Поиск геномных вариантов, ассоциированных с живой массой у овец, на основе анализа высокоплотных SNP генотипов
Автор: Денискова Т.Е., Петров С.Н., Сермягин А.А., Доцев А.В., Форнара М.С., Reyer H., Wimmers K., Багиров В.А., Brem G., Зиновьева Н.А.
Журнал: Сельскохозяйственная биология @agrobiology
Рубрика: Молекулярная структура генома
Статья в выпуске: 2 т.56, 2021 года.
Бесплатный доступ
Живая масса - один из важнейших экономически полезных признаков, характеризующийся сложным наследованием, поэтому поиск генетических механизмов, влияющих на ее формирование, вызывает повышенный научный интерес. В настоящей работе впервые представлены результаты анализа полногеномных ассоциаций в ресурсной популяции овец ( Ovis aries ) возвратных кроссов (романовская × катадин) × романовская, живая масса которых фиксировалась в возрастной динамике, а SNP-профили были получены с помощью высокоплотного ДНК-чипа. В результате были идентифицированы 38 SNP, достоверно ассоциированных с живой массой (p
Домашние овцы, ресурсная популяция, snp-маркеры, днк-чипы, gwas, живая масса, гены-кандидаты
Короткий адрес: https://sciup.org/142229475
IDR: 142229475 | УДК: 636.32: | DOI: 10.15389/agrobiology.2021.2.279rus
Текст научной статьи Поиск геномных вариантов, ассоциированных с живой массой у овец, на основе анализа высокоплотных SNP генотипов
Постнатальный рост организма животного — результат сложных взаимодействий между генетическими факторами, потреблением питательных веществ и функциями эндокринной системы (1). Углубление знаний о закономерностях роста и развития сельскохозяйственных животных имеет практическое значение для повышения их продуктивности (2, 3). Живая масса — один из важнейших экономически полезных признаков, характеризующийся сложным наследованием, поэтому поиск генетических механизмов, влияющих на ее формирование, вызывает повышенный научный интерес (4, 5).
Основываясь на результатах полногеномных ассоциативных исследований (genome-wide association study, GWAS) на мясных овцах с применением ДНК-чипа средней плотности Illumina OvineSNP50 («Illumina, Inc.», США), L. Zhang c соавт. (6) идентифицировали гены GRM1 , MBD5 , UBR2 , RPL 7 и SMC2 в качестве потенциальных кандидатов интенсивности роста ягнят в послеотъемный период. Поиск полногеномных ассоциаций на 1743 овцах выявил достоверно ассоциированный SNP (single nucleotide polymorphism) OAR6_41936490 на 6-й хромосоме OAR6 (7). В области, прилегающей к этому SNP, были локализованы три значимых гена-кандидата ( LAP3 , NCAPG и LCORL ), ассоциированные с признаками роста, строением каркаса, размером тела и живой массой у овец.
О. Matika с соавт. (8) провели полногеномный анализ ассоциаций на 600 ягнятах породы шотландская черноголовая. Фенотипические показатели, включая плотность костей, количество мышечной и жировой ткани, были получены с помощью компьютерной томографии, а генотипирование проводилось с использованием ДНК-чипа средней плотности Illumina OvineSNP50. На OAR6 был выявлен геномный регион, достоверно ассоциированный с массой костей (р < 5,55*10 - 8) и влияющий на плотность мышечных волокон и содержание жира. Кроме того, были идентифицированы QTL на OAR1, OAR3 и OAR24, отвечающие за развитие мышечных, жировых и костных компонентов туловища у ягнят. M. Ghasemi с соавт. (9) проводили поиск геномных ассоциаций с живой массой при рождении у 130 овец породы лори-бахтиари (Lori-Bakhtiari) с использованием 41323 SNP и идентифицировали потенциальные гены-кандидаты RAB6B , Tf serotransferrin и GIGYF2 на OAR1. Z. Lu с соавт. (10) на китайских тонкорунных овцах применили технологию ре-секвенирования для поиска ассоциаций с живой массой при рождении, при отъеме в возрасте 3,5 мес, в возрасте 12 и 30 мес.
В регионах, прилегающих к 113 SNP, которые достигают порогового уровня значимости полногеномных ассоциаций (р < 0,05), были идентифицированы и аннотированы гены AADACL3 , VGF , NPC1 и SERPINA12 , влияющие на развитие скелетных мышц и метаболизм липидов. Четыре гена, в том числе вовлеченный в регуляцию роста скелетных мышц ген MTPN , были предложены в качестве потенциальных кандидатов, ассоциированных с живой массой овец породы балучи (Baluchi) в возрасте 8 мес (11). Y. Cao с соавт. (12) фиксировали живую массу при рождении, при отбивке от маток, в полугодовом и годовом возрасте в двух поколениях двух популяций овец породы Ху (Hu). GWAS, проведенный на основе SNP-профилей средней плотности, и верификация выявленных полиморфизмов показали, что два SNP (OARX_76354330.1 и s64890.1) достоверно ассоциированы с живой массой (р < 0,05). С использованием модели множественных признаков были идентифицированы восемь новых локусов, расположенных вблизи генов FAM3C и WNT16 , которые ассоциированы с мясной продуктивностью овец (13).
Следует отметить, что исследования по картированию QTL могут приводить и к выявлению ложноположительных ассоциаций. Считается, что уменьшение частоты ложноположительных результатов (false discovery rate, FDR) и повышение точности картирования QTL может быть достигнуто при использовании специально созданных ресурсных популяций сельскохозяйственных животных (возвратные кроссы и F 2 ) (14), полученных от скрещивания родительских линий (или пород), которые имеют высокодивергентные фенотипы по интенсивности роста и живой массе. Кроме того, в ресурсной популяции возможно создать надежную базу фенотипов за счет понижения значимости человеческого фактора (в случае, если промеры или иные фенотипические показатели фиксирует один и тот же исполнитель).
В настоящей работе впервые представлены результаты анализа полногеномных ассоциаций в ресурсной популяции овец ( Ovis aries ) возвратных кроссов (романовская х катадин) х романовская, живая масса которых фиксировалась в возрастной динамике, а SNP-профили были получены с помощью высокоплотного ДНК-чипа. В результате идентифицированы 38 SNP, достоверно ассоциированных с живой массой (р < 0,00001), и функциональные гены-кандидаты, влияющие на рост скелетных мышц, формирование костного каркаса, липидный и углеводный метаболизм. Кроме того, показаны возрастные изменения в составе достоверно значимых SNP.
Нашей целью был поиск геномных вариантов, оказывающих влияние на живую массу у овец возвратных кроссов (романовская х катадин) х романовская из ресурсной популяции в разные возрастные периоды.
Методика . Исследования проводили на 95 овцах возвратных кроссов из ресурсной (кроссбредной) популяции в 2018-2021 годах. Ресурсная популяция овец входила в состав биоколлекции «Банк генетического материала домашних животных и птицы» (зарегистрирован Минобрнауки РФ, № 498808), созданной и поддерживаемой в ФГБНУ ФИЦ животноводства — ВИЖ им. академика Л.К. Эрнста.
Создание ресурсной популяции овец включало несколько этапов (15). На первом ярок романовской породы скрещивали с двумя баранами американской мясной породы катадин для получения F 1 кроссов. На втором этапе для получения возвратных кроссов (романовская х катадин) х романовская по одному баранчику F 1 от каждого барана-родоначальника скрещивали с романовскими матками, в результате чего было получено соответственно 36 и 32 потомков от каждого барана. F 1 ярочек от каждого барана-родоначальника скрещивали с двумя баранами романовской породы, в результате чего получили 16 и 10 потомков. То есть все возвратные кроссы по своему происхождению имели 25 % крови породы катадин и 75 % крови романовской породы.
Для экстракции геномной ДНК животных возвратных кроссов использовали ушные выщипы. ДНК выделяли с помощью наборов ДНК-Экстран-2 (ООО «Синтол», Россия). Препараты ДНК проходили контроль качества по концентрации (от 15 нг/мкл и выше, флуориметр Qubit 4.0, «Invitrogen/Life Technologies», США) и чистоте по соотношению поглощения OD 260 /OD 280 (от 1,8 и выше, спектрофотометр NanоDrop8000, «Thermo Fisher Scientific», США).
Исследуемых животных генотипировали с помощью ДНК-чипа высокой плотности Ovine Infinium® HD SNP BeadChip («Illumina, Inc.», США), содержащего ~ 600 тыс. SNP-маркеров. Фильтрацию SNP-маркеров осуществляли в программе PLINK v. 1.90
-
(16) . SNP, имеющие частоту минорного аллеля (MAF) ниже 3 % (maf 0.03), отклоняющиеся от равновесия по Харди-Вайнбергу при p < 10 - 6 (hwe 1e-6), находящиеся в неравновесии по сцеплению (indep-pairwise 50 5 0.5) и локализованные на половых хромосомах, исключались из анализа.
Живую массу измеряли с помощью платформенных весов МП 300 ВЕДА Ф-1 (50/100; 1400x700) Живой вес 12 ПМ (Московский весовой завод «МИДЛ», Россия) в возрасте 6 (ЖМ6), 42 (ЖМ42), 90 (ЖМ90), 180 (ЖМ180) и 270 сут (ЖМ270). Результаты измерений заносили в электронную базу данных в программе Microsoft Excel 2017. Рассчитывали средние арифметические значения ( M) , стандартные ошибки средних (±SEM), средние квадратические отклонения (± а ) и коэффициенты вариации ( Cv , %).
Для выявления геномных ассоциаций с живой массой применяли регрессионный анализ, реализованный в PLINK 1.90 (--assoc --adjust --qt-means). Для подтверждения достоверного влияния SNP и определения значимых регионов в геноме исследуемых овец был выполнен тест для проверки нулевых гипотез по Бонферрони при пороговом значении p < 1,09x10-7, 0’05/459868. Данные визуализировали в пакете R qqman (17) в программной среде R (18).
Поиск генов-кандидатов, локализованных в области идентифицированных SNP, осуществлялся с использованием инструмента VEP (Ensembl Variant Effect Predictor) (19) Ensembl genome browser 103 , дата обращения 12.01.2021) по версии сборки генома овцы Ovis_aries.Oar_v3.1. Онтология генов была проанализирована с помощью инструмента анализа микрочипов DAVID Functional Annotation Bioinformatics (20). Поиск вероятных совпадений с известными QTL проводился с использованием базы данных Sheep Quantitative Trait Locus Database (Sheep QTLdb) , дата обращения 12.01.2021) (21).
Результаты. Средние показатели живой массы в изучаемой выборке овец представлены в таблице 1. Для этого признака была выявлена довольно высокая вариация во все возрастные периоды ( Cv = 21,69-27,86 %).
1. Живая масса (кг) и коэффициент ее изменчивости в разные возрастные периоды у овец ( Ovis aries ) возвратных кроссов (катадин х романовская) х романовская из ресурсной популяции ( n = 95, ФГБНУ ФНЦ животноводства — ВИЖ им. академика Л.К. Эрнста, Московская обл., 2019-2021 годы)
|
Возраст |
M ±SEM |
Min-max |
I C v , % |
|
6 сут |
3,28±0,07 |
1,60-4,83 |
21,69 |
|
42 сут |
8,03±0,21 |
3,50-13,00 |
25,98 |
|
90 сут |
13,74±0,39 |
3,50-22,10 |
27,86 |
|
180 сут |
20,19±0,51 |
9,93-36,80 |
24,53 |
|
270 сут |
22,51±0,50 |
12,37-37,80 |
21,78 |
В результате GWAS-анализа было установлено, что в разные возрастные периоды набор SNP, ассоциированный с интегральным показателем скорости роста — живой массой, был неодинаковым (рис., табл. 2). Так, из 38 идентифицированных SNP 18 SNP, расположенных на OAR2, OAR4, OAR9 и OAR15, были достоверно ассоциированы с ЖМ6 (p < 0,00001), 3 SNP на OAR6 и OAR11 — с ЖМ42 (p < 0,00001), 2 SNP на OAR10 и OAR19 — с ЖМ90 (p < 0,00001), 6 SNP на OAR1 и OAR13 — с ЖМ180 (p < 0,00001), 6 SNP на OAR1 — с ЖМ270 (p < 0,00001). Кроме того, для 5 SNP, локализованных на OAR1 и OAR3, мы выявили достоверные ассоциации как с ЖМ180, так и с ЖМ270. На OAR1, OAR2, OAR4 и OAR5
были обнаружены блоки из 3-5 SNP. Нами идентифицированы шесть SNP - oar3_OAR4_87887519, oar3_OAR4_87889243, oar3_OAR9_89145258, oar3_OAR1_192662599, oar3_OAR13_31446454 и OAR1_208070059.1, у которых достоверность ассоциации превышала пороговое значение для GWAS (р < 1,09х10-7).
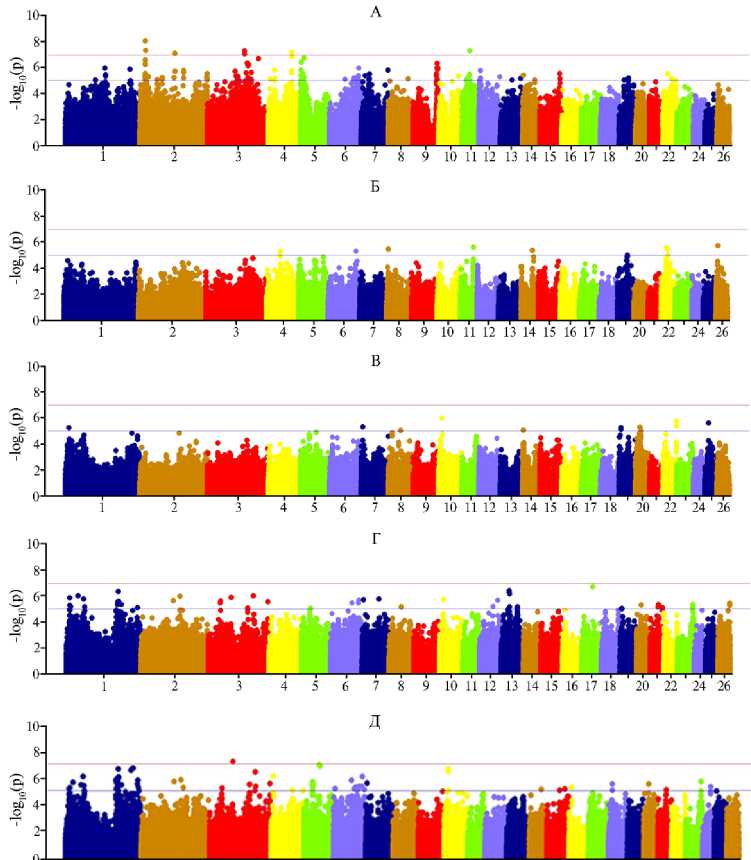
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 20 22 24 26
Хромосома
Результаты GWAS анализа для показателя живой массы у овец ( Ovis aries ) возвратных кроссов (катадин ½ романовская) ½ романовская из ресурсной популяции в разные возрастные периоды: А — 6 сут; Б — 42 сут; В — 90 сут; Г — 180 сут; Д — 270 сут. Верхняя горизонтальная линия — порог достоверности для полногеномных ассоциаций -log10(p) = 1,09х10-7, нижняя горизонтальная линия — порог достоверности для суггестивных ассоциаций -log10(p) = 1,02х10-5 ( n = 95, ФГБНУ ФНЦ животноводства — ВИЖ им. академика Л.К. Эрнста, Московская обл., 2019-2021 годы).
В возрасте 6 сут были выявлены достоверные ассоциации живой массы с SNP, находящимися внутри гена MBD5 и в непосредственной близости от генов ORC4 и ACVR2A на OAR2, которые были предложены в качестве функциональных кандидатов для постнатального роста у овец (6). Ген MBD5 участвует в регуляции многих эндокринных функций, включая гомеостаз глюкозы (22). Исследование, проведенное на мышах с нокаутом MBD5, выявило серьезную задержку роста, уменьшенный размер туловища, гипогликемию и снижение количества жира (22). Делеции в человеческом ортологе вызывают задержку развития (23) и различные пороки формирования скелета (24). То есть исследования свидетельствуют о значительной роли гена MBD5 в регуляции раннего постнатального роста у млекопитающих. Гены ORC4 и ACVR2A также участвуют в процессах роста. Мутации в гене ORC4 вызывают задержку роста и первичную остео-диспластическую карликовость (25, 26). Ген ACVR2A регулирует развитие костного аппарата (27).
-
2. Достоверно значимые (р < 0,00001) SNP, ассоциированные с живой массой у овец ( Ovis aries ) возвратных кроссов (катадин х романовская) х романовская из ресурсной популяции ( n = 95, ФГБНУ ФНЦ животноводства — ВИЖ им. академика Л.К. Эрнста, Московская обл., 2019-2021 годы)
|
Показатель |
OAR |
Число SNP |
SNP |
P |
Позиция |
Ген |
|
ЖМ6 |
2-я |
1 |
oar3 OAR2 19100665 |
5,41x10 - 6 |
19100665 |
SMC2 |
|
ЖМ6 |
2-я |
1 |
oar3 OAR2 30932797 |
5,52x10 - 6 |
30932797 |
PTCH1 |
|
ЖМ6 |
2-я |
1 |
oar3 OAR2 63406327 |
8,59x10 - 6 |
63406327 |
ALDH1A1 |
|
ЖМ6 |
2-я |
1 |
oar3 OAR2 126584456 |
6,05x10 - 6 |
126584456 |
PPP1R1C*, PDE1A |
|
ЖМ6 |
2-я |
5 |
oar3_OAR2_160277567 oar3_OAR2_160295110 oar3_OAR2_160299881 oar3_OAR2_160313744 oar3 OAR2 160316296 |
6,81x10 - 6 1,75x10 - 6 2,50x10 - 6 1,75x10 - 6 1,75x10 - 6 |
160277567 160295110 160299881 160313744 160316296 |
MBD5*, ORC4, ACVR2A |
|
ЖМ6 |
4-я |
1 |
oar3_OAR4_7542218 |
7,57x10 - 6 |
7542218 |
ABCA13 |
|
ЖМ6 |
4-я |
2 |
OAR4_24289280.1 oar3_OAR4_23286923 |
1,63x10 - 6 7,32x10 - 6 |
23180261 23286923 |
DGKB* |
|
ЖМ6 |
4-я |
3 |
oar3_OAR4_87887519 oar3_OAR4_87889243 oar3_OAR4_87932432 |
7,13x10 - 8 1,51x10 - 7 6,71x10 - 5 |
87887519 87889243 87932432 |
ASB15* |
|
ЖМ6 |
9-я |
1 |
S19680.1 |
4,46x10 - 6 |
86619418 |
RIPK2, MMP16 |
|
ЖМ6 |
9-я |
1 |
oar3 OAR9 89145258 |
4,95x10 - 7 |
89145258 |
ATP6V0D2* |
|
ЖМ6 |
15-я |
1 |
oar3 OAR15 74862644 |
7,80x10 - 6 |
74862644 |
ARHGAP1, F2 |
|
ЖМ42 |
6-я |
1 |
oar3_OAR6_103115026 |
4,87x10 - 6 |
103115026 |
EVC*, EVC2 |
|
ЖМ42 |
11-я |
2 |
S58053.1 oar3 OAR11 47604879 |
2,49x10 - 6 2,49x10 - 6 |
47604624 47604879 |
SCN4A*, ICAM2 |
|
ЖМ90 |
10-я |
1 |
oar3 OAR10 12704497 |
1,08x10 - 6 |
12704497 |
DGKH*, AKAP11 |
|
ЖМ90 |
19-я |
1 |
oar3 OAR19 7100087 |
5,95x10 - 6 |
7100087 |
CCR4 |
|
ЖМ180 |
1-я |
1 |
oar3 OAR1 43484960 |
1,03x10 - 6 |
43484960 |
WLS |
|
ЖМ180 |
1-я |
2 |
oar3_OAR1_192662599 OAR1 208070059.1 |
4,79x10 - 7 4,79x10 - 7 |
192662599 192689940 |
MB21D2 |
|
ЖМ180, ЖМ270 |
1-я |
4 |
oar3_OAR1_204348376 OAR1_220691763.1 oar3_OAR1_204368368 |
4,68x10 - 6/3,85x10 - 6 4,68x10 - 6/3,85x10 - 6 4,68x10 - 6/3,85x10 - 6 |
204348376 204351915 204368368 |
FXR1* |
|
ЖМ180, ЖМ270 |
1-я |
1 |
oar3_OAR1_14039930 |
6,91x10 - 6/5,38x10 - 6 |
14039930 |
HEYL |
|
ЖМ180, ЖМ270 |
3-я |
1 |
oar3_OAR3_220260675 |
2,86x10 - 6/3,12x10 - 6 |
220260675 |
WNT7B |
|
ЖМ180 |
13-я |
1 |
oar3 OAR13 31446454 |
6,84x10 - 7 |
31446454 |
MRC1* |
|
ЖМ180 |
13-я |
1 |
oar3 OAR13 61039017 |
7,75x10 - 6 |
61039017 |
PLAGL2 |
|
ЖМ180 |
13-я |
1 |
oar3 OAR13 61305745 |
7,17x10 - 6 |
61305745 |
DNMT3B |
|
ЖМ270 |
1-я |
5 |
oar3_OAR1_185310940 oar3 OAR1 185317664 |
7,76x10 - 6 2,52x10 - 6 |
185310940 185317664 |
PARP14* |
|
ЖМ270 |
1-я |
1 |
oar3 OAR1 185977490 |
6,28x10 - 6 |
185977490 |
ADCY5* |
|
ЖМ270 |
1-я |
1 |
oar3 OAR1 186482253 |
2,52x10 - 6 |
186482253 |
MYLK* |
Примечание. Звездочкой выделены гены, внутри которых локализованы идентифицированные SNP; расстояние от SNP, расположенных в межгенном пространстве, до ближайших генов составляло ±400 Кб. ЖМ6, ЖМ42, ЖМ90, ЖМ180 и ЖМ270 — соответственно живая масса в возрасте 6, 42, 90, 180 и 270 сут. OAR — хромосома. Через косую указаны значения для 180 сут и 270 сут.
Мы идентифицировали и другие известные функциональные гены-кандидаты овец. Гены SMC2 на OAR2 и RIPK2 на OAR9 были предложены в качестве потенциальных кандидатов, ассоциированных с живой массой и мясными качествами (6). Ген PARP14 влияет на состав жировой ткани (28).
Ген CCR4 входит в состав сложного комплекса, регулирующего энергетический обмен и метаболизм жирных кислот в скелетных мышцах у овец мясного типа (29).
Кроме того, мы идентифицировали гены-кандидаты признаков роста и развития, чьи функции хорошо описаны у крупного рогатого скота, в том числе гены, отвечающие за размеры и рост туловища — PTCH1 (30), DGKH (31), ассоциированные с формированием мышечной ткани — PPP1R1C , PDE1A (32), PLAGL2 (33), DNMT3B (34), участвующие в регуляции секреции инсулина — DGKB (35), липидного метаболизма — ALDH1A1 (36), MRC1 (37). Мутации в гене ABCA13 вызывают серьезные нарушения в остеогенезе у крупного рогатого скота, что, вероятно, подтверждает его значимость для формирования костного аппарата (38). Ген MB21D2 влияет на массу внутренних органов (в том числе почек) у крупного рогатого скота симментальской породы мясного типа (39).
В дополнении были найдены гены, отвечающие за признаки, прямо или косвенно связанные с показателем живой массы у млекопитающих. Так, гены MMP16 (40) и EVC (41) регулируют хондрогенез, а гены AKAP11 , ATP6V0D2 и WNT7B ассоциированы с остеогенезом у молодых активно растущих млекопитающих (42-44). Гены HACD2 , ADCY5 и WLS вовлечены в регуляцию липидного обмена (45-47). Гены ASB15 , FXR1 , HEYL и MYLK регулируют рост скелетных мышц (48-51).
В результате проведенного GWAS-анализа мы установили, что в разные возрастные периоды различные SNP были ассоциированы с показателем живой массы в исследованной популяции овец. В своей работе Y. Cao с соавт. (12) также обнаружили, что экспрессия гена CAPN6 значительно отличается в двуглавой мышце бедра и в длиннейшей мышце спины у овец в возрасте 60 сут и 6 мес. Более половины идентифицированных нами ассоциаций приходилось на самый ранний период онтогенеза. Вероятно, это связано с биологическими особенностями активного роста в последнюю фазу беременности и ранний постнатальный период жизни жвачных, в том числе овец. Например, третья волна активного миогенеза приходится на поздний эмбриональный или ранний постнатальный период (52), а число преадипоцитов бурой жировой ткани увеличивается до и после рождения ягнят, что особенно важно для их неонатального выживания (53).
Некоторые SNP, идентифицированные в процессе GWAS, попадали в регионы QTL, которые были обнаружены ранее другими исследователями. Так, шесть SNP (160,2-160,3 и 191,0 Мб) на OAR2 и один SNP (86,6 Мб) на OAR9 были локализованы вблизи от геномных областей, ассоциированных со среднесуточным приростом у овец (6). SNP (oar3_OAR15_74862644) на OAR15 располагался на расстоянии 400 Кб от QTL, влияющего на массу тела (74,4-74,4 Мб) (7). Кроме того, некоторые SNP попадали в регионы QTL, которые были картированы на основе микросателлитных маркеров. В частности, SNP OAR4_24289280.1 и oar3_OAR4_23286923 локализовались внутри QTL, потенциально ассоциированного с живой массой у овец на OAR4 (54), а oar3_OAR1_204348376 на OAR1, ассоциированный с ЖМ180 и ЖМ270, находился внутри QTL, регулирующего глубину мышц над третьим поясничном позвонком (55).
Большинство SNP на OAR1 (за исключением oar3_OAR1_14039930) попадали внутрь или находились в непосредственной близости от QTL, ассоциированных как с живой, так и с мышечной массой, процентом выхода постного мяса, содержанием жира в туше и массой костей в туше.
Примечательно, что эти QTL были картированы с использованием 189 мик-росателлитных маркеров в популяции возвратных кроссов (авасси х меринос) х меринос, происходящих от одного барана-родоначальника (56). В связи с тем, что мы проводили GWAS-анализ с использованием ДНК-чипов высокой плотности в популяции возвратных кроссов, происходящих от двух баранов (обеспечивается большая изменчивость показателя живой массы), вероятно, результаты нашей работы подтверждают данные C.R. Cavanagh с соавт. (56), и соответствующие SNP потенциально могут быть предложены как функциональные кандидаты живой массы у овец.
Следует отметить, что выявленные нами различия в наборах SNP, ассоциированных с живой массой в разные возрастные периоды, — экспериментальный факт, требующий фундаментального исследования обнаруженной закономерности. В частности, возникает вопрос, можно ли прогнозировать живую массу и мясную продуктивность овец в раннем возрасте посредством анализа желательных генотипов в генах-кандидатах, ассоциированных с этими показателями в более поздних возрастах. Положительный ответ позволит повысить эффективность овцеводства: можно будет разводить меньшее число животных с повышенной мясной продуктивностью, а мелких и медленно растущих овцы подвергать ранней элиминации, тем самым снижая затраты содержание и кормление. Кроме того, теоретически можно проводить отбор эмбрионов овец с желательным генотипами по генам, ответственным за усиленный ранний постнатальный рост ( MBD5 , ORC4 , ACVR2A , RIPK2 и SMC2 ), с последующим клонированием и пересадкой овцематкам-реципиентам. Кроме того, поскольку живая масса животного складывается из суммы масс скелетных мышц, костей с хрящевой тканью, жировой ткани, массы внутренних органов, направленное включение или, наоборот, нокаут генов, ответственных за формирование того или иного компонента живой массы, посредством геномного редактирования может быть использовано для повышения ориентированности овцеводческого производства в соответствии с потребительским спросом (например, более постное мясо или мясо с жировыми включениями).
Таким образом, GWAS-анализ полногеномных ассоциаций живой массы возвратных кроссов овец (романовская х катадин) х романовская позволил выявить 38 SNP, достоверно ассоциированных с живой массой (р < 0,00001), на OAR1, OAR2, OAR3, OAR4, OAR6, OAR9, OAR10, OAR11, OAR13, OAR15 и OAR19. Показано, что набор SNP, ассоциированный с живой массой, был неодинаковым в разные возрастные периоды. Наблюдаемый результат частично ожидаем, поскольку выявленная тенденция согласуется с биологической особенностью овец — активным ранним постнатальным ростом. Вероятно, это также объясняется неполным прекращением действия эмбриональных факторов роста. Мы идентифицировали SNP-маркеры и гены, которые могут быть разделены на три группы. Первая группа включала в себя функциональные гены-кандидаты, вовлеченные в регуляцию роста скелетных мышц, формирование костного каркаса, липидного и углеводного метаболизма. Эта группа генов после валидации на большем поголовье овец и выявлении желательных генотипов может быть рекомендованы для включения в селекционный процессов в ближайшее время. Вторая группа состояла из SNP-маркеров и генов, попадающих внутрь известных QTL, ассоциированных с живой массой у овец. Эти геномные регионы следует подвергнуть более тонкому картированию посредством секвенирования, чтобы полнее понимать характер взаимодействий локализованных в них генов, совокупность которых может быть предложена для разработки низкоплотных ДНК-чипов для анализа генетической предрасположенности к более активному росту и повышенной живой массе. Третья группа включала гены, о влиянии которых на живую массу у овец ранее не сообщалось. Изучение дополнительных SNP внутри и рядом с этими генами обеспечит лучшее понимание причинных мутаций, влияющих на живую массу у овец. Результаты нашей работы создают основу для отбора генов-кандидатов и SNP-маркеров для включения в программы маркерной и геномной селекции в овцеводстве.
Список литературы Поиск геномных вариантов, ассоциированных с живой массой у овец, на основе анализа высокоплотных SNP генотипов
- Jaquiery A.L., Oliver M.H., Bloomfield F.H., Harding J.E. Periconceptional events perturb postnatal growth regulation in sheep. Pediatric Research, 2011, 70(3): 261-266 (doi: 10.1203/PDR.0b013e3182242deb).
- Хайитов А.Х., Джураева У.Ш. Морфофизиологические закономерности роста костной и мышечной тканей у овец. Известия Санкт-Петербургского государственного аграрного университета, 2017, 3(48): 72-80.
- Сомова М.М., Мельникова Е.Е., Абрамова М.В., Коновалов А.В., Никитин С.А., Сермягин А.А. Зависимость показателей живой массы и скорости роста у ягнят романовской породы от различных паратипических факторов. Зоотехния, 2020, 8: 12-16.
- Трухачев В.И., Селионова М.И., Криворучко А.Ю., Айбазов А.М.М. Генетические маркеры мясной продуктивности овец (Ovis aries L.). Сообщение I. миостатин, кальпаин, кальпастатин (обзор). Сельскохозяйственная биология, 2018, 53(6): 1107-1119 (doi: 10.15389/agrobiology.2018.6.1107rus).
- Сермягин А.А., Белоус А.А., Требунских Е.А., Зиновьева Н.А. Показатели кормового поведения как новые селекционные признаки в разведении свиней. Сельскохозяйственная биология, 2020, 55(6): 1126-1138 (doi: 10.15389/agrobiology.2020.6.1126rus).
- Zhang L., Liu J., Zhao F., Ren H., Xu L., Lu J., Zhang S., Zhang X., Wei C., Lu G., Zheng Y., Du L. Genome-wide association studies for growth and meat production traits in sheep. PLoS ONE, 2013, 8(6): e66569 (doi: 10.1371/journal.pone.0066569).
- Al-Mamun H.A., Kwan P., Clark S.A., Ferdosi M.H., Tellam R., Gondro C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genetics Selection Evolution, 2015, 47(1): 66 (doi: 10.1186/s12711-015-0142-4).
- Matika O., Riggio V., Anselme-Moizan M., Law A.S., Pong-Wong R., Archibald A.L., Bishop S.C. Genome-wide association reveals QTL for growth, bone and in vivo carcass traits as assessed by computed tomography in Scottish Blackface lambs. Genetics Selection Evolution, 2016, 48: 11 (doi: 10.1186/s12711-016-0191-3).
- Ghasemi M., Zamani P., Vatankhah M., Abdoli R. Genome-wide association study of birth weight in sheep. Animal, 2019, 13(9): 1797-1803 (doi: 10.1017/S1751731118003610).
- Lu Z., Yue Y., Yuan C., Liu J., Chen Z., Niu C., Sun X., Zhu S., Zhao H., Guo T., Yang B. Genome-wide association study of body weight traits in Chinese fine-wool sheep. Animals, 2020, 10(1): 170 (doi: 10.3390/ani10010170).
- Pasandideh M., Gholizadeh M., Rahimi-Mianji G. A genome-wide association study revealed five SNP affecting 8-month weight in sheep. Animal Genetics, 2020, 51(6): 973-976 (doi: 10.1111/age.12996).
- Cao Y., Song X., Shan H., Jiang J., Xiong P., Wu J., Shi F., Jiang Y. Genome-wide association study of body weights in Hu sheep and population verification of related single-nucleotide polymorphisms. Frontiers in Genetics, 2020, 11: 588 (doi: 10.3389/fgene.2020.00588).
- Zlobin A.S., Nikulin P.S., Volkova N.A., Zinovieva N.A., Iolchiev B.S., Bagirov V.A., Borodin P.M., Aksenovich T.I., Tsepilov Y.A. Multivariate analysis identifies eight novel loci associated with meat productivity traits in sheep. Genes, 2021, 12(3): 367 (doi: 10.3390/genes12030367).
- Ledur M.C., Navarro N., Perez-Enciso M. Large-scale SNP genotyping in crosses between out-bred lines: how useful is it? Heredity, 2009, 105: 173-182 (doi: 10.1038/hdy.2009.149).
- Денискова Т.Е., Доцев А.В., Петров С.Н., Форнара М.С., Рейер Х., Виммерс К., Баги-ров В.А., Брем Г., Зиновьева Н.А. Геномная оценка и фенотипическая характеристика F2 ресурсной популяции овец. Аграрная наука Евро-Северо-Востока, 2019, 20(5): 498-507 (doi: 10.30766/2072-9081.2019.20.5.498-507).
- Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 2015, 4(7): 1-16 (doi: 10.1186/s13742-015-0047-8).
- Turner S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv (doi: 10.1101/005165).
- R Core Team (2018). R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. Режим доступа: https://www.R-project.org/. Без даты.
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biology, 2016, 17(1): 122 (doi: 10.1186/s13059-016-0974-4).
- Da Wei H., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 2009, 4(1): 44-57 (doi: 10.1038/nprot.2008.211).
- Hu Z.L., Park C.A., Reecy J.M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Research, 2019, 47(D1): D701-D710 (doi: 10.1093/nar/gky1084).
- Du Y., Liu B., Guo F., Xu G., Ding Y., Liu Y., Sun X., Xu G. The essential role of Mbd5 in the regulation of somatic growth and glucose homeostasis in mice. PLoS ONE, 2012, 7(10): e47358 (doi: 10.1371/journal.pone.0047358).
- Chung B.H., Stavropoulos J., Marshall C.R., Weksberg R., Scherer, S.W., Yoon G. 2q23 de novo microdeletion involving the MBD5 gene in a patient with developmental delay, postnatal microcephaly and distinct facial features. American Journal of Medical Genetics. Part A, 2011, 155(2): 424-429 (doi: 10.1002/ajmg.a.33821).
- Bravo-Oro A., Lurie I.W., Elizondo-Cardenas G., Pena-Zepeda C., Salazar-Martinez A., Correa-Gonzalez C., Castrillo J.L., Avila S., Esmer C. A novel interstitial deletion of 2q22.3 q23.3 in a patient with dysmorphic features, epilepsy, aganglionosis, pure red cell aplasia, and skeletal malformations. American Journal of Medical Genetics. Part A, 2015, 167(8): 1865-1871 (doi: 10.1002/ajmg.a.36806).
- Klingseisen A., Jackson A.P. Mechanisms and pathways of growth failure in primordial dwarfism. Genes & Development, 2011, 25(19): 2011-2024 (doi: 10.1101/gad.169037).
- de Munnik S.A., Hoefsloot E.H., Roukema J., Schoots J., Knoers N.V., Brunner H.G., Jackson A.P., Bongers E.M. Meier-Gorlin syndrome. Orphanet Journal of Rare Diseases, 2015, 10: 114 (doi: 10.1186/s13023-015-0322-x).
- Goh B.C., Singhal V., Herrera A.J., Tomlinson R.E., Kim S., Faugere M.C., Germain-Lee E.L., Clemens T.L., Lee S.J., DiGirolamo D.J. Activin receptor type 2A (ACVR2A) functions directly in osteoblasts as a negative regulator of bone mass. The Journal of Biological Chemistry, 2017, 292(33): 13809-13822 (doi: 10.1074/jbc.M117.782128).
- Rovadoscki G.A., Pertile S.F.N., Alvarenga A.B., Cesar A.S.M., Pertille F., Petrini J., Franzo V., Soares W.V.B., Morota G., Spangler M.L., Pinto L.F.B., Carvalho G.G.P., Lanna D.P.D., Coutinho L.L., Mourao G.B. Estimates of genomic heritability and genome-wide association study for fatty acids profile in Santa Ines sheep. BMC Genomics, 2018, 19(1): 375 (doi: 10.1186/s12864-018-4777-8).
- Arora R., Kumar N.S., Sudarshan S., Fairoze M.N., Kaur M., Sharma A., Girdhar Y., Sreesujatha R.M., Devatkal S.K., Ahlawat S., Vijh R.K., Manjunatha S.S. Transcriptome profiling of longissimus thoracis muscles identifies highly connected differentially expressed genes in meat type sheep of India. PLoS ONE, 2019, 14(6): e0217461 (doi: 10.1371/journal.pone.0217461).
- Randhawa I.A.S., Khatkar M.S., Thomson P.C., Raadsma H.W. Composite selection signals for complex traits exemplified through bovine stature using multibreed cohorts of European and African Bos taurus. G3 Genes\Genomes\Genetics, 2015, 5(7): 1391-1401 (doi: 10.1534/g3.115.017772).
- Widmann P., Reverter A., Fortes M.R.S., Weikard R., Suhre K., Hammon H., Albrecht E., Kuehn C. A systems biology approach using metabolomic data reveals genes and pathways interacting to modulate divergent growth in cattle. BMC Genomics, 2013, 14: 798 (doi: 10.1186/14712164-14-798).
- Doyle J.L., Berry D.P., Veerkamp R.F., Carthy T.R., Evans R.D., Walsh S.W., Purfield D.C. Genomic regions associated with muscularity in beef cattle differ in five contrasting cattle breeds. Genetics Selection Evolution, 2020, 52(1): 2 (doi: 10.1186/s12711-020-0523-1).
- Grigoletto L., Ferraz J., Oliveira H.R., Eler J.P., Bussiman F.O., Abreu Silva B.C., Baldi F., Brito L.F. Genetic architecture of carcass and meat quality traits in Montana Tropical® composite beef cattle. Frontiers in Genetics, 2020, 11: 123 (doi: 10.3389/fgene.2020.00123).
- Liu X., Guo X.Y., Xu X.Z., Wu M., Zhang X., Li Q., Ma P.P., Zhang Y., Wang C.Y., Geng F.J., Qin C.H., Liu L., Shi W.H., Wang Y.C., Yu Y. Novel single nucleotide polymorphisms of the bovine methyltransferase 3b gene and their association with meat quality traits in beef cattle. Genetics and Molecular Research, 2012, 11(3): 2569-2577 (doi: 10.4238/2012.June.29.1).
- Gan Q., Li Y., Liu Q., Lund M., Su G., Liang X. Genome-wide association studies for the concentrations of insulin, triiodothyronine, and thyroxine in Chinese Holstein cattle. Tropical Animal Health and Production, 2020, 52(4): 1655-1660 (doi: 10.1007/s11250-019-02170-z).
- Hu Z., Wu J., Qin L., Jin H., Lv Y., Zhang R., Xiao C., Cao Y., Zhao Y. ALDH1A1 effect on Yan Yellow Cattle preadipocyte differentiation. Animal Biotechnology, 2019: 1-10 (doi: 10.1080/10495398.2019.1679824).
- Clark D.L., Boler D.D., Kutzler L.W., Jones K.A., McKeith F.K., Killefer J., Carr T.R., Dilger A.C. Muscle gene expression associated with increased marbling in beef cattle. Animal Biotechnology, 2011, 22(2): 51-63 (doi: 10.1080/10495398.2011.552031).
- Zhang X., Hirschfeld M., Beck J., Kupke A., Köhler K., Schütz E., Brenig B. Osteogenesis imperfecta in a male holstein calf associated with a possible oligogenic origin. The Veterinary Quarterly, 2020, 40(1): 58-67 (doi: 10.1080/01652176.2020.1721611).
- An B., Xia J., Chang T., Wang X., Miao J., Xu L., Zhang L., Gao X., Chen Y., Li J., Gao H. Genome-wide association study identifies loci and candidate genes for internal organ weights in Simmental beef cattle. Physiological Genomics, 2018, 50(7): 523-531 (doi: 10.1152/physiol-genomics.00022.2018).
- Milner J.M., Rowan A.D., Cawston T.E., Young D.A. Metalloproteinase and inhibitor expression profiling of resorbing cartilage reveals pro-collagenase activation as a critical step for collagenol-ysis. Arthritis Research & Therapy, 2006, 8(5): R142 (doi: 10.1186/ar2034).
- Zhang H., Takeda H., Tsuji T., Kamiya N., Rajderkar S., Louie K., Collier C., Scott G., Ray M., Mochida Y., Kaartinen V., Kunieda T., Mishina Y. Generation of Evc2/Limbin global and conditional KO mice and its roles during mineralized tissue formation. Genesis, 2015, 53(9): 612-626 (doi: 10.1002/dvg.22879).
- Correa-Rodríguez M., Schmidt Rio-Valle J., Rueda-Medina B. AKAP11 gene polymorphism is associated with bone mass measured by quantitative ultrasound in young adults. International Journal of Medical Sciences, 2018, 15(10): 999-1004 (doi: 10.7150/ijms.25369).
- Oh J.H., Lee J.Y., Joung S.H., Oh Y.T., Kim H.S., Lee N.K. Insulin enhances RANKL-induced osteoclastogenesis via ERK1/2 activation and induction of NFATc1 and Atp6v0d2. Cellular Signalling, 2015, 27(12): 2325-2331 (doi: 10.1016/j.cellsig.2015.09.002).
- Chen J., Tu X., Esen E., Joeng K.S., Lin C., Arbeit J.M., Rüegg M.A., Hall M.N., Ma L., Long F. WNT7B promotes bone formation in part through mTORC1. PLoS Genetics, 2014, 10(1): e1004145 (doi: 10.1371/journal.pgen.1004145).
- Sawai M., Uchida Y., Ohno Y., Miyamoto M., Nishioka C., Itohara S., Sassa T., Kihara A. The 3-hydroxyacyl-CoA dehydratases HACD1 and HACD2 exhibit functional redundancy and are active in a wide range of fatty acid elongation pathways. The Journal of Biological Chemistry, 2017, 292(37): 15538-15551 (doi: 10.1074/jbc.M117.803171).
- Knigge A., Klöting N., Schön M.R., Dietrich A., Fasshauer M., Gärtner D., Lohmann T., Dreßler M., Stumvoll M., Kovacs P., Blüher M. ADCY5 gene expression in adipose tissue is related to obesity in men and mice. PLoS ONE, 2015, 10(3): e0120742 (doi: 10.1371/jour-nal.pone.0120742).
- Bagchi D.P., Li Z., Corsa C.A., Hardij J., Mori H., Learman B S., Lewis K.T., Schill R.L., Romanelli S.M., MacDougald O.A. Wntless regulates lipogenic gene expression in adipocytes and protects against diet-induced metabolic dysfunction. Molecular Metabolism, 2020, 39: 100992 (doi: 10.1016/j.molmet.2020.100992).
- McDaneld T.G., Hannon K., Moody D.E. Ankyrin repeat and SOCS box protein 15 regulates protein synthesis in skeletal muscle. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 2006, 290(6): R1672-R1682 (doi: 10.1152/ajpregu.00239.2005).
- Smith J.A., Curry E.G., Blue R.E., Roden C., Dundon S., Rodríguez-Vargas A., Jordan D.C., Chen X., Lyons S.M., Crutchley J., Anderson P., Horb M.E., Gladfelter A.S., Giudice J. FXR1 splicing is important for muscle development and biomolecular condensates in muscle cells. The Journal of Cell Biology, 2020, 219(4): e201911129 (doi: 10.1083/jcb.201911129).
- Fukuda S., Kaneshige A., Kaji T., Noguchi Y.T., Takemoto Y., Zhang L., Tsujikawa K., Ko-kubo H., Uezumi A., Maehara K., Harada A., Ohkawa Y., Fukada S.I. Sustained expression of HeyL is critical for the proliferation of muscle stem cells in overloaded muscle. eLife, 2019, 8: e48284 (doi: 10.7554/eLife.48284).
- Wadhwa R., Yaguchi T., Kaur K., Suyama E., Kawasaki H., Taira K., Kaul S.C. Use of a randomized hybrid ribozyme library for identification of genes involved in muscle differentiation. The Journal of Biological Chemistry, 2004, 279(49): 51622-51629 (doi: 10.1074/jbc.M407428200).
- Picard B., Lefaucheur L., Berri C., Duclos M.J. Muscle fibre ontogenesis in farm animal species. Reproduction Nutrition Development, 2002, 42(5): 415-431 (doi: 10.1051/rnd:2002035).
- Bonnet M., Cassar-Malek I., Chilliard Y., Picard B. Ontogenesis of muscle and adipose tissues and their interactions in ruminants and other species. Animal, 2010, 4(7), 1093-1109 (doi: 10.1017/S1751731110000601).
- Roldan D.L., Dodero A.M., Bidinost F., Taddeo H.R., Allain D., Poli M.A., Elsen J.M. Merino sheep: a further look at quantitative trait loci for wool production. Animal, 2010, 4(8):1330-1340 (doi: 10.1017/S1751731110000315).
- Matika O., Sechi S., Pong-Wong R., Houston R.D., Clop A., Woolliams J.A., Bishop S.C. Characterization of OAR1 and OAR18 QTL associated with muscle depth in British commercial terminal sire sheep. Animal Genetics, 2011, 42(2): 172-180 (doi: 10.1111/j.l365-2052.2010.02121.x).
- Cavanagh C.R., Jonas E., Hobbs M., Thomson P.C., Tammen I., Raadsma H.W. Mapping Quantitative Trait Loci (QTL) in sheep. III. QTL for carcass composition traits derived from CT scans and aligned with a meta-assembly for sheep and cattle carcass QTL. Genetics Selection Evolution, 2010, 42(1): 36 (doi: 10.1186/1297-9686-42-36).


