Повышение производительности цикла проектирование-создание-тестирование-обучение систем (DBTL) в синтетической биологии растений (обзор)
Автор: Prasad S.S., Das U.
Журнал: Сельскохозяйственная биология @agrobiology
Рубрика: Обзоры, проблемы
Статья в выпуске: 5 т.59, 2024 года.
Бесплатный доступ
Синтетическая биология растений - это молодая научная дисциплина, которая объединяет принципы инженерии с биологией с целью разработки уникальных систем на основе растений для различных применений, от производства биотоплива до улучшения сельскохозяйственных культур. Эта технология может революционно изменить традиционное сельское хозяйство, содействовать устойчивому развитию и решению глобальных проблем продовольственой безопасности, изменения климата и возобновляемых источников энергии. Цикл проектирование-создание-тестирование-обучение (Design-Build-Test-Learn, DBTL) обеспечивает основу для процесса планирования, разработки, оценки и совершенствования синтетических биологических систем. Он позволяет исследователям итеративно настраивать производительность биологических цепей, что делает этот цикл критически важным инструментом для построения сложных биологических систем с предсказуемым и надежным поведением. Однако на каждом этапе этого цикла могут проявиться узкие места из-за неэффективного проектирования, ограниченного запаса генетических компонентов, технических проблем при разработке и управлении биологическими системами и трудностей в корректном мониторинге производительности системы. Для преодоления узких мест в цикле DBTL можно использовать различные стратегии: совершенствование вычислительных технологий для эффективного проектирования, расширение набора генетических компонентов, повышение точности и масштабируемости приемов редактирования генома и внедрение методов высокопроизводительного скрининга для точного измерения производительности системы. В этой обзорной статье мы обсудим последние достижения в улучшении производительности цикла DBTL для преодоления его узких мест.
Цикл dbtl, мультиомика, биоортагональные реакции, микрофлюидика, изотопные трассеры, loica, crispr
Короткий адрес: https://sciup.org/142243772
IDR: 142243772 | УДК: 577.21 | DOI: 10.15389/agrobiology.2024.5.831rus
Текст обзорной статьи Повышение производительности цикла проектирование-создание-тестирование-обучение систем (DBTL) в синтетической биологии растений (обзор)
Селекция сельскохозяйственных видов создала множество высокоурожайных культур с улучшенными характеристиками — устойчивостью к болезням, засухоустойчивостью и более высокой пищевой ценностью. Повышение уровня продовольственной безопасности и расширение мировых запасов продовольствия напрямую связаны с успехами селекции. В настоя-шее время селекционеры сталкиваются с новыми сложными задачами из-за усугубления глобальных проблем, к которм относятся изменения климата, распространение вредителей и болезней и снижение плодородия почв. Экстремальные погодные явления (засухи и наводнения) происходят чаще из-за изменения климата, что может снизить урожайность и продовольственную безопасность (1). Плодородие почвы снижается из-за неправильного ее использования и других факторов, а вредители и болезни становятся все более устойчивыми к используемым методам их контроля. Создание культур, которые могут переносить последствия климатических изменений, не повреждаться вредителями, быть устойчивыми к болезням и сохранять высокую продуктивность на истощенных почвах, должно стать основной целью селекционных программ. Для эффективного формирования новых генетических характеристик и ускорения селекционного процесса потребуются новые технологии, в частности геномика, геномное редактирование и прецизионная селекция (2-4). Это возможно благодаря междисциплинарным подходам, таким как синтетическая неология, интегрирующая современные достижения в разных областях в качественно новую систему исслеований. Она широко используется в ряде направлений, включая науку об окружающей среде, энергетику, сельское хозяйство и медицину.
Синтетическая биология — междисциплинарная научная область, которая объединяет идеи из биологии, инженерии и компьютерных наук для создания новых биологических систем с различными свойствами и изменения уже существующих (5, 6). Она подразумевает разработку искусственных биологических систем и их элементов, включая гены, ферменты и пути, которые могут использоваться для выполнения определенных биологических функций. Одна из основных целей синтетической биологии — проектирование и создание биологических систем с желаемыми характеристиками и возможностями, например производящих биотопливо, синтезирующих новые лекарства и материалы или увеличивающих производство сельскохозяйственной продукции (7). Для этого требуется изменение уже существующих биологических систем (примеры — микроорганизмы, продуцирующие инсулин, или растения, устойчивые к вредителям и болезням). Кроме того, системная биологии помогает лучше понимать функционирование биологических систем и лежащие в их основе процессы.
Возможности синтетической биологии растений широки. Модифицируя и конструируя геномы растений, ученые создают новые сорта и гибриды с желаемыми характеристиками (устойчивость к болезням и засухе, улучшенное усвоение питательных веществ) (8, 9). Эти методы также использовались для получения растений с повышенной эффективностью процессов фотосинтеза, что также может значительно увеличить урожайность и помочь справиться с вызовами, связанными с проблемами продовольственной безопасности. Создание из растительного сырья новых материалов, таких как биоразлагаемые полимеры, помогает бороться с загрязнением окружающей среды, та же задача решается при использовании растений, которые эффективно удаляют токсины из почвы или воды (10). Производство биоматериалов и возобновляемого биотоплива из растительного сырья уменьшает зависимость от ископаемого топлива и помогает смягчить последствия изменения климата. Для выполнения таких разработок прежде всего требуется получение необходимого объема знаний о структуре и функциональной активности геномов растений, накопление такой информации в базах данных и ее анализ с помощью вычислительных инструментов для углубленного понимания процессов, происходящих в растениях. Решение таких залач возможно благодаря подходу на основе DBTL (Design–Build–Test– Learn). DBTL — это циклический процесс, используемый при разработке новых, генетичеки измененных биологических систем для тестирования и изучения механизмов, действующих в живых организмах, с помощью компьютерного моделирования и симулятора (11-14). DBTL включает три этапа. На первом этапе (проектирование) синтетические биологи применяют компьютерное моделирование и инструменты для дизайна новой биологической системы, например синтетического гена или метаболического пути. Это включает определение необходимых компонентов (генов, промоторов и других регуляторных элементов) и проектирование их взаимодействий и поведения (15). Далее следует второй этап — сборки, когда спроектированные компоненты физически собираются и интегрируются в живой организм, например в клетки бактерий или дрожжей. Для осуществления этого этапа выполняется синтез генов, редактирование генома и введение спроектированного генетического материала в организм-хозяин с помощью методов трансформации (16). На третьем этапе (тестирование) созданная биологическая система оценивается на предмет ее производительности и функциональности. Это может включать измерение выходных данных системы, таких как продукция определенного белка или метаболита, и сравнение фактических показателей с прогнозируемыми на этапе проектирования
-
(17) . На основе результатов тестирования конструкция может быть изменена, а цикл повторен для оптимизации производительности системы. Цикл DBTL в синтетической биологии позволяет исследователям быстро тестировать новые биологические системы и повторять их модификацию до тех пор, пока не будет достигнута желаемая функциональность.
DBTL помогает ускорить разработку новых биологических инструментов и систем для применений в различных областях — от биопроизводства до здравоохранения и экологии, тем не менее применение этого подхода связано с определенными ограничениями. Сложность биологических систем представляет собой одну из главных проблем. Синтетическая биология подразумевает проектирование, создание и тестирование систем, работающих в живых клетках и организмах, котрые невероятно сложны и динамичны. Это может привести к неожиданным результатам при тестировании. Разработка и тестирование биологических систем требуют точных и надежных технологий, что является еще одной трудностью. Синтез генов, редактирование генома и высокопроизводительный скрининг — это всего лишь несколько методов синтетической биологии, которые требуют как сложных инструментов, так и узкоспециализированных знаний, и то, и другое может быть дорогим и труднодоступным. Из-за большого объема «проб и ошибок» в цикле DBTL, итеративном по своей природе, его применение тоже может оказаться трудоемким и дорогим. Может потребоваться запустить цикл более одного раза, чтобы максимизировать производительность системы. Возможны сложности в управлении и координации. Наконец, создание и использование новых биологических систем может быть затруднено этическими ограничениями и вопросами безопасности, связанными с использованием генетически модифицированных организмов (ГМО) и синтетической биологии. Применение синтетической биологии может лимитироваться из-за нормативных ограничений и общественного недоверия.
Чтобы устранить препятствия, замедляющие цикл разработки, важно стандартизировать его элементы и протоколы, улучшить инструменты моделирования, имитации, автоматизации, поощрять сотрудничество и открытые принципы исследований, а также отдавать приоритет безопасности и этике. Улучшенные инструменты моделирования и имитации помогают точнее прогнозировать поведение биологических систем, сокращая время, необходимое для тестирования и обучения на каждой итерации. Повышение автоматизации сократит время, необходимое для создания и тестирования систем, а сотрудничество и открытые методы консолидируют силы исследователей для получения результата. Наконец, решение вопросов безопасности и этики предотвратит нанесение вреда исследователям или окружающей среде и гарантирует, что биологические системы производятся ответственно.
Цикл DBTL: последовательность действий. Цикл DBTL включает четыре итеративных шага: проектирование, сборка, тестирование и обучение. На этапе проектирования исследователи описывают спецификации биологической системы, определяют ее компоненты и решают, как они будут взаимодействовать для достижения желаемой цели. На этапе сборки выполняется физическое построение биологической системы. На этапе тестирования производительность системы проверяется для определения ее эффективности в выполнении желаемой функции. Наконец, на этапе обучения данные, полученные во время тестирования, изучаются для использования при корректировках в следующих итерациях цикла DBTL.
Проектирование: идентификация молекулы или пути. Это фундаментальный компонент цикла DBTL в биологической инженерии. Проектиро- вание включает в себя процесс создания биологических систем с учетом конкретной цели, например создание определенной молекулы или оптимизация метаболического пути. Это критический шаг, который обеспечивает целевой подход, повышает эффективность и предсказуемость и облегчает масштабируемость (18). Создавая биологическую систему с учетом конкретной цели, исследователи могут сократить число итераций, необходимых для получения предполагаемого результата, избежать ошибок и предсказать поведение системы на основе ее конструкции (15). В результате может снизиться вероятность непредвиденных событий, повысится общая производительность цикла DBTL. Интеграция этапа конструирования в цикл DBTL критически важна для повышения эффективности и успешности попыток проектирования биологических систем (19).
Сборка: этап разработки прототипа. На этом этапе происходит физическое построение биологической системы на основе дизайна ее конструкции. В цикле DBTL этот этап позволяет проверять и оптимизировать конструкцию для итерации цикла DBTL и масштабировать биологическую систему (20, 21). Физически создавая биологическую систему, исследователи должны убедиться, что она может соответствовать требованиям запланированного применения, протестировать и усовершенствовать систему для улучшения ее производительности и предложить физический прототип, который можно использовать для разработки крупномасштабных производственных процессов.
Тестирование: этап оценки и анализа . Третий шаг в цикле DBTL — это тестирование генно-инженерной биологической системы. Оценка производительности прототипа системы и анализ данных, собранных во время тестирования, дают информацию о будущих изменениях (22). Актуальность цикла DBTL заключается в возможности направлять процессинг биоинженерных систем в методическом аспекте, что позволяет постоянно улучшать производительность системы с течением времени. Применяя каждый из элементов цикла DBTL в своих исследованиях, синтетические биологи, создающие биологические системы для решения ряда социально значимых проблем, включая здравоохранение, энергетику и экологическую устойчивость, могут сделать свои изыскания более успешными.
Обучение: этап обратной связи и уточнения. На этом этапе исследователи изучают данные тестирования, чтобы получить информаацию о поведении биологической системы, что служит основой для корректировок в будущих итерациях (23). Ценность фазы обучения в цикле DBTL заключается в ее потенциале предоставления обратной связи для уточнения структуры и функций биологической системы. Изучая полученные данные, исследователи могут находить области для улучшения, оптимизировать эффективность системы и предлагать новые гипотезы для дальнейшего исследования. Информация, полученная на этапе обучения, может быть использована на этапах проектирования и сборки в цикле DBTL, что позволит создавать более эффективные и действенные биологические системы.
Расширение инструментария для повышения точности в цикле DBTL. Чтобы сократить время процедур в цикле DBTL, необходимо располагать соответствующими наборами данных и предиктивной информацией. Технологии предиктивного моделирования и имитации применимы на этапе проектирования. С помощью этих инструментов синтетические биологи могут лучше предсказывать поведение проектируемых биологических систем, что делает возможным создание более эффективных и действенных генетических конструкций. На этапе тестирования требуются высокопроизводительные методы скрининга, что позволяет синтетическим биологам анализировать огромное количество генетических конструкций за короткий период. Разработки в области автоматизации упростили создание и тестирование генетических конструкций, сократив время и работу, необходимые для завершения каждого цикла.
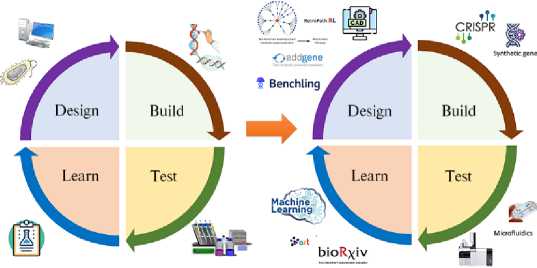
В общем виде представление о классическом наборе средств, применяемых при реализации цикла DBTL, и его современных усовершенствованных модификациях дает рисунок 1.
Classic method Advanced method
Рис. 1. Методы и средства, расширяющие возможности классического цикла DBTL (Design– Build–Test–Learn, проектирование–создание–тестирование–обучение). Компьютерное проектирование (Computer-Aided Design, CAD), проектирование и анализ последовательностей ДНК, РНК и аминокислот с помощью программного обеспечения Benching, проектирование биосинтетических путей с применением вычислительного инструмента RetroPath, использование CRISP технологий и синтетических генов при создании прототипов, масс-спектрометрии и микрофлюидики — на этапе тестирования, машинное обучение с привлечением различных баз данных.
Инструментарий . Изотопные трассеры широко применяются в цикле DBTL для измерения переноса атомов и молекул в биологических системах. Изотопные трассеры представляют собой атомы элемента с разной атомной массой (24, 25). С помощью изотопных трассеров исследователи могут отслеживать транспорт атомов и молекул по метаболическим путям и обнаруживать, какие пути активны и как они используются. Трассеры могут применяться для оптимизации производительности биологических систем посредством определения ключевых метаболических путей и ферментов как мишеней для оптимизации (17). Исследователи могут обнаруживать узкие места метаболизма и оптимизировать метаболический поток в циклах инжениринга, используя изотопные трассеры. Этот метод был применен для функциональной модификации микробиома (26) и ускорения биопроизводства (27).
Интеграция мультиомики с циклом DBTL объединяет данные из нескольких типов биологических образцов с итеративными инженерными циклами для оптимизации биологических систем (28-30). Интеграция с циклом DBTL позволяет выявить критические биологические пути и взаимодействия, которые можно модифицировать для улучшения производительности биологических систем. Объединяя данные нескольких типов, например геномики, транскриптомики, протеомики и метаболомики, исследователи могут находить возможные цели для проектирования и разрабатывать способы улучшения биологических систем (31). Вычислительные методы применялись при проектировании штаммов (32), для повышения продукции 3-гидроксипропионовой кислоты в Aspergillus pseudoterreus (33).
Машинное обучение помогает оптимизировать проектирование, автоматизировать тестирование, предсказать потребности в обслуживании и постоянно улучшать продукт благодаря автоматизации некоторых задач и анализа данных (15). Алгоритмы машинного обучения также могут оптимизировать проектирование продукта на основе критериев стоимости, эффективности и производительности. На основе информации от датчиков и других источников машинное обучение позволяет автоматизировать процесс тестирования. Более того, машинное обучение может анализировать данные этапов проектирования, разработки и тестирования, чтобы точно определить области, которые можно улучшить, гарантируя, что продукт будет постоянно развиваться и улучшаться в ответ на ввод пользователем уточняющих данных.
Жидкостная хроматография (ЖХ)—масс-спектроскопия в сочетании с циклом DBTL позволяют улучшить биологические системы (34). В ЖХ разделение и идентификация компонентов смеси на основе их химических и физических свойств осуществляется с использованием свойств жидкой подвижной фазы. Метод может применяться для выделения и идентификации метаболитов, их количественной оценки, определения изменений белков и анализа малых молекул с целью обнаружения потенциальных мишеней для оптимизации. Также сообщается об использовании этого метода при извлечении специализированных растительных метаболитов (35) и при метаболическом инжениринге у Streptomycetes (36).
Методы биоортогональной химии используют реакции, которые способны протекать внутри живых систем, не мешая естественным биохимическим процессам, так как участвующие в таких реакциях функциональные группы в живой клетке быстро и селективно реагируют друг с другом, но инертны по отношению к другим соединениям (37). В цикле DBTL этот подход эффективен при внесении изменений в биомолекулы и анализе их функции в биологических системах. С применением такого подхода при биологическом инжиниринге была повышена продукция липазы и эстеразы (38).
Микрофлюидика — это изучение и манипулирование жидкостями в микромасштабе, часто в каналах с размерами от десятков микрометров до нескольких миллиметров. Микрофлюидные устройства стали полезными инструментами для цикла DBTL, поскольку они предлагают различные преимущества по сравнению с традиционными методами (39). Также сообщалось об их автоматизации в синтетической биологии (40) и высокой пропускной способности (41).
Технология CRISPR обладает большим потенциалом для применения в DBTL. С помощью CRISPR можно проводить целевое редактирование генома, чтобы вводить определенные генетические модификации в организмы, что позволяет разрабатывать точные генетические схемы (20, 42). Более того, скрининг на основе CRISPR может использоваться для обнаружения и выделения благоприятных генетических вариаций, тем самым ускоряя процесс оптимизации в цикле DBTL. Технологии CRISPR также могут применяться для изучения функции генов и их сетей, способствуя общему пониманию биологических систем (25, 43).
Интеграция перечисленных методов существенно повысит эффективность циклов DBTL, позволяя точнее манипулировать биологическими системами, изучать их функционирование и повышать производительность, что в конечном итоге приведет к улучшению биологических систем.
Программное обеспечение. RetroPath и Selenzyme — два вычислительных инструмента, используемых в синтетической биологии и метаболической инженерии для проектирования и оптимизации метаболических путей в микробных клетках. RetroPath — вычислительный инструмент, который можно использовать для проектирования биосинтетических путей для 836
производства желаемого соединения. Он определяет потенциальные метаболические пути, которые могут производить целевое соединение, — от набора известных иходных компонентов до конечного продукта. RetroPath использует ретробиосинтетический подход, при котором целевое соединение разбивается на более мелкие молекулы, и определяются потенциальные биосинтетические пути, которые могут такие молекулы производить. Этот подход позволяет быстро и эффективно проектировать метаболические пути для производства широкого спектра соединений, включая натуральные продукты, фармацевтические препараты и биотопливо (44). Selenzyme роедтав-ляет собой вычислительный инструмент, который может быть использован при проектировании и оптимизации ферментных систем для производства сложных органических молекул. Принцип его работы основан на концепции направленной эволюции, когда мутации вводятся в последовательность гена белка для оптимизации его функции. Selenzyme использует комбинацию биоинформатики, молекулярного моделирования и методов машинного обучения для поиска мутаций, которые могут улучшить производительность ферментов в определенном метаболическом пути. Инструмент также может быть использован для выявления потенциальных новых ферментных систем для производства новых соединений (45).
PartsGenie был вычислительным программным инструментом при проектировании и оптимизации последовательностей ДНК для приложений синтетической биологии. Инструмент разработан, чтобы помочь исследователям эффективно создавать большие конструкции ДНК с помощью автоматизации многих трудоемких этапов, вовлеченных в процесс (46).
В недавно предложенном программном обеспечении SyncBio представлены современные платформы, включая Professional Synthesis Platform and High-throughput Synthesis Platform, что позволяет в случае сложных биосинтетических путей секвенировать гены, используя DBTL. SyncBio предлагает ряд инструментов и услуг для поддержки исследований в синтетической биологии, включая синтез ДНК, редактирование генов и экспрессию белков. Программное обеспечение также предоставляет доступ к базе данных генетических компонентов и биологических систем для проектирования и создания новых биологических систем (47).
Еще одно программное обеспечение — LOICA: Logical Operators for Integrated Cell Algorithms–Flapjack предназначено для запроса, построения и анализа генетической последовательности, что полезно для конструирования генетических схем и их контроллера (21, 48).
Цикл DBTL был полностью автоматизирован для проектирования биосистем с использованием интегрированной роботизированной системы и алгоритмов машинного обучения с роботизированной платформой BioAutomata (22). Это полностью автоматизированная роботизированная платформа, предназначенная для ускорения разработки биологических систем посредством автоматизации процесса DBTL. Платформа сочетает передовые технологии робототехники, синтетической биологии и машинного обучения, что позволяет проводить высокопроизводительные эксперименты и анализ данных. Она может применяться для автономного создания генетических конструкций, синтеза и сборки фрагментов ДНК, интегриции конструкции в организм хозяина и тестирования полученных биологических систем. Платформа использует методы машинного обучения для оптимизации проектирования генетических компонентов на основе данных предыдущих раундов. Автоматизируя процесс DBTL, можно значительно сократить время и усилия, необходимые для построения сложных биологических систем, что позволяет сосредоточиться на более новых и сложных элементах синтетической биологии.
Улучшенные конструкции. E. Orsi соавт. (49) протестировали DBTL для отбора синтетических модулей по ростовым парамерам с целью ускорения разработки клеточных биофабрик. Этот подход может успешно применяться, когда производство желаемого продукта или соединения напрямую связано с ростом и выживанием выбранного штамма микроорганизма. Результат достигается с помощью стратегии отбора модулей, благоприятствующих росту штаммов-продуцентов, но подавляющих рост не производящих целевой продукт штаммов. Если соответствующий метаболический путь удается сконструировать, то синтез целевого продукта становится необходимым для выживания штамма в определенной среде. Это создает селективные условия для роста штаммов, которые производят желаемый продукт, в то время как не производящие его штаммы элиминируются. Сочетание биопроизводительности и отбора по ростовым показателям имеет несколько преимуществ. Оно позволяет микробным штаммам, которые производят желаемый продукт, быстро и эффективно эволюционировать при этом минимизируя производство ненужных побочных продуктов. С помощью нового программного инструмента и автоматизированных лабораторных процедур для построения и тестирования метаболического пути был быстро создан прототип in vivo Escherichia coli для производства флавоноида (2S)-пиноцембрина (16).
Анализ методом Microflow targeted liquid chromatography-selected reaction monitoring (LC-SRM) эффективно идентифицировал целевые пептиды с соответствующими путями биосинтеза, ответственными за производство биотоплива в продуценте Pseudomonas putida . Метод помогает сократить время цикла DBTL (50). Мультиомный подход, используемый в цикле DBTL, объединяет несколько уровней омиксных данных (геномика, транскриптомика, протеомика, метаболомика и т.д.) для ускорения создания, улучшения и оценки биологических систем (51). Этот метод применяется в синтетической биологии и метаболической инженерии для повышения выхода желаемых веществ и метаболитов при лучшем понимании поведения биологических систем. Автоматизация сборки и тестирования применялась для ускорения разработки штаммов в исследованиях по биотопливу (52). Метод позволяет совершенствовать процесс DBTL, предоставляя доступ к передовым технологиям и знаниям. Благодаря этому инструменту ускоряется итеративное тестирование генетических конструкций, что позволяет в короткие сроки проверить множество гипотез. Также могут быть стандартизированы и оптимизированы экспериментальные методы, что повысит воспроизводимость результатов при испытаниях. Технологии машинного обучения и анализа данных применимы для обработки и оценки больших объемов экспериментальных данных, помогая выявлять тенденции и более эффективно улучшать генетические конструкции.
Автоматизацию некоторых действий и предоставление информации на основе анализа данных в сочетании с машинным обучением можно использовать для ускорения процесса. Проведены исследования по включению машинного обучения в DBTL для повышения производительности (15).
Метод автоматизированных рекомендаций (Automated Recommendation Tool) с машинным обучением разработан для получения прогнозов о штамме, рекомендуемом для следующего цикла, и уровне его производительности (53). Программа использует алгоритмы машинного обучения для оценки больших массивов экспериментальных данных и генерации автоматических рекомендаций для генетических конструкций, которые должны 838
быть протестированы. Эти рекомендации основаны на применении DBTL, где производительность каждой генетической конструкции в системе последовательно проверяется. Инструмент может рекомендовать изменения в текущих генетических конструкциях или предлагать совершенно новые конструкции, позволяя синтетическим биологам более эффективно исследовать новое пространство дизайна. Интеграция инструмента автоматических рекомендаций с DBTL может ускорить создание сложных биологических систем за счет минимизации времени и усилий, необходимых для этапов проектирования и тестирования процесса.
С помощью DBTL разработаны экологически чистые фармацевтические продукты (54). Ключевой фермент, отвечающий за сбалансированное производство тетрагидропапаверолина, был идентифицирован с помощью предиктивного программного обеспечения M-path. Метод M-path основан на линейном программировании для поиска возможных комбинаций векторов характеристик реакций, которые в сумме дают желаемый вектор характеристик пути, необходимого для получения целевого продукта из исходного соединения. То есть на основании известных исходных соединений и метаболических путей их превращений и химических характеристик на каждом этапе собирается подмножество химических реакций, позволяющее получить заданное соединение в одном метаболическом цикле (55).
Детектирование ферментов имеет решающее значение во многих областях — в медицине, биотехнологии, при мониторинге состояния окружающей среды. Метод M-path объединяет приемы анализа изображений с алгоритмами машинного обучения и осуществляет поиск доменов, определяющих функциональную активность ферментов в биологических образцах. Более того, можно одновременно обнаруживать множество ферментов, несущих определенные функциональные домены, правильно и быстро оценивая большие массивы данных. Этим обусловлено практическое приенение такого подхода в высокопроизводительных скрининговых приложениях, где необходимо быстро исследовать большое число образцов.
Последние разработки в области редактирования генома микроорганизмов. Исследование штамма Corynebacterium glutamicum методом DBTL с целью получения продуцента шикимовой кислоты было успешно проведено с использованием комбинации вычислительных, генетических и биохимических манипуляций (56). Примененная технология включала поиск данных литературы, в которых упоминаются определенные гены или генетические модификации, а затем анализ совместной встречаемости этих генов в литературе с другими генами или генетическими модификациями. Это помогает выявить связи и закономерности, которые могут быть не сразу очевидны при простом изучении отдельных исследований. ИИ-анализ литературы имеет множество применений в различных областях, включая биомедицину, генетику и биоинформатику. Его можно использовать для обнаружения новых связей между генами и заболеваниями, определения потенциальных мишеней для лекарственных препаратов и нашего лучшего понимания биологических процессов.
Escherichia coli , генетически модифицированная для продукции de novo (2s) флаванонов, была получена с применением DBTL (57). Разработана и оптимизирована в DBTL генетическая схема для производства пиноцембрина в E. coli . Процедура включала проектирование схемы, ее построение с использованием последовательностей ДНК, оценку ее производительности и использование полученной информации для улучшения схемы. Выход пиноцембрина был увеличен в повторных циклах
DBTL. DBTL использовали для получения новых метаболитов у генетически модифицированных стрептомицетов (36). Процесс включал идентификацию биосинтетического пути, проектирование генетической схемы, ее построение с помощью клонирования последовательностей ДНК, тестирование функции схемы и изучение результатов для оптимизации схемы. Аналогичным образом промышленные клетки бактерий (так называемые «шасси» — более или менее улучшенный хозяин для генетических конструкций) для производства биохимикатов получали с использованием DBTL (23).
Сообщалось о DBTL в сочетании с машинным обучением (ML) для успешного производства додеканола генетически модифицированной E. coli (18). Додеканол — это жирный спирт, который можно использовать в производстве моющих средств, средств личной гигиены и биотоплива. ML использовался для определения оптимальных комбинаций генов, прогнозирования поведения организма и автоматизации процесса проектирования и тестирования. Благодаря сочетанию DBTL с ML ускорилась разработка системы с более высоким выходом додеканола в E. coli .
Полученный с помощью синтетической инженерии штамм дрожжей, который производит артемизининовую кислоту, имеет потенциал для реорганизации производства артемизинина и расширения доступа к этому спасительному лекарству (58-61). Важнейший предшественник в создании противомалярийного лекарства артемизинин — это артемизиновая кислота. Однако растение Artemisia annua производит лишь относительно небольшое количество артемизинина, что делает его промышленное производство дорогим и сложным. Продукция артемизиновой кислоты штаммом дрожжей, созданным с использованием синтетической биологии, дает многообещающее решение этой проблемы. Синтетическая биология может помочь в решении проблемы малярии, обеспечив производство артемизиновой кислоты в больших масштабах устойчивым и экономически эффективным способом. Этап проектирования включал выявление ферментов и регуляторных элементов, необходимых для преобразования глюкозы в артемизиновую кислоту, и оптимизацию пути биосинтеза для максимальной эффективности. На этапе сборки спроектированный путь физически собирается и интегрируется в геном дрожжей. Это может включать использование таких методов, как редактирование генома и синтез генов, для введения необходимых генов и регуляторных элементов в геном дрожжей. Цель состоит в том, чтобы создать стабильный и функциональный синтетический геном, который может производить желаемый продукт. На этапе тестирования сконструированный штамм дрожжей оценивается на предмет его способности производить желаемый продукт. Это может включать измерение выхода и чистоты продукта, производимого дрожжами, и оценку производительности дрожжей в различных условиях роста. Этот этап может быть итеративным, а этапы проектирования и сборки будут совершенствоваться на основе результатов тестирования.
В таблице представлены стратегии и методы современной генной инженерии микроорганизмов.
Передовые стратегии и инструменты, используемые в цикле DBTL (Design– Build–Test–Learn, проектирование–создание–тестирование–обучение) в генной инженерии микроорганизмов
Этап цикла ] Стратегия Цель I Дополнительные инструменты ] Ссылка
Дизайн Построение базы Высокая продукция гента- Библиотека мутантов с машинным (62)
данных мицина обучением
|
Продолжение таблицы |
||
|
Мультиомика |
Идентификация генов, от- Создание библиотеки мутантов (63) ветственных за фотосинтез у эукариот Синтез природных продук- Омиксные технологии с машин- (64) |
|
|
Автоматизация |
тов ным обучением Полная автоматизация Искусственный интеллект с ма- (22) |
|
|
Дизайн–созда |
-Основана на по- |
цикла DBTL шинным обучением Проектирование in silico (65) Объединение биомодуля с Система компьютерного проекти- (66) |
|
ние |
лучаемых данных |
компьютерным проектиро- рования (Computer-Aided Design, |
|
Множественное |
ванием CAD) Разработка фиолетового Высокоэффективное наложение (67) |
|
|
наложение генов |
риса трансгенов с помощью векторов |
|
|
CRISPRa/i |
Улучшение синтеза белков CRISPRi-KRAB c ASGARD (68) |
|
|
Рибосвитч |
и липидов Повышение производитель- CRISPR/Cas9 (69) ности за счет замены промо тора Синтез большего количества Омиксные технологии, программа (70) |
|
|
Дизайн–созда-Мультиомика |
биотоплива BioFoundry Производство n-капроата Подход «сверху вниз» (12) |
|
|
ние–тестиро-вание |
Автоматизация |
Синтез автоматизированных Машинное обучение (17) |
|
CRISPR/dCpf1 |
бактериальных фабрик Увеличение производства Транскриптомика (71) |
|
|
Симуляция |
индигоидина Производство 3-гидрокси- Протеомика и метаболомика (33) пропионовой кислоты Автоматизация проектирова-Logical Operators for Integrated Cell (72) |
|
|
Тестирова- |
Клеточный ана- |
ния с использованием дан- Algorithms (LOICA) ных Инженерия изопропиноид- Рамановская спектроскопия (73) |
|
ние–обучение |
лиз |
ного пути |
|
Метаболическая |
Разработка сайта связыва- Регрессия гауссовского процесса с (74) ния рибосом машинным обучением Производство вакцин Метаболическая инженерия (75) |
|
|
модель Омиксные техно- |
Модифицирование для про- Флаксомика (76) |
|
|
логия |
изводства натурального про- |
|
|
MARL (Multi- |
дукта Оптимизация дизайна ИИ-обучение с подкреплением (77) |
|
|
Agent Reinforcement Learning) |
||
Современные возможности DBTL при манипуляциях с растительными клетками иллюстрирует рисунок 2.
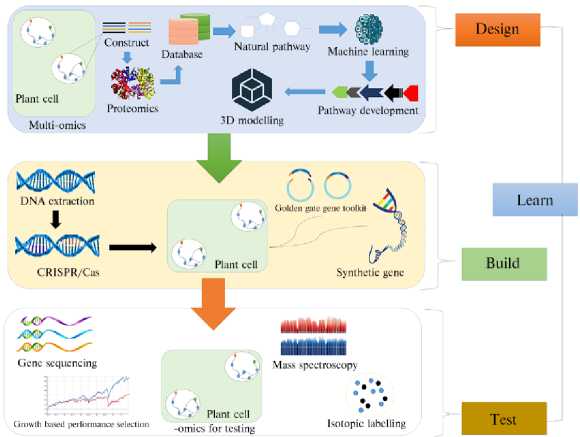
Рис. 2. Изменение метаболических путей в растительных клетках в цикле DBTL (Design–Build–
Test–Learn, проектирование–создание–тестирование–обучение). На этапе проектирования с помощью омиксных технологий, анализа баз данных, машинного обучения разрабатываются метаболические пути, которые сравниваются при 3D моделировании. Осуществляется требуемое геномное редактирование (CRISP/Cas), сиквенс наборов синтетических генов и контроль продукции желаемого метаболита. Итоговое тестирование системы включает селекцию по эффективности клеточного роста и выявление целевого продукта с помощью изотопной метки.
Таким образом, обсуждая перспективы DBTL (Design–Build–Test– Learn, цикл проектирование–создание–тестирование–обучение), следует признать, что этот подход является сильной парадигмой для создания биологических систем в синтетической биологии и уже привел к нескольким заметным достижениям. Будущие перспективы применения DBTL обнадеживают. Существует значительный потенциал для дополнительных прорывов в проектировании, разработке и тестировании биологических систем с помощью цикла DBTL, особенно с учетом растущей доступности высокопроизводительных технологий, машинного обучения и аналитики больших данных. Подход на основе DBTL может трансформировать здравоохранение, возобновляемую энергетику, способствовать экологической устойчивости, позволяя проектировать и разрабатывать эффективные биологические системы. Продолжающееся развитие и совершенствование DBTL, безусловно, приведет к новым открытиям и достижениям в синтетической биологии.
Список литературы Повышение производительности цикла проектирование-создание-тестирование-обучение систем (DBTL) в синтетической биологии растений (обзор)
- Sala A., Woodruff D.R., Meinzer F.C. Carbon dynamics in trees: feast or famine? Tree Physiology, 2012, 32(6): 764-75 (doi: 10.1093/TREEPHYS/TPR143).
- Liang Z., Chen K., Li T., Zhang Y., Wang Y., Zhao Q., Liu J., Huawei Z., Liu C., Ran Y., Gao C. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nature Communications, 2017, 8(1): 14261 (doi: 10.1038/ncomms14261).
- Dlugosz E.M., Lenaghan S.C., Stewart C.N. Jr. A robotic platform for high-throughput protoplast isolation and transformation. Journal of Visualized Experiments, 2016, NA(115): 54300-NA (doi: 10.3791/54300).
- Mortimer J.C. Plant synthetic biology could drive a revolution in biofuels and medicine. Experimental Biology and Medicine, 2019, 244(4): 323-331 (doi: 10.1177/1535370218793890).
- Liu W., Stewart C.N. Plant synthetic biology. Trends in Plant Science, 2015, 20(5): 309-317 (doi: 10.1016/j.tplants.2015.02.004).
- Llorente B., Williams T.C., Goold H.D. The multiplanetary future of plant synthetic biology. Genes, 2018, 9(7): 348 (doi: 10.3390/genes9070348).
- Tothill I.E. Biosensors developments and potential applications in the agricultural diagnosis sector. Computers and Electronics in Agriculture, 2001, 30(1-3): 205-218 (doi: 10.1016/S0168-1699(00)00165-4).
- Boyle P.M., Burrill D.R., Inniss M.C., Agapakis C.M., Deardon A., DeWerd J.G., Gedeon M.A., Quinn J.Y., Paull M.L., Raman A.M., Theilmann, M.R., Wang L., Winn J.C., Medvedik O., Schellenberg K., Haynes K.A., Viel A., Brenner T.J., Church G.M., Shah J.V., Silver P.A. A BioBrick compatible strategy for genetic modification of plants. Journal of Biological Engineering, 2012, 6(1): 8 (doi: 10.1186/1754-1611-6-8).
- Sainsbury F., Lomonossoff G.P. Transient expressions of synthetic biology in plants. Current Opinion in Plant Biology, 2014, 19: 1-7 (doi: 10.1016/j.pbi.2014.02.003).
- Yang Y., Chaffin T.A., Ahkami A.H., Blumwald E., Stewart C.N. Plant synthetic biology innovations for biofuels and bioproducts. Trends in Biotechnology, 2022, 40(12): 1454-1468 (doi: 10.1016/j.tibtech.2022.09.007).
- Chao R., Mishra S., Si T., Zhao H. Engineering biological systems using automated biofoundries. Metabolic Engineering, 2017, 42: 98-108 (doi: 10.1016/J.YMBEN.2017.06.003).
- Kim B.-C.C., Moon C., Jeon B.S., Angenent L.T., Choi Y., Nam K. Shaping a reactor microbiome generating stable n-caproate productivity through Design-Build-Test-Learn approach. Chemical Engineering Journal, 2021, 425: 131587 (doi: 10.1016/j.cej.2021.131587).
- Pham H.L., Ho C.L., Wong A., Lee Y.S., Chang M.W. Applying the design-build-test paradigm in microbiome engineering. Current Opinion in Biotechnology, 2017, 48: 85-93 (doi: 10.1016/j.copbio.2017.03.021).
- Pouvreau B., Vanhercke T., Singh S. From plant metabolic engineering to plant synthetic biology: The evolution of the design/build/test/learn cycle. Plant Science, 2018, 273: 3-12 (doi: 10.1016/j.plantsci.2018.03.035).
- Liao X., Ma H., Tang Y.J. Artificial intelligence: a solution to involution of design–build–test–learn cycle. Current Opinion in Biotechnology, 2022, 75: 102712 (doi: 10.1016/j.copbio.2022.102712).
- Carbonell P., Jervis A.J., Robinson C.J., Yan C., Dunstan M., Swainston N., Vinaixa M., Hollywood K.A., Currin A., Rattray N.J.W., Taylor S., Spiess R., Sung R., Williams A.R., Fellows D., Stanford N.J., Mulherin P., Le Feuvre R., Barran P., Goodacre R., Turner N.J., Goble C., Chen G.G., Kell D.B., Micklefield J., Breitling R., Takano E., Faulon J.L., Scrutton N.S. An automated Design-Build-Test-Learn pipeline for enhanced microbial production of fine chemicals. Communications Biology, 2018, 1(1): 66 (doi: 10.1038/s42003-018-0076-9).
- Gurdo N., Volke D.C., McCloskey D., Nikel P.I. Automating the design-build-test-learn cycle towards next-generation bacterial cell factories. New Biotechnology, 2023, 74: 1-15 (doi: 10.1016/j.nbt.2023.01.002).
- Opgenorth P., Costello Z., Okada T., Goyal G., Chen Y., Gin J., Benites V., de Raad M., Northen T.R., Deng K., Deutsch S., Baidoo E.E.K., Petzold C.J., Hillson N.J., Garcia Martin H., Beller H.R. Lessons from two design–build–test–learn cycles of dodecanol production in Escherichia coli aided by machine learning. ACS Synthetic Biology, 2019, 8(6): 1337-1351 (doi: 10.1021/acssynbio.9b00020).
- Rizzo P., Chavez B.G., Leite Dias S., D'Auria J.C. Plant synthetic biology: from inspiration to augmentation. Current Opinion in Biotechnology, 2023, 79: 102857 (doi: 10.1016/j.copbio.2022.102857).
- Cummins B., Vrana J., Moseley R.C., Eramian H., Deckard A., Fontanarrosa P., Bryce D., Weston M., Zheng G., Nowak J., Motta F.C., Eslami M., Johnson K.L., Goldman R.P., Myers C.J., Johnson T., Vaughn M.W., Gaffney N., Urrutia J., Gopaulakrishnan S., Biggers V., Higa T.R., Mosqueda L.A., Gameiro M., Gedeon T., Mischaikow K., Beal J., Bartley B., Mitchell T., Nguyen T.T., Roehner N., Haase S.B. Robustness and reproducibility of simple and complex synthetic logic circuit designs using a DBTL loop. Synth. Biol. (Oxf.), 2023, 8(1): ysad005 (doi: 10.1093/synbio/ysad005).
- Vidal G., Vidal-Céspedes C., Rudge T.J. LOICA: Logical Operators for Integrated Cell Algorithms. In: bioRxiv, Cold Spring Harbor Laboratory, 2021 (doi: 10.1101/2021.09.21.460548).
- HamediRad M., Chao R., Weisberg S., Lian J., Sinha S., Zhao H. Towards a fully automated algorithm driven platform for biosystems design. Nature Communications, 2019, 10(1): 5150 (doi: 10.1038/s41467-019-13189-z).
- Yang Y., Geng B., Song H., Hu M., He Q., Chen S., Bai F., Yang S. [Progress and perspective on development of non-model industrial bacteria as chassis cells for biochemical production in the synthetic biology era]. Sheng wu gong cheng xue bao = Chinese Journal of Biotechnology, 2021, 37(3): 874-910 (doi: 10.13345/j.cjb.200626).
- Feith A., Schwentner A., Teleki A., Favilli L., Blombach B., Takors R. Streamlining the analysis of dynamic 13C-labeling patterns for the metabolic engineering of Corynebacterium glutamicum as l-histidine production host. Metabolites, 2020, 10(11): 458 (doi: 10.3390/metabo10110458).
- Czajka J.J., Banerjee D., Eng T., Menasalvas J., Yan C., Munoz Munoz N., Poirier B.C., Kim Y.-Mo, Baker S.E., Tang Y.J., Mukhopadhyay A. Optimizing a high performing multiplex- CRISPRi P. putida strain with integrated metabolomics and 13C-metabolic flux analyses. In: bioRxiv, Cold Spring Harbor Laboratory, 2021 (doi: 10.1101/2021.12.23.473729).
- Lawson C.E. Retooling microbiome engineering for a sustainable future. mSystems, 2021, 6(4), e0092521 (doi: 10.1128/mSystems.00925-21).
- Vavricka C.J., Hasunuma T., Kondo A. Dynamic metabolomics for engineering biology: accelerating learning cycles for bioproduction. Trends in Biotechnology, 2020, 38(1): 68-82 (doi: 10.1016/j.tibtech.2019.07.009).
- Amer B., Baidoo E.E.K. Omics-driven biotechnology for industrial applications. Frontiers in Bioengineering and Biotechnology, 2021, 9: 613307 (doi: 10.3389/fbioe.2021.613307).
- Kim Y.-M., Petzold C.J., Kerkhoven E.J., Baker S.E. Editorial: Multi-omics technologies for optimizing synthetic biomanufacturing. Frontiers in Bioengineering and Biotechnology, 2021, 9: 818010 (doi: 10.3389/fbioe.2021.818010).
- Pathania R., Srivastava A., Srivastava S., Shukla P. Metabolic systems biology and multi-omics of cyanobacteria: Perspectives and future directions. Bioresource Technology, 2022, 343: 126007 (doi: 10.1016/j.biortech.2021.126007).
- Gurdo N., Volke D.C., Nikel P.I. Merging automation and fundamental discovery into the design–build–test–learn cycle of nontraditional microbes. Trends in Biotechnology, 2022, 40(10): 1148-1159 (doi: 10.1016/j.tibtech.2022.03.004).
- St. John P.C., Bomble Y.J. Approaches to computational strain design in the multiomics era. Frontiers in Microbiology, 2019, 10: 597 (doi: 10.3389/fmicb.2019.00597).
- Pomraning K.R., Dai Z., Munoz N., Kim Y.-M., Gao Y., Deng S., Kim J., Hofstad B.A., Swita M..S, Lemmon T., Collett J.R., Panisko E.A., Webb-Robertson B.M., Zucker J.D., Nicora C.D., De Paoli H., Baker S.E., Burnum-Johnson K.E., Hillson N.J., Magnuson J.K. Integration of proteomics and metabolomics into the design, build, test, learn cycle to improve 3-hydroxypropionic acid production in Aspergillus pseudoterreus. Frontiers in Bioengineering and Biotechnology, 2021, 9: 603832 (doi: 10.3389/fbioe.2021.603832).
- Dinglasan J.L.N., Reeves D.T., Hettich R.L., Doktycz M.J. Liquid chromatography coupled to refractive index or mass spectrometric detection for metabolite profiling in lysate-based cell-free systems. Journal of Visualized Experiments, 2021, (175): 10.3791/62852 (doi: 10.3791/62852).
- Garagounis C., Delkis N., Papadopoulou K.K. Unraveling the roles of plant specialized metabolites: using synthetic biology to design molecular biosensors. New Phytologist, 2021, 231(4): 1338-1352 (doi: 10.1111/nph.17470).
- Whitford C.M., Cruz-Morales P., Keasling J.D., Weber T. The Design-Build-Test-Learn cycle for metabolic engineering of Streptomycetes. Essays in Biochemistry, 2021, 65(2): 261-275 (doi: 10.1042/EBC20200132).
- Lawson C.E., Harcombe W.R., Hatzenpichler R., Lindemann S.R., Löffler F.E., O'Malley M.A., García Martín H., Pfleger B.F., Raskin L., Venturelli O.S., Weissbrodt D.G., Noguera D.R., McMahon K.D. Common principles and best practices for engineering microbiomes. Nature Reviews Microbiology, 2019, 17(12): 725-741 (doi: 10.1038/s41579-019-0255-9).
- Gamboa-Melendez H., Larroude M., Park Y.K., Trebul P., Nicaud J.-M., Ledesma-Amaro R. Synthetic biology to improve the production of lipases and esterases (review). Methods Mol. Biol., 2018, 1835: 229-242 (doi: 10.1007/978-1-4939-8672-9_13).
- Kothamachu V.B., Zaini S., Muffatto F. Role of digital microfluidics in enabling access to laboratory automation and making biology programmable. SLAS Technology, 2020, 25(5): 411-426 (doi: 10.1177/2472630320931794).
- Shih S.C.C., Moraes C. Next generation tools to accelerate the synthetic biology process. Integrative Biology, 2016, 8(5): 585-588 (doi: 10.1039/C6IB90017H).
- Sohrabi S., Kassir N., Keshavarz Moraveji M. Droplet microfluidics: fundamentals and its advanced applications. RSC Advances, 2020, 10(46): 27560-27574 (doi: 10.1039/D0RA04566G).
- Arazoe T., Kondo A., Nishida K. Targeted nucleotide editing technologies for microbial metabolic engineering. Biotechnology Journal, 2018, 13(9): 1700596 (doi: 10.1002/biot.201700596).
- Baltes N.J., Voytas D.F. Enabling plant synthetic biology through genome engineering. Trends in Biotechnology, 2015, 33(2): 120-131 (doi: 10.1016/j.tibtech.2014.11.008).
- Delépine B., Duigou T., Carbonell P., Faulon J.-L. RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metabolic Engineering, 2018, 45: 158-170 (doi: 10.1016/j.ymben.2017.12.002).
- Carbonell P., Wong J., Swainston N., Takano E., Turner N.J., Scrutton N.S., Kell D.B., Breitling R., Faulon J.L. Selenzyme: enzyme selection tool for pathway design. Bioinformatics, 2018, 34(12): 2153- 2154 (doi: 10.1093/bioinformatics/bty065).
- Swainston N., Dunstan M., Jervis A.J., Robinson C.J., Carbonell P., Williams A.R., Faulon J.L., Scrutton N.S., Kell D.B. PartsGenie: an integrated tool for optimizing and sharing synthetic biology parts. Bioinformatics, 2018, 34(13): 2327-2329 (doi: 10.1093/bioinformatics/bty105).
- Fields-Johnson C., Fike J., Galbraith J., Maguire R., Day S., Zedaker S., Mathis J. Pine sawdust biochar as a potential amendment for establishing trees in Appalachian mine spoils. REFORESTA, 2018, (6): 1-14 (doi: 10.21750/REFOR.6.01.54).
- Yáñez Feliú G., Earle Gómez B., Codoceo Berrocal V., Muñoz Silva M., Nuñez I.N., Matute T.F., Arce Medina A., Vidal G., Vitalis C., Dahlin J, Federici F, Rudge TJ.Flapjack: data management and analysis for genetic circuit characterization. ACS Synthetic Biology, 2021, 10(1): 183-191 (doi: 10.1021/acssynbio.0c00554).
- Orsi E., Claassens N.J., Nikel P.I., Lindner S.N. Growth-coupled selection of synthetic modules to accelerate cell factory development. Nature Communications, 2021, 12(1): 5295 (doi: 10.1038/s41467-021-25665-6).
- Gao Y., Fillmore T.L., Munoz N., Bentley G.J., Johnson C.W., Kim J., Meadows J.A., Zucker J.D., Burnet M.C., Lipton A.K., Bilbao A., Orton D.J., Kim Y.M., Moore R.J., Robinson E.W., Baker S.E., Webb-Robertson B.M., Guss A.M., Gladden J.M., Beckham G.T., Magnuson J.K., Burnum-Johnson K.E. High-throughput large-scale targeted proteomics assays for quantifying pathway proteins in Pseudomonas putida KT2440. Frontiers in Bioengineering and Biotechnology, 2020, 8: 603488 (doi: 10.3389/fbioe.2020.603488).
- Roy S., Radivojevic T., Forrer M., Marti J.M., Jonnalagadda V., Backman T., Morrell W., Plahar H., Kim J., Hillson N., Garcia Martin H. Multiomics data collection, visualization, and utilization for guiding metabolic engineering. Frontiers in Bioengineering and Biotechnology, 2021, 9: 612893 (doi: 10.3389/fbioe.2021.612893).
- Zhang J., Chen Y., Fu L., Guo E., Wang B., Dai L., Si T. Accelerating strain engineering in biofuel research via build and test automation of synthetic biology. Current Opinion in Biotechnology, 2021, 67: 88-98 (doi: 10.1016/j.copbio.2021.01.010).
- Radivojević T., Costello Z., Workman K., Garcia Martin H. A machine learning Automated Recommendation Tool for synthetic biology. Nature Communications, 2020, 11(1): 4879 (doi: 10.1038/s41467-020-18008-4).
- Kamran S., Shahid I., Baig D.N., Rizwan M., Malik K.A., Mehnaz S. Contribution of Zinc solubilizing bacteria in growth promotion and zinc content of wheat. Frontiers in Microbiology, 2017, 8: 2593 (doi: 10.3389/fmicb.2017.02593).
- Araki M., Cox R.S., Makiguchi H., Ogawa T., Taniguchi T., Miyaoku K., Nakatsui M., Hara K.Y., Kondo A. M-path: a compass for navigating potential metabolic pathways. Bioinformatics, 2015, 31(6): 905-911, (10.1093/bioinformatics/btu750).
- Nakazawa S., Imaichi O., Kogure T., Kubota T., Toyoda K., Suda M., Inui M., Ito K., Shirai T., Araki M. History-driven genetic modification design technique using a domain-specific lexical model for the acceleration of DBTL cycles for microbial cell factories. ACS Synthetic Biology, 2021, 10(9): 2308-2317 (doi: 10.1021/acssynbio.1c00234).
- Dunstan M.S., Robinson C.J., Jervis A.J., Yan C., Carbonell P., Hollywood K.A., Currin A., Swainston N., Feuvre R.L., Micklefield J., Faulon J.L., Breitling R., Turner N., Takano E., Scrutton N.S. Engineering Escherichia coli towards de novo production of gatekeeper (2S)- flavanones: naringenin, pinocembrin, eriodictyol and homoeriodictyol. Synthetic Biology, 2020, 5(1): ysaa012 (doi: 10.1093/synbio/ysaa012).
- Ro D.-K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M., Ho K.A., Eachus R.A., Ham T.S., Kirby J., Chang M.C., Withers S.T., Shiba Y., Sarpong R., Keasling J.D. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature, 2006, 440(7086): 940-943 (doi: 10.1038/nature04640).
- Westfall P.J., Pitera D.J., Lenihan J.R., Eng D., Woolard F.X., Regentin R., Horning T., Tsuruta H., Melis D.J., Owens A., Fickes S., Diola D., Benjamin K.R., Keasling J.D., Leavell M.D., McPhee D.J., Renninger N.S., Newman J.D., Paddon C.J. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proceedings of the National Academy of Sciences, 2012, 109(3): E111-118 (doi: 10.1073/pnas.1110740109).
- Chen R., Yang S., Zhang L., Zhou Y.J. Advanced strategies for production of natural products in yeast. iScience, 2020, 23(3): 100879 (doi: 10.1016/j.isci.2020.100879).
- Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell, 2016, 164(6): 1185-1197 (doi: 10.1016/j.cell.2016.02.004).
- Zhu X., Du C., Mohsin A., Yin Q., Xu F., Liu Z., Wang Z., Zhuang Y., Chu J., Guo M., Tian X. An efficient high-throughput screening of high gentamicin-producing mutants based on titer determination using an integrated Computer-Aided Vision Technology and Machine Learning. Analytical Chemistry, 2022, 94(33): 11659-11669 (doi: 10.1021/acs.analchem.2c02289).
- Liu J.-M., Chen L., Jensen P.R., Solem C. Food grade microbial synthesis of the butter aroma compound butanedione using engineered and non-engineered Lactococcus lactis. Metabolic Engineering, 2021, 67: 443-452 (doi: 10.1016/j.ymben.2021.08.006).
- Ramzi A.B., Baharum S.N., Bunawan H., Scrutton N.S. Streamlining natural products biomanufacturing with omics and machine learning driven microbial engineering. Frontiers in Bioengineering and Biotechnology, 2020, 8: 608918 (doi: 10.3389/fbioe.2020.608918).
- Carbonell P., Le Feuvre R., Takano E., Scrutton N.S. In silico design and automated learning to boost next-generation smart biomanufacturing. Synthetic Biology, 2020, 5(1): ysaa020 (doi: 10.1093/synbio/ysaa020).
- Freemont P.S. Synthetic biology industry: data-driven design is creating new opportunities in biotechnology. Emerging Topics in Life Sciences, 2019, 3(5): 651-657 (doi: 10.1042/ETLS20190040).
- Zhu Q., Yu S., Zeng D., Liu H., Wang H., Yang Z., Xie X., Shen R., Tan J., Li H., Zhao X., Zhang Q., Chen Y., Guo J., Chen L., Liu Y.G. Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Molecular Plant, 2017, 10(7): 918-929 (doi: 10.1016/j.molp.2017.05.008).
- Lin J., Lin W., Ng I. CRISPRa/i with Adaptive Single Guide Assisted Regulation DNA (ASGARD) mediated control of Chlorella sorokiniana to enhance lipid and protein production. Biotechnology Journal, 2022, 17(10): 2100514 (doi: 10.1002/biot.202100514).
- Kuivanen J., Holmström S., Lehtinen B., Penttilä M., Jäntti J. A high-throughput workflow for CRISPR/Cas9 mediated combinatorial promoter replacements and phenotype characterization in yeast. Biotechnology Journal, 2018, 13(9): 1700593 (10.1002/biot.201700593).
- Liu Z., Wang J., Nielsen J. Yeast synthetic biology advances biofuel production. Current Opinion in Microbiology, 2022, 65: 33-39 (doi: 10.1016/j.mib.2021.10.010).
- Czajka J.J., Banerjee D., Eng T., Menasalvas J., Yan C., Munoz N.M., Poirier B.C., Kim Y.M., Baker S.E., Tang Y.J., Mukhopadhyay A. Tuning a high performing multiplexed-CRISPRi Pseudomonas putida strain to further enhance indigoidine production. Metabolic Engineering Communications, 2022, 15: e00206 (doi: 10.1016/j.mec.2022.e00206).
- Vidal G., Vidal-Céspedes C., Rudge T.J. LOICA: integrating models with data for genetic network design automation. ACS Synthetic Biology, 2022, 11(5): 1984-1990 (doi: 10.1021/acssynbio.1c00603).
- Li X.X., Lan C., Li X.X., Hu Z., Jia B. A review on design-build-test-learn cycle to potentiate progress in isoprenoid engineering of photosynthetic microalgae. Bioresource Technology, 2022, 363: 127981 (doi: 10.1016/j.biortech.2022.127981).
- Zhang M., Holowko M.B., Hayman Zumpe H., Ong C.S. Machine learning guided Batched design of a bacterial ribosome binding site. ACS Synthetic Biology, 2022, 11(7): 2314-2326 (doi: 10.1021/acssynbio.2c00015).
- Kamminga T., Slagman S.-J., Martins dos Santos V.A.P., Bijlsma J.J.E., Schaap P.J. Risk-based bioengineering strategies for reliable bacterial vaccine production. Trends in Biotechnology, 2019, 37(8): 805-816 (doi: 10.1016/j.tibtech.2019.03.005).
- Ando D., García Martín H. Genome-scale 13C fluxomics modeling for metabolic engineering of Saccharomyces cerevisiae. Methods Mol. Biol., 2019, 1859: 317-345 (doi: 10.1007/978-1-4939-8757-3_19).
- Sabzevari M., Szedmak S., Penttilä M., Jouhten P., Rousu J. Strain design optimization using reinforcement learning. PLOS Computational Biology, 2022, 18(6): e1010177 (doi: 10.1371/journal.pcbi.1010177).


