, Formononetin, Afromosin, Chrysosplenol B, Chrysosplenol C, Axillarin and antiviral Remdesivir with SARS coronavirus proteins (PDB D: 5N5O and 6LU7) by Autodock vina 1.1.2 (Trott et al., 2010). Protein Data Bank (http:/ has been employed to obtain SARS coronavirus main proteases (PDB D: 5N5O) and (PDB D: 6LU7) crystal structures. Chem3D Pro 12.0 and ChemDraw Ultra 12.0 programs were utilized to sketch structures of the inhibitors and too energy minimization. The AutoDock Software 1.5.6 application bundle was used to build Autodock Vina input data. Discovery studio 2019
program package was utilized for binding pocket prediction of main protease (PDB D: 5N5O and 6LU7) via co-crystallized ligands. The 5N5O Protein Quest Grid has been recognized as centre x,y,z: -23.002, -3.023, 4.681 with measurements x,y,z: 24 through 1.0 Å interval. 6LU7 protein quest grid was defined as centre x,y,z:-10.656, 17.223, 67.024 in dimension x,y,z: 20 in 1.0 Å interval. The meaning of completeness were fixed to 8. The other restrictions have been fixed and not specified by default for Autodock Vina. The compound which devotes the smallest inhibitory value is the main inhibitor, and the consequences were visually examined by Discovery studio 2019.
Molecular property and ADME prediction
Here in, Lipinski's 'Rule of five' being utilized for the theoretical prediction of silicon ADMEs and the toxicity of Quercetin , Quercetagetin , Rutin , Volkensiflavone , Ternatin , Meliternatin , Formononetin , Afromosin , Chrysosplenol B , Axillarin and Antiviral Remdesivir compounds (Lipinski et al ., 2001). A Swiss ADME online tool was also used to estimate the Lipinski parameters (Swiss ADME). To predict the biocompatibility and transportation of an active compound through a bloodbrain obstacle, the topological polar surface (tPSA) has been cast-off (Ertl et al ., 2000). Bioavailability is too multidimensional but mainly concerned with the absorption of the digestive system (Daina et al ., 2016) Absorption percentage was determined from the formulas: percent ABS = 109 (TPSA x 0.345). Further predictions involved CYP2D6, P-glycoprotein inhibition, phospholipidosis (PLD), CYP2D9 and water solubility.
RESULTS AND DISCUSSION
In-silico docking
Docking simulations remained carried out to progress kind of the conceivable progression of biological activity. The natural analogues Quercetin, Quercetagetin, Volkensiflavone, Ternatin, Meliternatin, Formononetin, Afromosin, Chrysosplenol B, Chrysosplenol C and Axillarin, as well as Antiviral Remdesivir, were evaluated for their inhibition capability concerning SARS coronavirus proteins 5N5O and 6LU7 through the software Autodock Vina. All these checked inhibitors demonstrate negative binding energy. The natural flavonoid derivative Volkensiflavone demonstrates astonishing inhibition capability with inhibition ability of -8.9 kcal/mol over former derivatives Quercetin (-7.3kcal/mol), Quercetagetin (-7.6 kcal/mol), Ternatin (-7.5 kcal/mol), Meliternatin (-7.7 kcal/mol), Formononetin (-7.0 kcal/mol), Afromosin (-6.7 kcal/mol), Chrysosplenol B (-7.5 kcal/mol), Chrysosplenol C (-7.8 kcal/mol), Axillarin (-7.7 kcal/mol) and antiviral drug Remdesivir (-7.4 kcal/mol) in 5N5O receptor individually. The ability of hydrogen bonding is an essential aspect of bonding equilibrium between ligand and protein, and the supporting bonding gap between atoms H-acceptor and H-donor is less than 3.5 Å (Taha et al., 2015). The related hydrogen bonding distances for the specific inhibitors in respective protein remained fewer than 3.5 Å, demonstrating the strong hydrogen link amongst ligands and receptor. Volkensiflavone demonstrates two associations between hydrogen bonding and 5N5O receptor. Asn142 and Gln189 amino acid residues were associated with bond lengths of 2.84 and 2.36 Å in contact with hydrogen. The residues of His41, Met49, Ser144, Cys145 and Met165 amino acids came in contact with hydrophobics. Fig. 2 indicates the hydrophobic and hydrogen bonding interaction of residues of amino acids with compound Volkensiflavone in 5N5O protein. The antiviral Remdesivir treatment demonstrates two associations by hydrogen bonding through the 5N5O receptor. The amino acids Cys44 and Glu166 are entangled through associations between hydrogen and the bond lengths 2.20 and 2.49 Å. The amino acid residues Thr25, Met165, Leu167, Pro168 and Gln189, were mixed within hydrophobic encounters. Fig. 3 indicates the hydrogen and hydrophobic interaction of Remdesivir antiviral medication with 5N5O protein. Table-1 displays the molecular interactions of natural analogues on target protein 5N5O. Volkensiflavone natural flavonoid analogue demonstrates impressive inhibition ability through binding energy of -9.0 kcal/mol relative to other compounds Quercetin (-7.4 kcal/mol), Quercetagetin (-7.6 kcal/mol), Ternatin (-7.1 kcal/mol), Meliternatin (-7.9 kcal/mol), Formononetin (-7.3 kcal/mol), Afromosin (-7.0 kcal/mol), Chrysosplenol B (-7.4
kcal/mol), Chrysosplenol C (-7.4 kcal/mol), Axillarin (-7.4 kcal/mol) and antiviral drug Remdesivir (-7.7 kcal/mol) in 6LU7 protein individually. Compound Volkensiflavone displays three interactions of hydrogen bonding with receptor 6LU7. His41, Met49 and Phe140 were hydrogen bonding amino acid residues with bond lengths of 2.76, 2.96 and 1.86 Å. The amino acid residues Cys145, Met165 and Glu166, have been entangled in hydrophobic encounters. Fig. 4 demonstrated the bonding of hydrogen and hydrophobic constructions of amino acid residues with Volkensiflavone compound in the 6LU7 protein. The antiviral medication Remdesivir demonstrates six associations between hydrogen and receptor 6LU7. The residues of Thr26, Cys44, Gly143, Cys145 and Glu166 amino acids were entangled in hydrogen bonding to the 2.08, 2.87, 2.16 & 2.73, 3.00 and 2.38 Å bonding ranges. The residues of the His41, Met49 and Met165 amino acids were in touch with hydrophobia. The hydrogen bonding and hydrophobic connexions with Remdesivir are seen in Fig. 5 of amino acid residues in the 6LU7 protein. The findings revealed that the compound Volkensiflavone had exceptional inhibition capacity in their respective target proteins 5N5O and 6LU7 relative to other compounds. The findings of natural antiviral analogues against SARS coronavirus (PDB D: 6LU7) have been shortened in Table - 2 .
3.2 Molecular property and ADME prediction
The progress of bioactive components as healers is driven by high oral bioavailability (Newby et al ., 2015). The key forecasters of this analysis demonstrated, for example, the low polar surface region, hydrogen bonding ability, decreased molecular versatility and intestinal absorption (Azam et al ., 2012). The natural antiviral analogues Quercetin , Quercetagetin , Ternatin , Meliternatin , Formononetin , Afromosin , Chrysosplenol B , Chrysosplenol C and Axillarin agrees Lipinski's "Rule of 5" with 0 infringement and compounds Volkensiflavone and Remdesivir Fails "Rule of 5" with two infringement HBA, MW>500, HBD and RoB ( Table - 3 ). The molecular conformational changes were defined by the number of revolving ties and the potential for the receptor binding. The compounds Quercetin , Quercetagetin ,
Volkensiflavone, Ternatin, Meliternatin, Formononetin, Afromosin, Chrysosplenol B, Chrysosplenol C and Axillarin were under ten rotatable bonds except for Remdesivir (14 rotatable bonds), which are formed without the chirality core and have poor oral bioavailability conditions. The property of tPSA (topological Polar Surface Area) revealed the passive molecular transport across membranes and blood-brain barrier penetration (Ertl et al., 2000) Checked substances except for compounds Quercetagetin, Volkensiflavone, and Remdesivir with tPSA values < 140Å2 fulfil the requirements for subsequent oral administration for gastrointestinal absorption. n comparison, all of the compounds studied except for Quercetin (tPSA = 131.36Å2), Quercetagetin (tPSA = 151.59 Å2), Volkensiflavone (tPSA = 177.89 Å2), Ternatin (tPSA = 107.59 Å2), Chrysosplenol B (tPSA = 107.59 Å2), Chrysosplenol C (tPSA = 118.59 Å2), Axillarin (tPSA = 129.59 Å2), and Remdesivir (tPSA = 213.36 Å2) have lower blood-brain barrier (tPSA > 90 Å2), which reveals detrimental possessions of Central nervous system (CNS).
O
Afromosin
Epstein-Barr virus
OCH
Axillarin Rhinovirus
Chrysosplenol B Rhinovirus
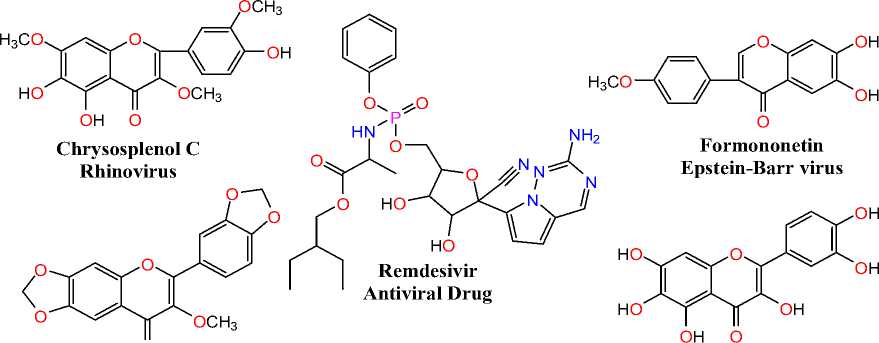
O
Meliternatin HSV-1, HSV-2, VSV type 2, Poliovirus type 2 and adenovirus type 2
Quercetagetin HIV and Rauscher murine leukemia (RLV)
OH O
Quercetin Potato virus X (PVX)
Ternatin
HSV-1, HSV-2, VSV type 2, Poliovirus type 2 and adenovirus type 2
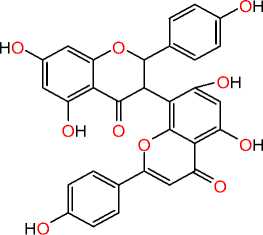
Volkensiflavone influenza B virus
Figure 1. Natural antiviral derivatives
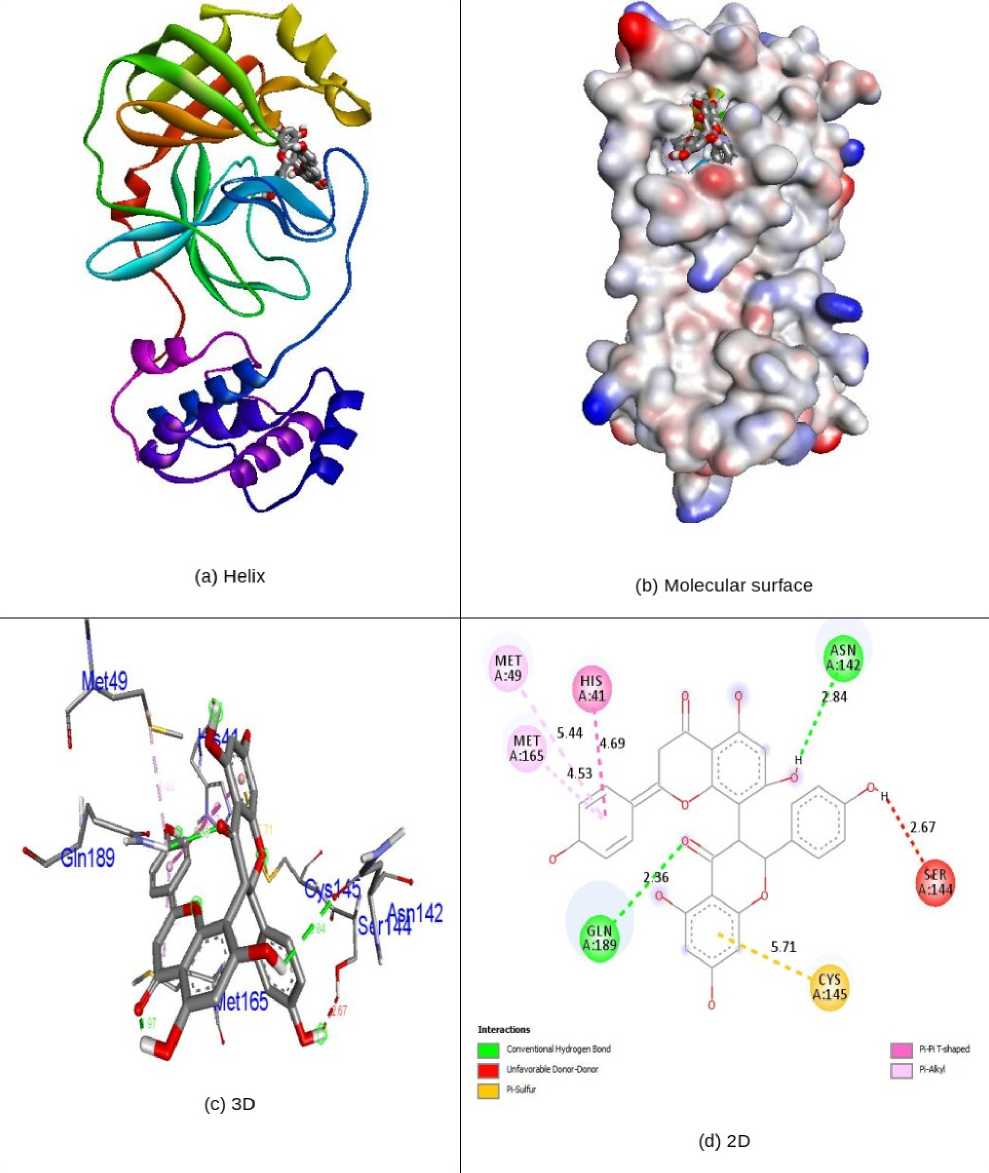
Figure 2. nteractions of Volkensiflavone inside the binding pocket of 5N5O protein
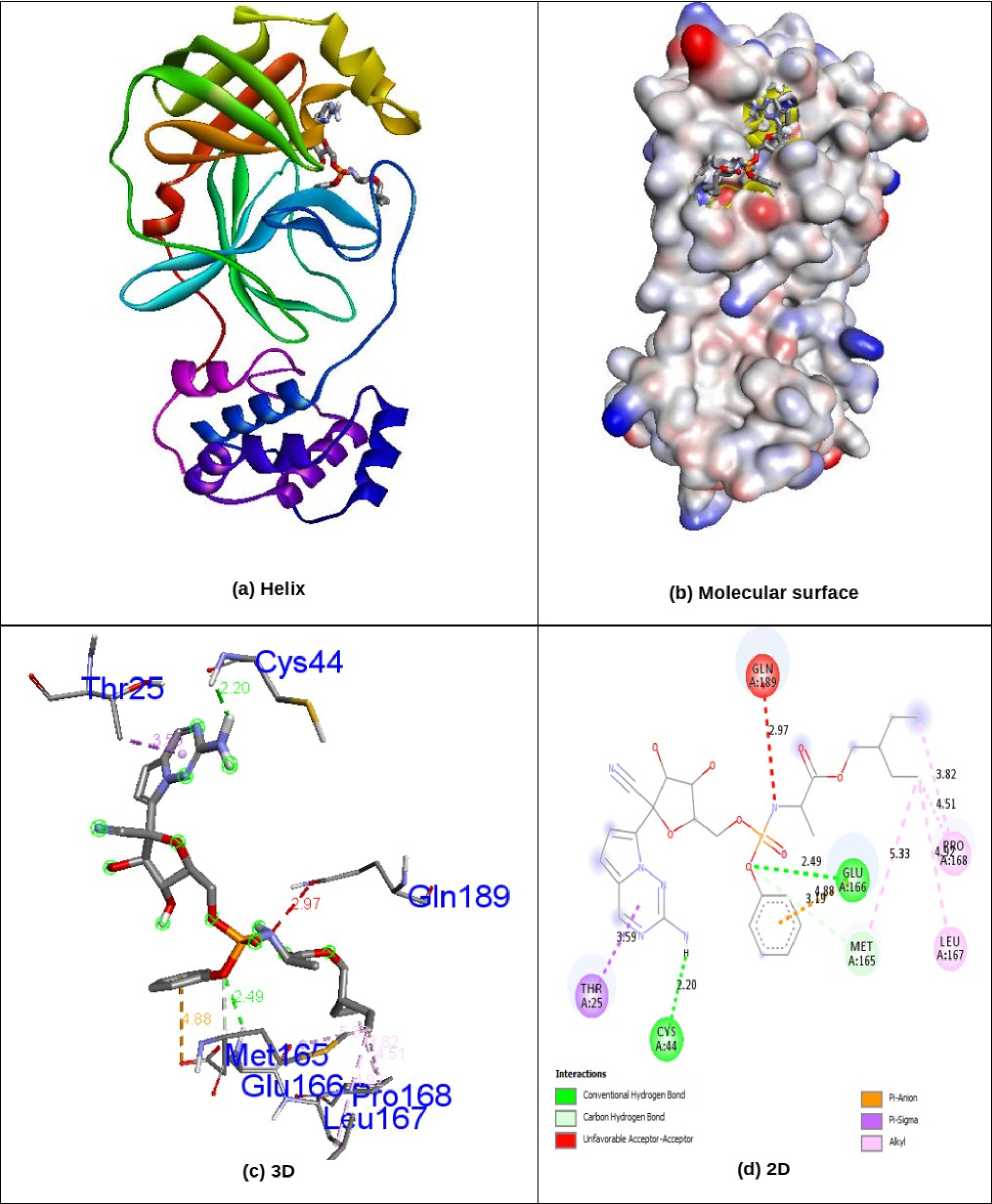
Figure 3. nteractions of antiviral drug Remdesivir inside the binding pocket of 5N5O protein
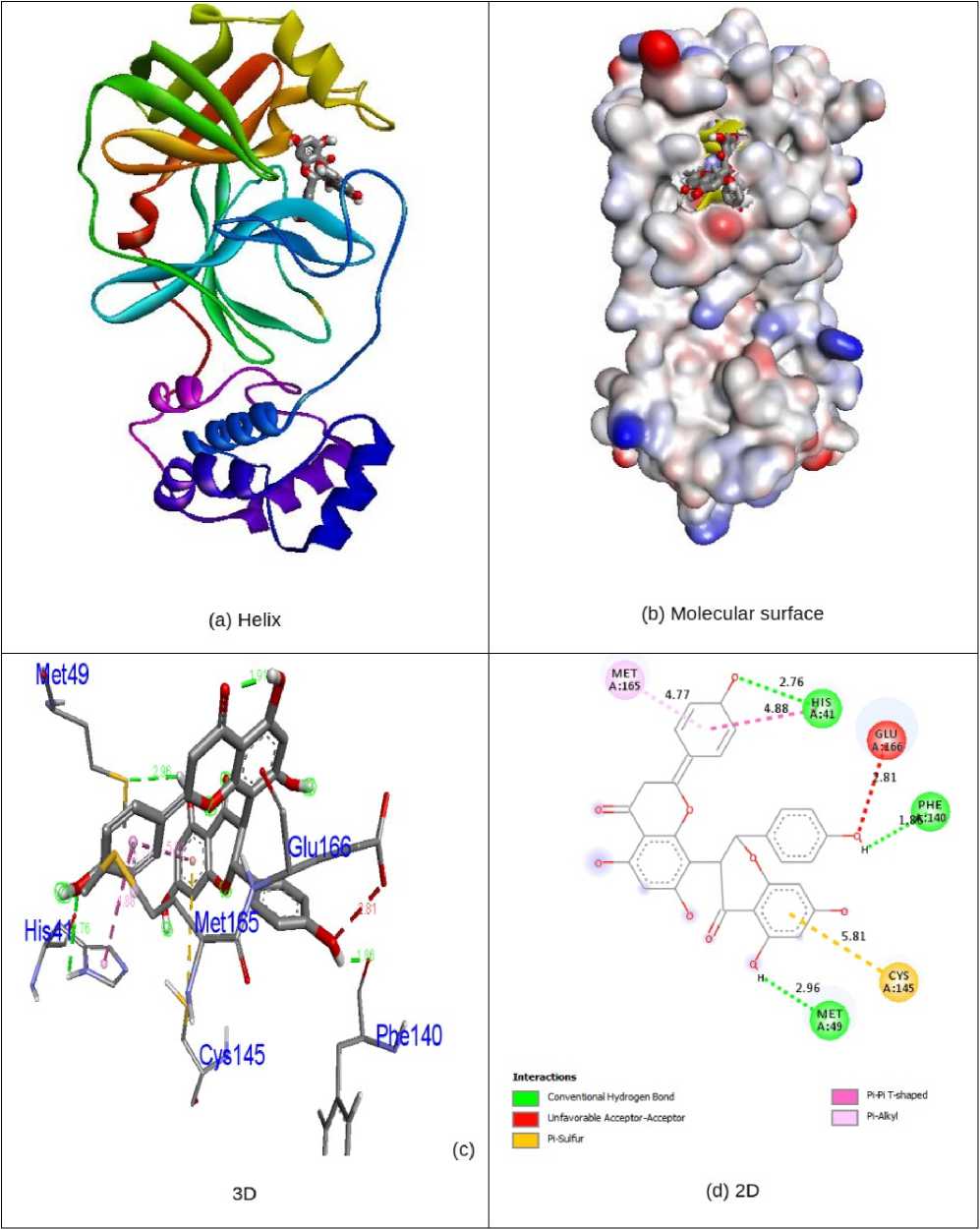
Figure 4. nteractionsof Volkensi flavones inside the binding site of 6LU7 protein
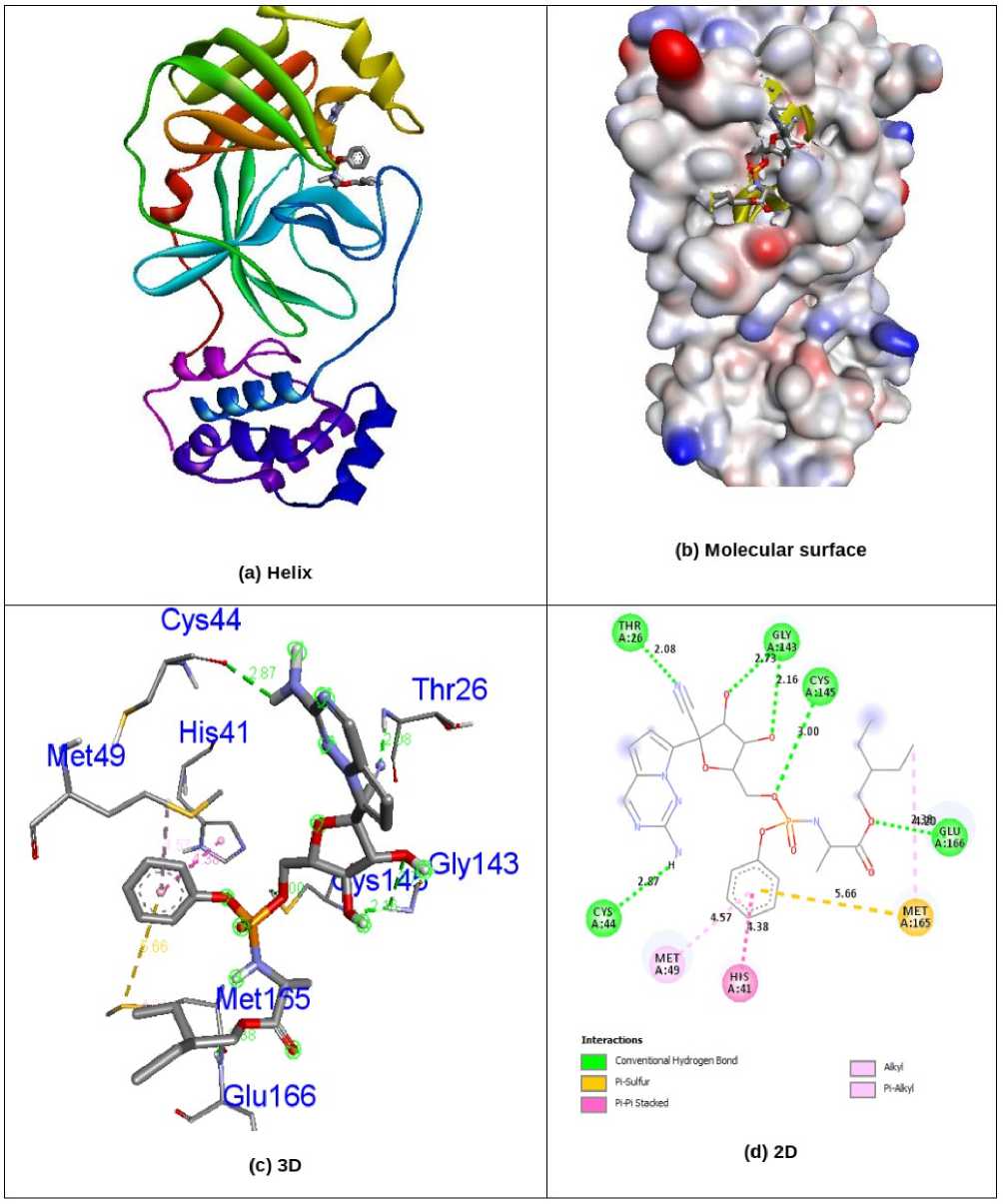
Figure 5. nteractionsof antiviral drug Remdesivir inside the binding pocket of 6LU7 protein
Table 1 : nteractionsof compounds (1a-1k) against SARS coronavirus protease (PDB D : 5N5O)
|
nhibitors |
Binding energy (kcal/mol) |
Number of H-bonds |
Hydrogen bonding residues |
|
Quercetin (1a) |
-7.3 |
3 |
Leu141, Cys145, Glu166 |
|
Quercetagetin (1b) |
-7.6 |
3 |
Gly143, Ser144 |
|
Volkensiflavone (1c) |
-8.9 |
2 |
Asn142, Gln189 |
|
Ternatin (1d) |
-7.5 |
2 |
Gly143, Cys145 |
|
Meliternatin (1e) |
-7.7 |
0 |
- |
|
Formononetin (1f) |
-7.0 |
2 |
Asn142, Arg188 |
|
Afromosin (1g) |
-6.7 |
2 |
Asn142, Gln189 |
|
Chrysosplenol B (1h) |
-7.5 |
1 |
Gly143 |
|
Chrysosplenol C (1i) |
-7.8 |
3 |
Gly143, Cys145 |
|
Axillarin (1j) |
-7.7 |
5 |
Leu141,Gly143, Ser144, Cys145 |
|
Remdesivir (1k) |
-7.4 |
2 |
Cys44, Glu166 |
Table - 2. nteractionsof compounds (1a-1k) against SARS coronavirus protease (PDB D: 6LU7)
|
nhibitors |
Binding energy (kcal/mol) |
Number of H-bonds |
Hydrogen bonding residues |
|
Quercetin (1a) |
-7.4 |
0 |
- |
|
Quercetagetin (1b) |
-7.6 |
3 |
Met49, Leu141, Asn142 |
|
Volkensiflavone (1c) |
-9.0 |
3 |
His41, Met49, Phe140 |
|
Ternatin (1d) |
-7.1 |
3 |
Ser46, Leu141, Cys145 |
|
Meliternatin (1e) |
-7.9 |
1 |
Gly143 |
|
Formononetin (1f) |
-7.3 |
1 |
Asn142 |
|
Afromosin (1g) |
-7.0 |
1 |
Phe140 |
|
Chrysosplenol B (1h) |
-7.4 |
4 |
Gly143, Ser144, Cys145 |
|
Chrysosplenol C (1i) |
-7.4 |
4 |
Ser46, Leu141, Cys145 |
|
Axillarin (1j) |
-7.4 |
2 |
His41, Thr190 |
|
Remdesivir (1k) |
-7.7 |
6 |
Thr26, Cys44, Gly143, Cys145, Glu166 |
Abbreviations: a Topological polar surface area; b Absorption; c Molecular weight; d Number of rotatable bonds; e Number of hydrogen bond donors; f Number of hydrogen bonds acceptors; g Molar refractivity; h Logarithm of compound partition coefficient between n-octanol and water; i Logarithm of water solubility.
Table 3 Molecular property prediction and Virtual ADME (absorption, distribution, metabolism, excretion) of the potent compounds (1a-1k).
|
Comp. |
tPSA a |
%Abs b |
MWc |
RoB d |
HBD e |
HBA f |
MR g |
logP h (MlogP) |
LogS i |
CYP2D6 nhibitor |
|
Rule |
≤140 ´Å2 |
>50 |
≤500 |
≤10 |
≤5 |
≤10 |
40–130 |
<5 |
>-4 |
- |
|
Quercetin (1a) |
131.36 |
63.68 |
302.24 |
1 |
5 |
7 |
78.03 |
1.63 (-0.56) |
-3.16 |
Yes |
|
Quercetagetin (1b) |
151.59 |
56.70 |
318.24 |
1 |
6 |
8 |
80.06 |
1.73 (-1.08) |
-3.01 |
No |
|
Volkensiflavon e (1c) |
177.89 |
47.62 |
540.47 |
3 |
6 |
10 |
143.60 |
2.19 (0.41) |
-6.55 |
No |
|
Ternatin (1d) |
107.59 |
71.88 |
374.34 |
5 |
2 |
8 |
97.93 |
3.41 (-0.12) |
-4.24 |
No |
|
Meliternatin (1e) |
76.36 |
82.65 |
340.28 |
2 |
0 |
7 |
86.54 |
3.29 (0.98) |
-4.18 |
Yes |
|
Formononetin (1f) |
79.90 |
81.43 |
284.26 |
2 |
2 |
5 |
78.46 |
2.39 (0.77) |
-3.57 |
Yes |
|
Afromosin (1g) |
68.90 |
85.22 |
298.29 |
3 |
1 |
5 |
82.93 |
2.97 (1.01) |
-3.77 |
Yes |
|
Chrysosplenol B (1h) |
107.59 |
71.88 |
374.34 |
5 |
2 |
8 |
97.93 |
3.35 (-0.12) |
-4.24 |
No |
|
Chrysosplenol C (1i) |
118.59 |
68.08 |
360.31 |
4 |
3 |
8 |
93.46 |
2.90 (-0.35) |
-4.02 |
Yes |
|
Axillarin (1j) |
129.59 |
64.29 |
346.29 |
3 |
4 |
8 |
89.00 |
2.37 (-0.59) |
-3.81 |
Yes |
|
Remdesivir (1k) |
213.36 |
35.39 |
602.58 |
14 |
4 |
12 |
150.43 |
2.74 (0.18) |
-4.12 |
No |
Abbreviations: a Topological polar surface area; b Absorption; c Molecular weight; d Number of rotatable bonds; e Number of hydrogen bond donors; f Number of hydrogen bonds acceptors; g Molar refractivity; h Logarithm of compound partition coefficient between n-octanol and water; i Logarithm of water solubility.
The tested compounds demonstrated absorption percentage (percentage Abs = > 50), suggesting strong bioavailability except Volkensiflavone (% Abs = 47.62) and Remdesivir (% Abs = 35.39). Bioavailability by oral route was appropriate (> 50 percent). The compounds Quercetin, Quercetagetin, Formononetin, Afromosin and Axillarin were very water-soluble (-logS > -4) excluding compounds Volkensiflavone (-logS-6.55), Ternatin (-logS -4.24) Meliternatin (-logS -4.18), Chrysosplenol B (-logS -4.24), Chrysosplenol C (logS -4.02), and Remdesivir (-logS -4.12) have moderate water solubility. The side effects of liver impairment doesn’t suspected in the case of compounds Quercetagetin, Volkensiflavone, Ternatin, Chrysosplenol B and Remdesivir since they were predicted to be CYP2D6 non-inhibitors. A part of the Pgp (P-glycoprotein) family transporter ATP-binding cassette (ABC) comprises the intestinal absorption, brain penetration and pharmaceutical metabolism; the aforementioned caginess may significantly modify the bioavailability and defence of the drug (Fromm et al., 2000). Phospholipidosis convinced medication is a condition known for the further growth of phospholipids in flesh and medication-related poisonousness (Nonoyama et al., 2008). The findings indicate that the studied compounds Quercetin, Quercetagetin, Volkensiflavone, Ternatin, Meliternatin, Formononetin, Afromosin, Chrysosplenol B, Chrysosplenol C and Axillarin were not a part of P-gp substrate and phospholipidosis was not promoted. The checks for P-gp-phospholipidosis was anticipated only in Remdesivir. The overall findings of ADME and toxicity indicate respectable pharmacological profile and rapid gastrointestinal ingestion through blood-brain blood barrier penetration in the isolated compounds were Meliternatin and Afromosin. All tested compounds assessed were recognised by way of drug-like and passed "Rule of 5" of Lipinski except compounds Volkensiflavone and Remdesivir. The restrictions predicted are all within the context of accepted principles.
CONCLUSION
COV D-19 has arisen in the humanoid community, in China, and is a possible risk to healthiness internationally. However, there is not at all precisely approved medication to delight the situation and the already accessible COV D-19 medicines coping with essential protease. The goal of the current study was to inspect some natural analogues extracted from medicinal plants that might remain tossed off to combat COV D-19. The utmost frequently proposed compounds in healing plants that may function as important COV D-19 inhibitors of key protease (PDB D: 5N5O, 6LU7) were Quercetin , Quercetagetin ,
Volkensiflavone , Ternatin , Meliternatin ,
Formononetin , Afromosin , Chrysosplenol B , Chrysosplenol C and Axillarin with negative binding energies. Studies in molecular docking have revealed that natural flavonoid derivative Volkensiflavone displayed exceptional inhibition with a binding energy value of -8.9,-9.0kcal/mol of 5N5O and 6LU7 enzyme, relative to the other compounds and Remdesivir antiviral medication (Binding energy -7.4 and -7.7 Kcal/mol). However, the advance study is necessary to inspect the possible application of these compounds in medicinal plants.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.


