Разнообразие алкалоидов и вирулентность спорыньи Claviceps purpurea (fries) Tulasne: эволюция, генетическая диверсификация, метаболическая инженерия (обзор)
Автор: Волнин А.А., Савин П.С.
Журнал: Сельскохозяйственная биология @agrobiology
Рубрика: Обзоры, проблемы, итоги
Статья в выпуске: 5 т.57, 2022 года.
Бесплатный доступ
Спорынья Claviceps purpurea (Fries) Tulasne имеет важнейшее хозяйтвенное значение: это продуцент большого количества биологически активных соединений - алкалоидов, уникальная модель системы паразит-хозяин, а также патоген, наносящий значительный экономический ущерб сельскому хозяйству. Место происхождения спорыньи - Южная Америка (в палеоцене), возраст Claviceps оценивается в 20,4 млн лет (K. Píchová с соавт., 2018). Внутривидовое разнообразие и дивергенция генов кластера синтеза индольных алкалоидов у спорыньи происходили согласно эволюционной модели «песочных часов» (M. Liu с соавт., 2021). Выделены и охарактеризованы основные эргоалкалоиды C. purpurea - эргометрин, эргозин, эрготамин, a-эргокриптин, эргокорнин, эргокристин и их 8-S(-инин-) эпимеры (они составляли не менее 50 % от общего извлеченного метаболома алкалоидов) (S. Uhlig с соавт., 2021). Показано разное число генов алкалоидного кластера у Claviceps , наличие двух-трех копий генов dmaW , easE , easF , а также факты частых приобретений и потерь генов (M. Liu с соавт., 2021). Различия в метаболомных профилях алкалоидов C. purpurea коррелировали с различиями в гене lpsA : разнообразие алкалоидов спорыньи обусловлено вариабельностью последовательностей в тандемно дублированной области easH / lpsA (C. Hicks с соавт., 2021). Гены lpsA1 и lpsA2 были результатом события рекомбинации (S. Wyka с соавт., 2022). Предполагается, что гены lpsA подвергаются рекомбинационному перетасовыванию (C. Hicks с соавт., 2021). Для C. purpurea показаны высокие скорости рекомбинации (ρ = 0,044), относительно большой акцессорный геном (38 %) и транспозон-опосредованная дупликация генов (S. Wyka с соавт., 2022). Разработана трансгенная линия дрожжей, синтезирующая энантиочистую D-лизергиновую кислоту до титра 1,7 мг/л (G. Wong с соавт., 2022). Генно-инженерные культуры Metarhizium brunneum дают относительный процент выхода D-лизергиновой кислоты 86,9 % и дигидролизиргиновой кислоты 72,8 % (K. Davis с соавт., 2020). Экспрессия генов trpE , а также dmaW количественно связана с интенсивностью синтеза алкалоидов у сапрофитных культур спорыньи (M. Králová с соавт., 2021). Пектин служит основной мишенью CAZymes белков, ответственных за деградацию клеточной стенки при инфицировании растения C. purpurea и C. paspali (B. Oeser с соавт., 2017; H. Oberti с соавт., 2021). Значительный вклад в вирулентность спорыньи вносят полигалактуроназа, MAP-киназа, фактор регуляции транскрипции CPTF1 (ген Cptf1 ), малая GTP-аза (ген Cdc42 ) (B. Oeser с соавт., 2017; E. Tente с соавт., 2021). Спорынья влияет на гормональные пути растения с участием ауксина, этилена и цитокинина (эффект дифференцирован относительно типа ткани и времени после заражения) (E. Tente, 2020, Tente с соавт., 2021). У пшеницы устойчивость к спорынье связана с мутациями в белках DELLA (E. Tente, 2020; A. Gordon с соавт., 2020), у ржи - с активностью пектинэстеразы и метаболическими процессами модификации клеточной стенки и роста пыльцевых трубок (COBRA-подобный белок и ингибитор пектинэстеразы) (K. Mahmood с соавт., 2020).
Claviceps purpurea, спорынья, алкалоиды, пути биосинтеза, токсичность, вирулентность, генотип, генные кластеры, claviceps
Короткий адрес: https://sciup.org/142236375
IDR: 142236375 | УДК: 632.4.01/.08:579.64:577 | DOI: 10.15389/agrobiology.2022.5.852rus
Текст обзорной статьи Разнообразие алкалоидов и вирулентность спорыньи Claviceps purpurea (fries) Tulasne: эволюция, генетическая диверсификация, метаболическая инженерия (обзор)
Недавние достижения в геномике и молекулярной биологии позволили идентифицировать гены, кодирующие белки, которые катализируют последовательные этапы биосинтеза алкалоидов у грибов (1). Разнообразие вторичных метаболитов грибов может быть расширено за счет активации молчащих кластеров генов у искусственно культивируемых штаммов или пополнения существующих коллекций грибных культур с целью получения новых натуральных биологических продуктов (1, 2).
Спорынья — паразитарный гриб Claviceps purpurea (Fries) Tulasne — служит важным источником биологически активных веществ для производства лекарственных препаратов (3-5). Природные эргоалкалоиды, а также их полусинтетические производные используются в качестве фармацевтических средств в современной медицине, например в акушерстве и гинекологии (6) в
∗ Работа выполнена в рамках темы 10.6. Защита и биотехнология растений (FGUU-2022-0014).
качестве ингибиторов высвобождения пролактина (7, 8), а также для лечения ряда неврологических заболеваний (9), в частности паркинсонизма (9, 10), деменции (9, 11) и гипертонии (9, 12). Недавние исследования указывают на высокую перспективность алкалоидов спорыньи (метерголиновые производные) для создания новых антимикробных средств (13). В России при участии ФГБНУ ВИЛАР разработан ряд лекарственных препаратов на основе алкалоидов спорыньи: абергин ( α , β-эргокриптины), новокристин (дигидроэргокристин), беллатаминал (эрготамин), малеат эргометрина (эргометрин) (14).
Коллекция ФГБНУ ВИЛАР включает пять штаммов паразитарной культуры спорыньи Claviceps purpurea (Fries) Tulasne (продуценты α , β-эр-гокриптинов, эрготамина, эрготоксина, эргокристина и эргокорнама). Все паразитарные штаммы поддерживаются в активном состоянии, в том числе проходят этап сезонного культивирования в полевых условиях на посевах озимой ржи (сорт Московская 12) и промежуточный этап in vitro в виде ак-сенической культуры. Кроме того, зарегистрированы два сапрофитных штамма спорыньи (продуцируют эргокриптин и эрготамин), которые также поддерживаются в условиях живой коллекции (15-17).
В сельском хозяйстве из-за спорыньи происходят значительные экономические потери. Спорынья представляет опасность для здоровья не только животных, но и человека (18, 19). В мировой практике загрязнение продукции, кормов для животных, пастбищных культур токсинами спорыньи — довольно частое явление, а токсический эффект от воздействия эргоалкалоидов на обмен веществ животного достаточно хорошо изучен (20-22), в том числе in vitro (23, 24). Кроме того, токсическим эффектом могут обладать не только алкалоиды, но и другие вторичные метаболиты спорыньи, например, эргопигменты (25), леканоровая кислота и ее производные (24) индол-дитерпены (25-28), эпиполитиодикетопиперазины (29).
Количественные характеристики и состав метаболома эргоалкалои-дов могут сильно варьироваться в зависимости от конкретных условий развития гриба и его вида, что затрудняет мониторинг содержания эргоалка-лоидов и, следовательно, оценку степени контаминации сельскохозяйственной продукции эрготоксинами (30, 31). У растений пшеницы и ячменя алкалоиды проникают и в здоровое зерно, которое формируется выше и ниже инфицированных участков соцветия (32).
Вопросы токсикологии и вторичного метаболизма спорыньи в последнее десятилетие все более актуализируются. В научных обзорах рассматриваются технологические и физиологические особенности культивирования спорыньи (15, 16), функциональные характеристики ключевых генов и генных кластеров, участвующих в биосинтезе алкалоидов у разных родов семейства Спорыньевые ( Clavicepitaceae ) (30, 33-35), обсуждаются идентификация и классификация промежуточных продуктов синтеза (36, 37), конструкции путей образования эргоалкалоидов у гриба и различных организмов-гетерологов, стратегии оптимизации создания и использования рекомбинантных штаммов-продуцентов. Описаны факторы вирулентности спорыньи (32, 38, 39). Публикуемые аналитические материалы (39) уточняются и дополняются (40, 41).
В представляемом обзоре мы детализировали и систематизировали результаты изучения механизмов биосинтеза алкалоидов и вирулентности спорыньи у разных представителей рода Claviceps с учетом новых данных о роли генов кластера биосинтеза алкалоидов и вирулентности в эволюции и распространении спорыньи, уделив внимание методологическим аспек- там, в том числе метаболической инженерии. Такой комплексный подход к пониманию генетической диверсификации спорыньи, связанного с этим разнообразия алкалоидов, токсинов и знание молекулярных механизмов их биосинтеза расширяют представления о биологии рода Claviceps и формируют базу для решения практических задач как в сельском хозяйстве, так и при создании лекарственных препаратов на основе метаболитов спорыньи.
Эволюция, распространение и роль токсических метаболитов спорыньи. Молекулярно-генетические технологии позволяют изучать биологию спорыньи на уровне генных кластеров, геномов, транскрип-томов, метаболомов, используя широкий арсенал средств. Для анализа генома спорыньи применяют пиросеквенирование методом одно- или парноконцевых прочтений (single/paired-end pyrosequencing) (42), секвенирование с парноконцевыми прочтениями (paired-end sequencing, PEs) (43, 44), секвенирование по Сэнгеру (Sanger sequencing) (45), пиросеквенирование методом дробовика (shotgun pyrosequencing) и секвенирование спаренных концов (matepair sequencing) (46). Так, PEs применяли для анализа транскриптома при сигнальных взаимодействиях C. purpurea с хозяином Secale cereale (47) и секвенирования генома Claviceps paspali (48), мультиплексное секвенирование с одноконцевыми прочтениями (single-end multiplexed sequencing) — для анализа транскриптома пшеницы при заражении C. purpurea (49).
Citrinae , Paspalorum , Pusillae характеризуются узким диапазоном распространения и низкой токсичностью, в то время как секция Claviceps считается эволюционно более успешной и способной к адаптации, поскольку обладает самым широким кругом растений-хозяев и ареалом (43, 50, 53). В настоящее время классификации и систематическое положение природных паразитарных штаммов спорыньи продолжают уточнять (54-56).
Популяция спорыньи в США отличается от европейских изолятов (57). Утверждается, что в США отсутствуют субпопуляции этого паразитического гриба, формируемые в зависимости от растения-хозяина (54, 57, 58). В США отмечаются умеренные уровни генотипического разнообразия (H = 3,43-4,23) и генного разнообразия (Hexp = 0,45-0,57) (59). Наблюдается генетическая дифференциация между североамериканскими популяциями от разных растений-хозяев (22 %), но она смешана по географическому принципу. Стандартизированный индекс ассоциации варьировался от 0,007 до 0,122 для четырех групп (два региона и два растения-хозяина — мятлик кентукийский и райграс пастбищный) (59). Ранее были выявлены три гене- тические линии C. purpurea (60), отражающие экологическую дифференциацию и адаптацию (53, 61, 62). Линии, обозначенные как экотипы G1, G2 и G3, были дифференцированы на основе морфологии конидий, профилей алкалоидов, RAPD (random amplified polymorphic DNA, случайно амплифи-цированная полиморфная ДНК) маркеров и способности или неспособности склероциев сохранять плавучесть (53, 61, 63). В настоящее время экотипы C. purpurea G1, G2 и G3 описаны как три самостоятельных вида — соответственно C. purpurea sensu stricto, C. humidiphila и C. spartinae (53, 60).
Причины эволюционного успеха и высокой адаптивности секции Claviceps до конца не ясны (43). До некоторой степени это можно объяснить продукцией большого количества токсичных метаболитов, что ограничивает поедание пораженных грибом растений позвоночными и беспозвоночными животными и важно для мутуалистических отношений паразита и хозяина, обеспечивающих защиту растения от фитофагов (50). Однако этот вопрос остается предметом дискуссии (40).
По результатам филогенетического анализа и анализа полиморфизма ДНК во внутривидовом разнообразии спорыньи для дивергенции генов кластера индольных алкалоидов (EAS) установлено соответствие эволюционной модели «песочных часов» (hourglass model, HGM) (44), позаимствованной из описания процессов в онтогенезе, когда морфологические расхождения на средних стадиях развития эмбриона более консервативны, чем на более ранних и поздних стадиях (44, 64, 65). Так, скорость эволюции генов ранних стадий синтеза алкалоидов dmaW и easF намного выше, чем генов промежуточных стадий easA , easС , easD и easЕ (44). Этот паттерн, подтвержденный геномными исследованиями, совпадает с моделью песочных часов (44, 66). Модель HGM предполагает наличие ограничений для развития (44, 67). Метаболические пути синтеза алкалоидов рассматривались как необычайно неэффективные из-за того, что многие промежуточные соединения накапливаются в большем количестве, чем необходимо для образования конечных продуктов (44, 68). Однако такая избыточность промежуточных соединений, вероятно, может служить фактором стабилизации биосинтетического пути и затруднять отбор вариантов с изменениями в соответствующих генах (44).
Разнообразие алкалоидов Claviceps было сформировано тремя основными процессами: приобретением генов, утратой генов и изменением последовательности генов биосинтеза алкалоидов (35, 46).
Изменения архитектуры и пластичность генома могут формировать направление эволюционного процесса грибов и их адаптивность (43). Предположительно именно вторичные метаболиты спорыньи служат факторами первичного влияния на диверсификацию и продвижение вида в новые экологические ниши и помогают поддерживать его глобальное распространение и широкий круг хозяев (42). Состав кластера генов синтеза алкалоидов и уникальные полиморфизмы показывают, что в настоящее время C. purpurea претерпевает процесс адаптации, следствие которой — большое разнообразие пептидных алкалоидов (45). Таким образом, эволюция спорыньи направлена от специализированных геномов (Citrinae и Paspalorum) к адаптивным (Pusillae и Claviceps). Этому способствует совместная локализация транспонируемых элементов вокруг эффекторов. Также предполагается, что в секции Claviceps произошла утрата точечной мутации RIP (repeat-indused point mutation), что привело к неограниченному тандемному дублированию генов, которое соответствует увеличению потенциала расширения круга хозяев и видообразования (43). Результаты показывают отсутствие дуплика- ции генов в секциях Citrinae и Paspalorum, вероятно, из-за наличия RIP-подобных механизмов, что соответствует высокой специфичности к растению-хозяину и низкому видовому разнообразию Paspalorum (43). Неясно, дали ли такие перестройки преимущество Claviceps при переходе на новых хозяев и в новые климатические условия при разделении секций и выходе из Южной Америки или были следствием этого события (43).
Для C. purpurea показаны достоверно высокие скорости рекомбинации (ρ = 0,044), относительно большой акцессорный геном (38 %) и транспозон-опосредованная дупликация генов (42). Важно отметить, что при этом общее содержание транспонзируемых элементов в геноме спорыньи относительно низкое (8,8 %), размер генома не варьирует (42), а рекомбинация не является основным фактором дупликации у C. purpurea (42). Псевдогенизация и неофункционализация также могут быть существенными процессами. Из-за отсутствия RIP-мутаций увеличение количества транспозируемых элементов, вероятно, контролируется высокими скоростями рекомбинации (42).
Мы считаем, что модель «песочных часов» в эволюционной траектории кластера генов алкалоидной продуктивности спорыньи с большой долей вероятности указывает на то, что паразитарные штаммы, перспективные с точки зрения биопродукции алкалоидов, могут быть ослаблены под действием естественного отбора и утрачены.
Идентификация штаммов спорыньи. Для быстрой идентификации штаммов C. purpurea разработаны SSR (simple sequence repeats, короткие простые повторы, микросателлиты) маркеры. Помимо различения изолятов C. purpurea , эти SSR могут дифференировать изоляты трех других видов рода Claviceps — C. pusilla , C. paspali и C. fusiformis . (69). Для генотипирования может применяться мультилокусное сиквенс-типирование (multilocus sequence typing, MLST) в сочетании с количественной полимеразной цепной реакцией в реальном времени (qPCR) (70).
Некоторые из алкалоидов специфичны для определенных штаммов C. purpurea и могут использоваться в качестве хемотаксономических маркеров при идентификации (71).
Алкалоиды спорыньи и их продуценты. В настоящее время производство алкалоидов спорыньи с различной фармакологической активностью основано преимущественно на ферментации с использованием сапрофитных культур (9, 17, 36). Приблизительно 60 % производимых в промышленных масштабах эргоалкалоидов получают методом глубинного (погружного) культивирования специально созданных мутантных или рекомбинантных штаммов C. purpurea или организмов-гетерологов на жидких питательных средах, остальные 40 % — при культивировании спорыньи в поле (9, 72, 73).
Основные проблемы производства эргоалкалоидов — большое разнообразие синтезируемых алкалоидов, что усложняет последующую экстракцию и очистку от побочных продуктов и увеличивает стоимость производства (9, 36), а также нестабильность и склонность к деградации сапрофитных штаммов в процессе культивирования и хранения (9, 36, 72, 73). Сапрофитные штаммы утрачивают склероциоподобную клеточную морфологию мицелия с необратимой потерей способности продуцировать алкалоиды (72, 73). Современные методы химического синтеза D-лизергиновой кислоты включают длительные многостадийные реакции в жестких условиях и не всегда энантиоселективны (3, 9, 74, 75).
Паразитический этап жизненного цикла спорыньи характеризуется формированием склероциев — единственного вегетативного покоящегося органа гриба, в котором синтезируются алкалоиды (76). Склероции спорыньи, собранные в разных географических зонах, могут служить ценным исходным материалом для селекции высокопродуктивных штаммов (77).
Для культивирования спорыньи крайне важное значение имеет чередование паразитической стадии жизненного цикла и аксенической культуры (пересев склероция на питательную среду in vitro с возможностью получения сапрофитного мицелия со склероциеподобной морфологией и пурпурной пигментацией) (40). Морфологический отбор в аксенической культуре позволяет получить плектенхиматическую форму мицелия, которая напоминает раннюю склероциальную стадию инфицирования ржи, продуцирующую лизергиновую кислоту и пептидные алкалоиды в условиях погружного культивирования (40, 78, 79). Лизергиновую кислоту in vitro продуцируют также изоляты паразитарных штаммов, отобранных в нетипичных условиях обитания (40, 80). Однако это редкое явление: отбор по признакам морфологии требует культивирования большого количества склероциев в аксенической культуре, обнаруженный мицелий плохо отделяется, а полученные изоляты крайне чувствительны к условиям культивирования, что делает процесс очень трудоемким и зависящим от большого числа различных факторов (17, 40).
Для получения ауксотрофных мутантных штаммов спорыньи или мутантов с измененной продукцией эргоалкалоидов (7) может применяться УФ-мутагенез (7, 16, 81-83) или химические мутагены (7, 16), в частности азотистая кислота (7, 84) или N-метил-N´-нитро-N-нитрозогуанидин (7, 85).
Среди способов получения рекомбинантных штаммов-продуцентов спорыньи отметим технологию геномного редактирования CRISPR/Cas9 (7, 86, 87), полиэтиленгликоль (PEG)-опосредованную трансформацию (7, 88), агробактериальную трансформацию с использованием Agrobacterium tumefaciens (ATMT) (27). Совершенствование методов генной инженерии, основанных на гомологичной рекомбинации (HR) (89-91), позволяет с достаточно высокой эффективностью получать дизайнерские линии спорыньи (8, 47) и организмов-гетерологов (73), в том числе с повышенным уровнем синтеза целевых алкалоидов (8, 73).
Методы отбора проб, выделения, очистки, обнаружения, количественной оценки алкалоидов спорыньи в продуктах питания, кормах, растительных материалах и тканях животных очень подробно описаны, стандартизированы и валидированы (92-94). Для количественного определения используют жидкостную хроматографию с флуоресцентным (LC-FLD) или масс-пектрометрическим (LC-МС/МС) детектированием (92, 94). Реже применяется жидкостная хроматография с УФ-детекцией (LC-UV) (92) и имму-ноферментный анализ (ELISA) (92, 94, 95). Также применяются экспресс-методы — количественный анализ с реактивом Ван Урка (van Urk reagent, колориметрическое неселективное определение суммы индольных алкалоидов) (40, 94) и тонкослойная хроматография (40).
Разнообразие и биологию синтеза алкалоидов спорыньи важно учитывать при их обнаружении в сельскохозяйственной продукции (96). Количественное сравнение продукции эргоалкалоидов у 13 видов Claviceps, образцы которых были собраны в природных сообществах и в агроценозах трав и зерновых в Европе, Северной Америке, Новой Зеландии и Южной Африке (25), привело авторов к выводу о необходимости пересмотреть представления о дозах и механизмах токсичности вторичных метаболитов спорыньи. По разным данным, у C. purpurea s. l. среднее содержание алкалоидов в склероциях варьирует от 0,01 -1,3 мг/г (61, 96-98) до 2,887,26 мг/г (99), но иногда достигает значения 5-10 мг/г (61, 96, 97, 100).
Получены масс-спектры 67 пептидных алкалоидов спорыньи (71). Выявлены основные эргоалкалоиды: эргометрин, эргозин, эрготамин, а -эр-гокриптин, эргокорнин, эргокристин и их 8-8(-инин-) эпимеры, которые составляли не менее 50 % от общего выделенного метаболома алкалоидов (25, 71, 95). Матричная лазерная десорбционно-ионизационная масс-спектрометрическая визуализация (MALDI-MSI) позволила оценить распределение двух репрезентативных алкалоидов (эргокристина и эргометрина), продуцируемых разными штаммами C. purpurea при заражении ржи. Эргометрин демонстрирует относительно равномерное распределение по всему склероцию, в то время как эргокристин сконцентрирован в проксимальной области (101).
Кластер генов биопродукции алкалоидов. О кластерной организации генов биосинтеза алкалоидов у спорыньи впервые сообщили в 1999 году, и в частности было показано значение гена dmaW для биосинтеза (8, 102). Кластеры генов биосинтеза эргоалкалоидов обнаружены у различных грибов (30), например у Clavicipitaceae (30, 103, 104), в частности у
Claviceps (30, 102, 103), Epichloe (20, 30), Periglandula (3), Metarhizium brun-neum (86, 105), Neotyphodium lolli (106), Balansia cyperi, Balansia obtecta (30); у Aspergillus (107, 108), в частности у Aspergillus fumigatus (107, 110), A. leporis, A. homomorphus, A. hancockii (111) и A. japonicus (112, 113); у Clavulinopsis fusiformis (106); у Arthroderma benhamiae (114, 115); у Penicillium camemberti и Penicillium biforme (116).
Эргоалкалоиды представлены тремя основными классами (34, 36, 37). У алкалоидов клавинового типа (эргоклавины) структурный каркас сформирован альдегидом ханоклавина-I. Ханоклавин-I, а также некоторые другие эргоклавины, например агроклавин и элимоклавин, тоже служат промежуточными продуктами синтеза эргоамидов и эргопептидов. Второй класс — это энантиомеры лизергиновой кислоты и эргоамиды: D-лизергиновая кислота и ее амиды, в том числе эргометрин. Лактамные алкалоиды и эргопептиды — самые сложные соединения, представляющие самый многочисленный и разнообразный класс эргоалкалоидов (34, 36, 37). Фармакологический эффект алкалоидов спорыньи связывают с молекулярным сходством между их эрголиновым скелетом и моноаминовыми нейротрансмиттерами — адреналином, дофамином и серотонином (9, 117, 118). Все эр-гоалкалоиды имеют общую структуру тетрациклической системы (эрголин), содержащей индольное ядро, в которой кольца А и В образуются из триптофана, а кольца С и D — в результате циклизации диметилаллилпирофос-фата и триптофана (25, 36, 72).
Кластер генов синтеза алкалоидов у спорыньи представлен 12-14 генами (8, 44): cloA , dmaW , easA , easC , easD , easE, easF , easG easH , lpsA , lpsB , lpsC ; ген lpsA имеет два гомолога — lpsA1 и lpsA2 , ген easH представлен как easH1 и псевдоген — редуцированный easH2 (8, 44-46). Гены кластера биосинтеза алкалоидов отвечают за работу всех ферментов, необходимых для образования конечных продуктов биосинтеза — эрготамина и эргокриптина из триптофана у некоторых штаммов C. purpurea (44, 46). Два дополнительных гена easP и easO обнаружены у C. paspali (44).
Гены dmaW, easFи easC экспрессируются на четырех начальных этапах биосинтеза алкалоидов спорыньи, кроме того, ген easE отвечает за замыкание кольца-С эрголина, затем следует образование тетрациклических клавинов, для чего необходима активность генов easD, easA, easG и cloA, и более поздние стадии — синтез амидов лизергиновой кислоты, дигидроалкалоидов и сложных пептидов, с участием генов lpsA—C и easH (рис. 1) (34, 35, 44).
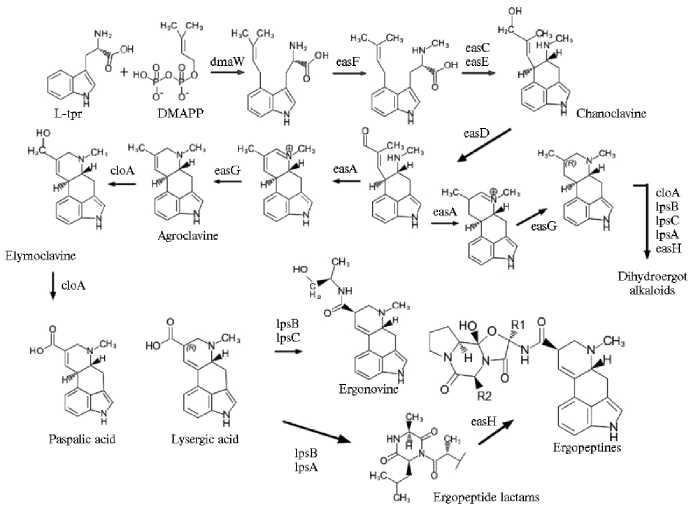
Рис. 1. Метаболический путь биосинтеза эргоалкалоидов Claviceps spp. (с дополнениями S. Robinson и C. Young в редакции M. Liu) (34, 35, 44).
Результаты исследований показали присутствие разного числа генов индольного кластера у представителей Claviceps (44,119). Установлено наличие двух-трех копий генов dmaW , easE , easF , а также выявлена в целом высокая частота приобретения и утраты генов (44). Гомологи девяти генов кластера обнаружены у C. fusiformis (44) . У некоторых изолятов C. paspali ген easG может отсутствовать (55), а easE может быть нефункциональным (44). У C. paspali кластер генов биоинтеза алкалоидов нестабилен и подвержен частичной элиминации, и выявлены изоляты, способные продуцировать предсказанные, но еще не идентифицированные алкалоиды (55). У некоторых штаммов C. africana обнаружено восемь генов биосинтеза алкалоидов (отсутствуют cloA , easH2 , lpsB и lpsC ), у C. lovelessii — десять генов, в частности отсутствуют lpsC и easH2 , а easH1 и lpsB несли мутации, приводящие к появлению стоп-кодонов (44). У некоторых штаммов C. maximensis и C. citrina ( Citrinae ) при отсутствии продукции определенных алкалоидов не выявлялись одни и те же гены (44). Только представители Claviceps имели lpsC и easH2 , хотя у представителей C. perihumidiphila , C. ripicola и C. arundinis отсутствовал lpsC , а C. capensis , C. cyperi , C. humidiphila и C. monticola имели частично совпадающую последовательность гена lpsC . Три штамма C. purpurea и три штамма C. quebecensis не содержали easH2 (44).
Пути биосинтеза алкалоидов и их генетический контроль. Метаболомные профили алкалоидов спорыньи состоят из сложных смесей второстепенных стереоизомеров, конституциональных изомеров и переходных продуктов (45). Изменения в работе кластера генов биосинтеза алкалоидов могут привести к накоплению различных промежуточных и побочных продуктов метаболического пути и повлиять на скорость накопления алкалоидов, особенно тех, синтез которых регулируется промежуточ- ными продуктами по принципу обратной связи (8, 34, 35, 120). Метаболический путь биосинтеза эргоалкалоидов имеет несколько точек ветвления. Исключающий такие точки оптимизированный путь биосинтеза на примере образования D-лизергиновой кислоте из триптофана приведен на рисунке 2 (9). Накопление промежуточных и конечных продуктов у разных видов и штаммов спорыньи осуществляется неодинаково: образуются клавиновые производные, дигидропроизводные, происходит изомеризация лизергиновой кислоты и синтез лизергин-амидов, пептидных алкалоидов и их эпимеров (34, 121, 122).
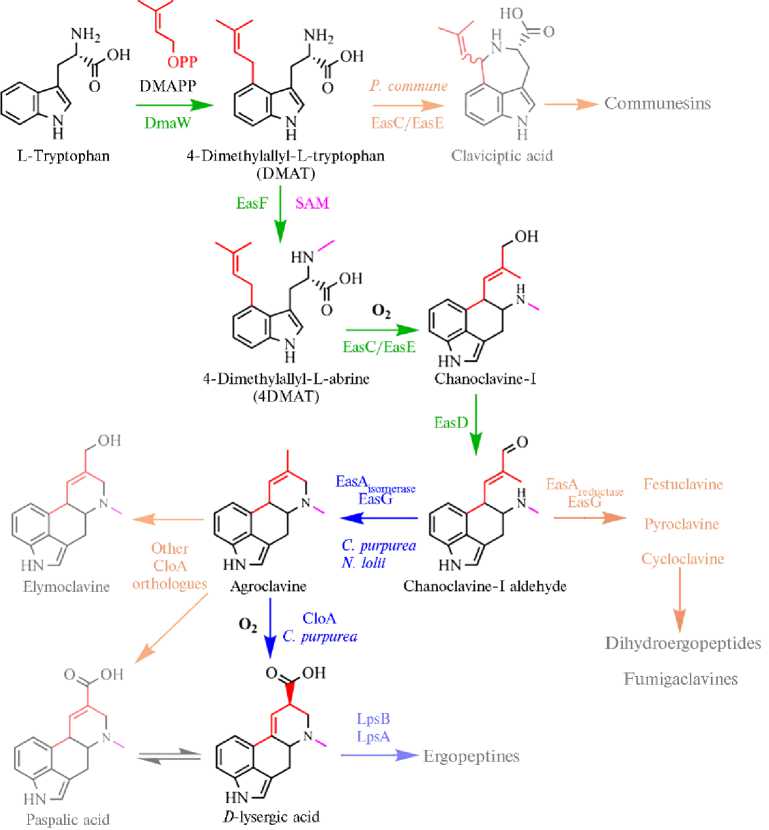
Рис. 2. Искусственно воспроизведенный оптимальный путь биосинтеза D-лизергиновой кислоте из триптофана, исключающий точки ветвления (9).
Считается (37), что ранние этапы биосинтеза эргоалкалоидов консервативны. Более поздние этапы контролируются уникальными генами, что служит причиной модификаций и обеспечивает большое разнообразие и видоспецифичность алкалоидов у разных таксономических групп спорыньи (37, 114, 123).
Синтез триптофана у C. purpurea (также как у других грибов) включает пять стадий, осуществляемых тремя ферментативными комплексами, которые находятся под контролем четырех разных генов (8, 124, 125). Наиболее важен комплекс антранилатсинтазы, состоящий из двух субъединиц — AAS-I (α-субъединица, кодируемая геном TrpE) и AAS-II (трифункциональ-ный пептид, содержащий β-субъединицу антранилатсинтазы, фосфорибози-лантранилатизомеразы и индол-3-глицеринфосфатсинтазы, ген TrpC) (8, 124). В то время как α-субъединица, содержащая разные сайты для связывания субстрата (хоризмат) и ингибирования триптофаном по принципу обратной связи, обепечивает синтез антраниловой кислоты непосредственно из хоризмата в присутствии большого количества аммиака, β-субъединица вместе с AAS-I катализирует превращение хоризмата в антранилат в присутствии глутамина (8, 126).
Биосинтез алкалоидов у видов Claviceps начинается с пренилирования L-триптофана диметилаллилдифосфатом (DMAPP), что приводит к образованию 4-( γ , γ )-диметилаллилтриптофана (4-DMAT) (8, 127). Ген 4-( γ , γ )-диметилаллилтриптофансинтазы (DMATS) — dmaW кодирует фермент первого этапа биосинтетического пути (109), DMAT-синтаза ограничивает скорость образования эрголина и положительно регулируется триптофаном, отрицательно — промежуточными продуктами агроклавином и элимокла-вином (8, 36, 127). В промоторной области активного гена dmaW были идентифицированы сайты связывания для CreA (8, 128) — регуляторного белка, участвующего в репрессии углеродного катаболита, регулятора метаболизма азота AREA (8, 129), фактора регуляции транскрипции PacC, который модулируется в зависимости от рН и связан с вирулентностью у Aspergillus (8, 130), а также фактора транскрипции NUC-1, индуцируемого недостатком фосфора (8, 102, 131). Следующий этап биосинтеза эргоалкалоидов катализирует DMAT-N-метилтрансфераза (ген easF ) (8, 132). Последующие реакции, приводящие к образованию простейшего клавина — ханоклавина-I, катализируют ханоклавин-I-синтаза (ген easE ) (8) и бифункциональная ка-талаза/декарбозилаза (ген easC ) (8, 106, 133). Ген easЕ содержит два экзона и один интрон и имеет длину 1503 п.н., кодируемый им белок состоит из 483 аминокислотных остатков (36). Участок белка, образованный 14-161-м аминокислотными остатками, определен как домен, связывающий флави-надениндинуклеотид (FAD), что позволяет предположить, что EasE может работать FAD-зависимо (36, 110). Алкалоиды спорыньи лизергинового и пептидного класса образуются из альдегида ханоклавина-I как общего предшественника (122). Окисление ханоклавина-I в альдегид ханоклавина-I в присутствии НАД+ катализируется ханоклавин-I-дегидрогеназой, кодируемой геном easD (134).
После пяти консервативных этапов образования альдегида ханокла-вина-I путь биосинтеза алкалоидов спорыньи разветвляется: из альдегида ханоклавина-I синтезируется либо фестуклавин (путь дигидроалкалоидов), либо агроклавин (путь D-лизергина) (34). Этот процесс регулируется геном easA , который кодирует флавинзависимую оксидоредуктазу (8, 34, 108), и геном easG , кодирующим редуктазу (8). Фестуклавин образуется у C. af-ricana , агроклавин — у C. paspali , C. purpurea и C. fusiformis. Затем агрокла-вин у C. purpurea превращается через элимоклавин в D-лизергиновую кислоту, у C. paspali — в паспаловую кислоту (8, 34).
Фермент CLOA (clavine oxidase, аллель cloA, цитохром P450-зависи-мая монооксигеназа) играет ключевую роль в окислении своего субстрата агроклавина до D-лизергиновой кислоты (135). Это процесс, требующий кумулятивного шестиэлектронного окисления и изомеризации двойной связи (34, 136). Делеция cloA вызывает блокировку превращения элимокла- вина в D-лизергиновую кислоту (мутант в значительных количествах накапливает агроклавин, элимоклавин и ханоклавин, но не эргопептиды). CLOA действует как критический фермент и связывает пути биосинтеза двух разных групп алкалоидов спорыньи (137): агроклавин окисляется в паспаловую кислоту через элимоклавин с помощью цитохром P450-зависимой монооксигеназы в присутствии НАДФН и кислорода, а паспаловая кислота может спонтанно изомеризоваться в D-лизергиновую кислоту (36, 137).
D-лизергиновая кислота впоследствии превращается в амиды лизергиновой кислоты и эргопептиды при участии четырех лизергилпептидсин-тетаз (LPS) — тримодулярной LPS1 (LpsA1) с тремя доменами, обладающими каталитическими свойствами (ген lpsA1 ), мономодулярной LPS2 (LpsВ, ген lpsB ), мономодулярной LPS3 (LpsС, ген lpsC ) и тримодулярной LPS4 (LpsA2, ген lpsA2 ). Комплекс LPSB/LPSC катализирует образование эргометрина, комплекс LPSB с LPS1 или с LPS4 опосредует сборку эрго-пептидов (8, 138-141). Биосинтез эргоамидов и эргопептидов инициируется ферментом LpsB. D-лизергиновая кислота после распознавания в качестве субстрата активируется с образованием АМФ-эфира и связывается с белком-носителем LpsB. LpsA или LpsC конкурируют за связывание с LpsB (141). LpsC способен присоединять одну аминокислоту с продукцией эрго-амидов, LpsA — три аминокислотных остатка с прогрессивным удлинением молекулы и образованием эргопептамов, которые монооксигеназа негемовая железодиоксигеназа EasH (ген easH1 ) (8, 142) преобразует в эргопеп-тиды посредством окисления с последующей спонтанной циклизацией (34). Различия в метаболомных профилях алкалоидов C. purpurea коррелировали с изменениями в гене lpsA (45). Изучение делеционного мутанта lpsA1 стало основанием для предположения, что LpsA1 ответствен за образование фенилаланинсодержащих эрготаминовых и эргокристиновых эрго-пептидов (45, 140).
Биохимический анализ экстрактов C. purpurea sclerotia , полученных при заражении четырех зерновых культур, разделил изоляты на два класса на основе содержания алифатических гидрофобных остатков (I) и фенилаланинсодержащих алкалоидов (II) в метаболомных профилях (45). Эрготамин и эргокристин оказались преобладающими эргопептидами склероциев в классе II, эргокриптин и эргокорнин — в классе I. Как эргокорнин, так и эргокриптин были обнаружены как в классе I, так и II в образцах от всех четырех хозяев, однако склероции класса II накапливали эргокорнин и эр-гокриптин в значительно большем количестве (45). Профили алкалоидов спорыньи, специфичные для каждого из этих классов, — результат вариабельности нуклеотидных последовательностей в тандемно дублицирован-ной области easH / lpsA (45, 103, 137).
В биосинтетическом кластере эрголина гены lpsA1 и lpsA2 были результатом события рекомбинации (42). Присутствие мобильных транспози-руемых элементов (TE), сходных с ДНК-транспозонами семейств MULE и TcMar и обнаруженных в межгенном пространстве генов lpsA , убедительно свидетельствует о том, что TE-опосредованная транспозиция или мутации, связанные с TE-вставками, вносят потенциальный вклад в вариабельность гена lpsA (45). Богатые повторяющимися элементами высокополиморфные межгенные пространства lpsA1 / lpsA2 , ассоциированные с разными штаммами C. purpurea , тесно связаны с дивергентными lpsA2 . Доменоспецифические и сильно варьирующие по частоте мутаций области lpsA1 / lpsA2 , обнаруженные при сравнении штаммов, позволяют предположить, что гены lpsA , вероятно, подвергаются рекомбинационному перетасовыванию (45, 143, 144).
Метаболическая инженерия биосинтеза алкалоидов. Получены варианты штамма C. purpurea, у которых сверхэкспрессиро-ваны мутантный вариант S76L гена trpE, кодирующего антранилатсинтазу (с целью преодоления ингибирования избытком триптофана) (8, 145), а также ген dmaW. кодирующий диметилаллилтриптофансинтазу — фермент биосинтеза ключевого промежуточного продукта образования алкалоидов у спорыньи: каждая из манипуляций приводила к значительному (до 7 -кратного) увеличению продукции алкалоидов спорыньи в погруженных культурах (8).
Одно из направлений метаболической инженерии спорыньи — реконструкция целевых путей биосинтеза в удобных гетерологических хозяевах, например в Aspergillus nidulans (104, 146). Воссоздание метаболического пути спорыньи с помощью ферментов, направляющих метаболический поток к желаемым точкам ветвления в гетерологичной системе, гарантирует отсутствие вариаций в профиле продуцируемых алкалоидов спорыньи (9, 136).
В конструировании путей синтеза эргоалкалоидов у различных гетерологичных хозяев, таких как Aspergillus nidulans , Aspergillus fumigatus, был достигнут значительный прогресс: получены важные промежуточные и конечные продукты или новые производные алкалоидов спорыньи (112, 123, 136, 146).
Разработана трансгенная линия дрожжей, несущая 8 генов, ответственных за синтез D-лизергиновой кислоты ( dmaW, easF , easC , easE , easD , easA , easG и cloA ) (см. рис. 2) и в биореакторе синтезирована энантиочистая D-лизергиновая кислота в концентрации до 1,7 мг/л (9).
С применением геномного редактирования (CRISPR/Cas9) получены рекомбинантные культуры Metarhizium brunneum (86). У них относительный выход D-лизергиновой кислоты (86,9 %) и дигидролизергиновой кислоты (72,8 %) был намного выше, чем у сконструированных штаммов Neosartorya fumigata (соответственно 2,6 и 2,0%) (86). Для C. purpurea разработана система геномного редактирования CRISPR/Cas9 с эффективностью редактирования от 50 до 100 %. С ее помощью были успешно нокаутированы три гена-мишени, которые тесно связаны с биосинтезом уридина ( ura5), морфологией гиф ( rac ) и продукцией эргоалкалоидов ( easA ), что позволило получить мутанта, ауксотрофного по уридину (Д ura5) , мутанта с измененным фенотипом в аксенической культуре (Д rac ) и мутанта, который не продуцировал алкалоиды (A easA ) (87).
Мы обращаем внимание на важность нескольких критических точек биосинтетического пути, которые могут повлиять на количество образуемых эргоалкалоидов in vitro. Это синтез триптофана из предшественников и его вовлечение в биосинтетическую цепочку на первых стадиях метаболического пути, образование клавинов, этап агроклавин—D-лизергиновая кислота, а также три точки ветвления и последующие реакции с выходом целевых компонентов (синтез производных клавина, дигидропроизводных алкалоидов, изомеризация D-лизергиновой кислоты и этап D-лизергиновая кислота—эргопептиды).
Вирулентность спорыньи и взаимодействие с растени-ем-хозяином. Разрабатывая приемы снижения ущерба от поражения спрыньей и обеспечения токсикологической безопасности важно учитывать не только способность гриба синтезировать эргоалкалоиды, но также его вирулентность и механизмы взаимодействия с растением-хозяином. Как уже отмечалось, эргоалкалоиды сыграли существенную роль в распространении спорыньи и расширении ареала этого аскомицета. C. purpurea пора- жает ряд экономически значимых зерновых культур, в том числе рожь, пшеницу и ячмень (19, 49, 147). Спорынья инфицирует злаки во время цветения, заражая ткани неоплодотворенных соцветий женских растений и заменяя семена склероциями (53, 147). Таким образом, перекрестноопыляю-щиеся злаки, которые демонстрируют открытое цветение, такие как рожь, особенно подвержены риску заражения (49, 148). Спорынья инфекционна и для гибридных форм ячменя или пшеницы, получивших чувствительность как побочный результат селекции (49, 149, 150). Инфекции C. purpurea подвержены гексаплоидная мягкая пшеница (Triticum aestivum L.) и тетраплоидная твердая пшеница T. turgidum subsp. durum (Desf.) Husn. (151, 152). Чувствительность к C. africana описана у сорго, чувствительность к C. africana — у проса (39, 153, 154). Взаимодействие гриба с хозяином может варьироваться от антагонистического до мутуалистического (155, 156). Спорынья считается биотрофным организмом (47), однако высказываются предположения о некротрофических свойставх (49).
Основными количественными показателями вирулентности спорыньи считают количество медвяной росы, которым характеризуется эффективность заражения на стадии конидиоспоры, размер склероция и общее число склероциев на колос (эффективность паразитирования и уровень продукции аскоспор) (152, 157, 158). Отмечена положительная линейная зависимость между размером склероциев и количеством вырабатываемых стром, содержащих аскоспоры (159).
В ряде случаев C. purpurea проявляет слабую хозяйскую специфичность (61, 160). Дикие изоляты, паразитирующие на представителях Dactylis , при заражении ржи не вызывали обычного сфацелиального плодоношения, но поддерживали рост тонких склероциев, иногда по два в соцветии, с низким содержанием алкалоидов, однако склероции приобретали привычный вид после двух циклов аксенического (in vitro) и паразитарного (на ржи) культивирования с последовательным отбором плектенхиматозного аксени-ческого мицелия (40). В аксенической культуре непигментированный мицелий был непатогенным, но пигментированная (пурпурная) плектенхима-тозная форма проявляла способность паразитировать на соцветиях ржи (40). Хозяйская специфичность спорыньи неодинакова в разные фазы развития гриба: на сфацелиальной стадии и при формировании пади специфичность шире, а образование зрелых склероциев сильнее ограничено кругом хозяев (161, 162).
Предпологается, что склероции и аскоспоры могут быть первичными инокулятами (59, 163, 164). Выявлены различия в относительном вкладе аскоспор и конидий в распространение спорыньи (59). Анализ популяций C. purpurea из разных ареалов показал, что на изоляты, собранные из одной семенной головки, приходится 66 % генетической изменчивости. Это рассматривают как указание на инфицированность значительной части семенных головок, несущих множественные склероции, именно аскоспорами, но не конидиями. В то же время большинство клональных изолятов (они имели идентичные мультилокусные генотипы) также были собраны из одной семенной головки, что указывает на роль конидий (пади) во вторичных инфекциях внутри семенных головок (59).
Спорынья — гомозиготный организм (40). Аскоспоры (этап полового размножения) распространяются преимущественно ветром (59, 164). Конидии (вегетативный этап цикла) попадают на здоровые соцветия с брызгами дождя или при орошении, механически (при контакте растений друг с другом) или через насекомых, что может привести к множественным 864
циклам заражения во время цветения (19, 59, 165, 166). Для североамериканских изолятов было показано, что при широком круге хозяев (167) растения с более ранним сроком цветения (например, рожь), которые заражаются спорыньей ранней весной, способны служить резервуаром конидий и источником инфекции для растений с более поздним цветением (59, 164).
Снижает заражение спорыньей немедленное опыление женских соцветий, закрытое цветение (клейстогамия) и физиологическая устойчивость (39). Стерильность пыльцы — один из основных факторов, облегчающих инфицирование спорыньей (102, 168). Гаметоцидная обработка ржи повышает продуктивность спорыньи при культивировании in planta в полевых условиях (169). Распространено мнение, что C. purpurea вызывает инфекцию, имитируя рост пыльцевых трубок (39, 102). На этой стадии рост C. purpurea происходит в основном межклеточно, но также обнаруживаются инвазивные гифы, которые полностью покрыты плазматической мембраной хозяина (47, 102).
У пшеницы выявлены несколько локусов частичной устойчивости к спорынье (152, 157, 170, 171). Так, гены резистентности обнаружены на хромосоме 6В у сорта Kenya Farmer и на хромосомах 1В, 3В, 4В и 5В у сорта Carleton (172). Два QTL устойчивости к спорынье идентифицированы у гек-саплоидной пшеницы сорта Robigus (расположенны на хромосомах 2А и 4В, QCp.niab.2A и QCp.niab.4B) и два — у сорта Solstice (хромосомы 4D и 6A, QCp.niab.6A и QCp.niab.4D) (171). Четыре локуса устойчивости к спорынье (QCp.aafc.DH-1B, QCp.aafc.DH-2A, QCp.aafc.DH-5A, QCp.aafc.DH-5B) детектированы на хромосомах 1B, 2A, 5A и 5B у сорта твердой пшеницы Greenshank (152), наибольший вклад в устойчивость вносят QCp.aafc.DH-2A (значительно уменьшает образование медвяной росы) и QCp.aafc.DH-5B (снижает общее число склероциев на колос) (152). Линии сорго с мужской стерильностью проявляют небольшую, но стабильную устойчивость к C. af-ricana (173). У сорго идентифицированы девять локусов, которые влияют на процент заражения этим грибом, из них наибольший вклад в резистентность вносят QTL, расположенные на хромосомах SBI-01, SBI-02, SBI-06, SBI-07 и SBI-08 (174). У ячменя описана генетическая изменчивость устойчивости к спорынье. Сообщалось, что сорта Laurier, Maskot и Sabina наиболее устойчивы к заражению (содержание склероциев менее 0,1 %), тогда как сорта Albany, Leger, Symko и Morrison наиболее восприимчивы (149). Частичная устойчивость пшеницы к спорынье сочетается с аллелями гена карликовости, которые расположены в локусах Rht , кодирующих белки DELLA ( Rht-B1 и Rht-D1 , хромосомы соответственно 4B и 4D) (32, 171). Rht-B1b и Rht-D1b — мутации, определяющие утрату чувствительности DELLA к гибберелловой кислоте (32). Уменьшение количества медвяной росы, а также размеров и массы склероциев выявлено у линий, несущих мутантные аллели карликовости и полукарликовости Rht-D1b , Rht-D1c , Rht-B1c (32). Связь между Rht-B1b и Rht-D1b и устойчивостью к спорынье указывает на роль гибберелловой кислоты в инфекции C. purpurea (32).
Установлено перепрограммирование гормональных путей растения-хозяина и дифференциальная экспрессия ауксина, этилена и цитокинина в зависимости от типа ткани и времени после инокуляции C. purpurea (32, 49, 57). Обнаружено увеличение содержания гиберрелловой кислоты, ауксина и цитокининов дигидрозеатинового типа (DHZ) (32). Показано, что деградация белков DELLA, опосредованная гибберелловой кислотой, и подавление сигнальных путей жасмоновой кислоты повышают процент инфицирования пшеницы спорыньей (175). При заражении спорыньей гены пшеницы, связанные с ауксином, становятся наиболее дифференциально экспрессируемыми на ранних стадиях инфекции. Гены семейства AUX/IAA (ауксин/индол-3-уксусная кислота) кодируют известные репрессоры транскрипции генов ответа на ауксин, в то время как семейство генов GH3 (гликозидгидролаза 3) кодирует ферменты, конъюгирующие с ауксином, которые регулируют пул ауксинов посредством отрицательной обратной связи. И AUX/IAA, и GH3 отвечают за ранний ответ на ауксин. Подавление передачи сигналов ауксина за счет повышения экспрессии генов AUX/IAA и связывания избыточного количества ауксина белками семейства GH3 служит прямым ответом растения-хозяина на инфицирование C. purpurea (49, 175). Среди генов, связанных с этиленом, наибольшая активация обнаружена у генов 1-аминоциклопропан-1-карбоксилатоксидазы (ACO), 1-ами-ноциклопропан-1-карбоксилатсинтазы (ACS). Ряд генов биосинтеза и сигнальных путей жасмоновой кислоты (JA) в ответ на инфекцию экспрессировались по-разному. Так, для 12-оксофитодиеноатредуктазы (OPR) и ал-леноксидсинтазы (AOS), которые катализируют первую стадию образования JA, отмечали, что в случае OPR экспрессия повышалась, AOS — усиливалась только у одного гена, остальные подавлялись (49). Цитокинины тоже вовлечены в инфицирование пшеницы C. purpurea с активацией цитокини-ноксидазы/дегидрогеназы и цитокинингликозилтрансферазы в тканях растения (49). Механизмы, регулирующие метаболизм гиберелловой кислоты (GA), индуцируются при заражении: ген гиббереллин-2-бета-оксидазы активируется на ранней стадии инфекции, ген GA-рецептора GID1 также активируется в течение 24 ч после заражения, затем в течение 48 и 72 ч подавляется (49). Салициловая кислота, цитокинин и ауксин вовлечены во взаимодействие C. purpurea и трахинии двуколосковой Brachypodium distachyon (176, 177). Другие защитные механизмы также были активны при инфицировании пшеницы C. purpurea (в частности, происходило устойчивое усиление экспрессии генов хитиназы) (49).
У ржи выявлены значительные различия по устойчивости к спорынье (178-180). Считается, что основной вклад в резистентность ржи обусловлен активностью пектинэстеразы и метаболическими процессами модификации клеточной стенки и роста пыльцевых трубок (180). Сравнени-тельный анализ транскриптома у гибридов ржи, чувствительных (DH372) и умеренно устойчивых (Helltop) к спорынье показал, что в ответ на инфекцию у гибридов по-разному активируются соответственно 12 и 8 генов (180). Среди них шесть генов ( XLOC_059237 , XLOC_1003867 , XLOC_118963 , XLOC_1220465 , XLOC_1387037 и XLOC_386424 ) модификации клеточной стенки и пектинэстеразных метаболических путей (180). COBRA-подобный белок ( XLOC_1432429 ) и вероятный ингибитор пектинэстеразы ( XLOC_118963 ) проявляют наивысшую дифференциальную экспрессию (180). В путях модификации клеточной стенки три гена XLOC_1343481 , XLOC_1343482 и XLOC_145869 связаны с полигалактуроназой (180). Полигалактуроназа известна как фактор патогенности при взаимодействии C. purpurea и ржи (180, 181), а ее ингибирование — как один из основных мехаизмов устойчивости растений к грибным патогенам (182). Показано, что активность обоих полигалактуроназ C. purpurea (гены cppg1 и cppg2 ) снижается при заражении трансгенных растений с высокой степенью метилэтерификации пектина (183, 184). Защитные реакции, связанные с клеточной стенкой, имеют решающее значение для базальной устойчивости растений к грибным патогенам (180, 185). Также интересно отметить, что COBRA-подобный протеин, который вовлечен в реакцию растений ржи на инфицирование C. purpurea , опосредует направленный рост пыльцевых трубок у Arabidopsis thaliana (186).
Нокаут гена COBRA-подобного белка вызывал гаметофитную мужскую стерильность (186). Мутантные штаммы спорыньи с ослабленной вирулентностью индуцируют экспрессию других генов ржи, связанных с защитой (47), в частности гена хитиназы ( Sc2Loc00083431.2 ), гена с высокой гомологией с геном-ингибитором грибных ксиланаз ( Sc4Loc00580338.2 ), а также предполагаемого белка устойчивости к инфекции ( Sc4Loc01458017.2 ) (47).
У спорыньи наблюдаются значительные различия в вирулентности в связи со скоростью роста мицелия (157, 187, 188). Удлинение гиф определяется метаболизмом полисахаридов (189). Делеция гена Mid1 приводит к снижению темпов роста и полной потере вирулентности. На определенном этапе инфекционного процесса эти признаки коррелируют (190). При культивировании у одних видов Claviceps мицелий растет быстрее, чем у других: так, у японского изолята C. sorgicola скорость составляет в среднем 1 мм/сут (191), у индийских изолятах C. africana и C. sorghi — 0,1 мм/сут (192).
Спорынья секретирует ферменты, разрушающие клеточную стенку растений, сигнальные моллекулы и соединения-эффекторы (177, 193). Примерно 90 % генов спорыньи с наиболее высокой экспрессией — это гены, которые кодируют белки, участвующие в росте и развитии (47). Геном C. purpurea содержит более 400 генов, кодирующих предполагаемые эффекторы развития инфекции, многие из этих генов кластеризованы и сильно избыточны. Высказывалось предположение (47), что высокая избыточность генов в кластерах синтеза эффекторных соединений служит механизмом, предотвращающим потери генов (47). Функциональный анализ некоторых эффекторных метаболитов спорыньи показал, что по крайней мере один из них накапливается в апопласте растения (47). Спорынья имеет обширную эффекторную сеть, влияющую на течение инфекции, однако значительный вклад в вирулентность какого-то определенного эффекторного соединения пока не установлен (47). Спорынья обладает ферментами (CAZymes), разрушающими клеточную стенку растения (47, 194). Десять из наиболее экспрессируемых генов C. purpurea — это гены CAZyme белков (47). Изоляты C. paspali секретируют CAZyme-подобные белков (195), основная мишень которых — пектины (47, 195). К сигнальным компонентам относятся MAP-киназы (47, 181, 196, 197), НАДФН-оксидазы, генерирующие АФК (активные формы кислорода) (ген Cpnox2 ; контролирует процесс заражения и снижает повреждения у хозяина) (198), а также цитокинины, необходимые для успешного развития инфекции (199, 200). Цитокинины требуются для установления и поддержания взаимодействия растений и грибов (199, 200). Усиление поглотительной активности и приток питательных веществ к очагам инфекции — наиболее важная роль цитокининов спорыньи (177). Дефицит цитокининов, снижающий вирулентность, может быть достигнут делетированием гена изопентенилтрансферазы, который также участвует в регуляции трансляции (177, 200). Однако важно учитывать, что установлено наличие альтернативных путей активации цитокининов (177). Ген полигалактуроназы спорыньи (фактор вирулентности при взаимодействии C. purpurea и ржи) (47) cppg1 ( cp6977 ), ген MAP-киназы cpmk1 ( cp1700 ) и ген супероксиддисмутазы cpsod1 ( cp7438 ) относятся к группе генов с высоким уровнем экспрессии, но при этом роль супероксиддисмутазы в развитии инфекции несуществена (201).
Два фактора вирулентности оказались особенно интересными с точки зрения взаимодействия C. purpurea и ржи. Это фактор регуляции транскрипции, кодируемый геном спорыньи Cptf1 (гомолог дрожжевого гена Ap1), и малая GTP-аза, кодируемая геном Cpcdc42. Делеции этих генов снижают вирулентность спорыньи (47, 202, 203). Малая GTP-аза (ген Cdc42) участвует в организации цитоскелета у грибов. У дикого штамма C. purpurea экспрессия гена гетерологичной GTP-азы Colletotrichum trifolii (Ctcdc42) имела значительное влияние на вегетативную дифференцировку спорыньи. Экспрессия доминантно-активного алелля Ctcdc42 (DA) приводила к потере конидий и аберрантной форме клеток, доминантно-негативный (DN) аллель Ctcdc42 (при таких мутациях измененный продукт нарушает соотношение белков дикого типа при совместной экспрессии генов) стимулировал ветвление и конидиогенез. Делеция гена Cpcdc42 не была летальной и приводила к фенотипу, сравнимому с фенотипом негативных трансформантов, мутанты DeltaCpcdc42 были непатогенными и не вызывали симптомов заболевания (инвазивный рост останавливался на ранней стадии) (203).
Мутантные линии спорыньи, ауксотрофные по триптофану, неин-фекционны для ржи, вероятно, из-за сниженной продукции растительных ауксинов, которые синтезируются из индол-3-глицеролфосфата по трипто-фан-зависимому и триптофан-независимому путям биосинтеза и помогают грибу колонизировать растение-хозяина (7).
Стоит еще раз подчеркуть, что определение паразитарных грибов как растительных мутуалистов или патогенов остается весьма размытым, периодически публикуются новые данные, свидетельствующие в пользу как первого, так и второго утверждения (204). Экспериментально показано, что инфицирование спорыньей у овсяницы красной ( Festuca rubra ), образующей симбиоз с эндофитным грибом Epichloe festucae , в 4,5 раза снижает поражение растений тлей ( Sitobion sp.) (204, 205). При этом растения, содержащие симбиотический эндофит, были более подвержены инфекции Claviceps (45 % растений E+ против 31 % растений E - ) (204). В то же врем присутствие Epichloe occultans в Lolium multiflorum и Epichloe gansuensis в Achnatherum inebrians коррелировало со снижением частоты инфицирования C. purpurea , то есть, вероятно, у некоторых видов растений представители рода Epichloe опосредуют защиту от C. purpurea (204, 206 -208) . В этих случаях рост устойчивости к патогенам может быть результатом повышения иммунокомпетентности хозяина или прямой конкуренции между спорыньей и эндофитным штаммом Epichloe (204, 209, 210). Отсутствие различий в количестве и таксономическом составе микобиоты между растениями, инфицированными и не инфицированными спорыньей, частично подтверждает гипотезу о пользе этого патогена для растений (211).
Наблюдения за 25-летней динамикой чувствительности к спорынье у основных сельскохозяйственных культур на территории Канады показали, что сильнее всего поражается спорыньей рожь, далее следуют хлебная и твердая пшеницы, затем ячмень и овес (212). Интересным оказался тот факт, что заболеваемость и тяжесть вспышек ежегодно менялись, причем со временем инфекция все более распространялась, но ее вредоносность и степень поражения не изменялись (212).
Важным предметом изучения остается связь между вирулентностью спорыньи и содержанием эргоалкалоидов (213, 214). У генотипов ржи, менее восприимчивых к спорынье, снижено содержание эргоалкалоидов (213), однако гаметоцидная обработка ржи не влияла на это (169). Выявлена отрицательная зависимость между массой одного склероция и накоплением эр-гоалкалоидов у озимой ржи и яровой пшеницы (214). В России выявлены два сорта пшеницы (Новосибирская 18 и Т-66), иммунных к спорынье, и 13 относительно устойчивых (с поражением не более 5,2 % и примесью склероциев в зерне не более 0,3 %), у ржи относительную устойчивость де-868
монстрируют 10 сортов (поражение варьируется от 5,8 до 33,0 %, засоренность зерна склероциями — от 0,3 до 1,4 %) (214). Отмечены генотипы, у которых устойчивость к поражению спорыньей потенциально сочетается с отсутствием накопления эргоалкалоидов в склероциях: это сорта озимой ржи Румба, Симфония, Гармония и яровой пшеницы Epos (214).
Гибридная селекция ржи позволяет увеличить урожайность зерна, одновременно усиливая чувствительность к спорынье, связанную с цитоплазматической мужской стерильностью (CMS), которая наследуется по материнской линии (215, 216). Тесная связь между наличием фертильной пыльцы и чувствительностью к спорынье создает проблему для селекции (213, 215, 217).
Таким образом, на сорвеменном этапе исследований C. purpurea — это, с одной стороны, важнейший продуцент большого количества биологически активных соединений (алкалоидов) и уникальная модель системы паразит-хозяин (218-221), с другой — патоген, наносящий значительный экономический ущерб производителям зерна, кормовой и животноводческой отрасли по всему миру (222, 223). В то время как медицинские и биотехнологические исследования сосредоточены на положительных эффектах алкалоидов (224) и других вторичных метаболитов спорыньи и особенностях их получения (224-228), в растениеводстве, животноводстве и пищевой промышленности опасность эргоалкалоидов остается острой и вызывающей беспокойство общемировой проблемой (229-232). Ее серьезность усугубляется тем, что в меняющихся условиях окружающей среды у Claviceps изменяются профили и интенсивность продукции алкалоидов (231).
В последние годы в качестве перспективного подхода можно рассматривать управление симбиозами растений и эндофитов для оптимизации профиля и концентрации продуцируемых вторичных метаболитов (в том числе токсичных для человека и сельскохозяйственных животных) (231, 233). Значимыми остаются филогенетические исследования и изучение эволюционной изменчивости спорыньи (234). Выявленные закономерности могут быть полезны при получении новых высокопродуктивных рекомбинантных сапрофитных штаммов-продуцентов C. purpurea , при селекции устойчивых к спорынье сортов и при оптимизации приемов защиты растений.
Анализ публикаций свидетельствует, что разные растения демонстрируют аналогичный ответ на инфекцию C. purpurea , однако механизмы устойчивости (например, у ржи и пшеницы) при этом, по всей видимости, различаются. В отношении исторически и эволюционно обусловленных хозяев с открытым цветением (например, ржи) C. purpurea — биотроф, а партнерские взаимодействия носят мутуалистический характер. В случае нетипичных хозяев (например, пшеницы) и новых селекционных форм, чувствительных к спорынье, вероятны некротрофия и классическое паразитирование C. purpurea .
Итак, в представленном обзоре мы рассмотрели данные о генных кластерах, регулирующих механизмы вирулентности и биосинтеза алкалоидов у возбудителя спорыньи Claviceps purpurea, в контексте эволюционной изменчивости, видообразования и идентификации штаммов Claviceps, а также в связи с достижаниями и перспективами генной инженерии. Возбудитель спорыньи C. purpurea охарактеризован как очень изменчивый и способный к адаптации организм, чем обусловлено большое разнообразие синтезируемых им алкалоидов, широкий спектр растений-хозяев и, в итоге, обилие штаммов этого гриба. Для путей биосинтеза алкалоидов спорыньи характерно наличие нескольких точек ветвления и накопление промежуточных метаболитов. При промышленном культивировании такая избыточность про- дуктов требует дополнительной очистки, а у диких штамов повышает токсичность для сельскохозяйственных животных и человека. Знание закономерностей и генетического контроля вторичного метаболизма у C. purpurea важно для эффективного управления технологическими процессами получения эргоалкалоидов. Конечная практическая цель проводимых геномных, транскриптомных и метаболомных исследований C. purpurea и системы C. purpurea—растение-хозян — снижение токсичности и вирулентности гриба, ограничение распространения спорыньи в новые ареалы и на новые растения, в том числе за счет создания резистентных селекционных форм.
Список литературы Разнообразие алкалоидов и вирулентность спорыньи Claviceps purpurea (fries) Tulasne: эволюция, генетическая диверсификация, метаболическая инженерия (обзор)
- Jamieson C.S., Misa J., Tang Y., Billingsley J.M. Biosynthesis and synthetic biology of psychoactive natural products. Chemical Society Reviews, 2021, 50(12): 6950-7008 (doi: 10.1039/d1cs00065a).
- Rämä T., Quandt C.A. Improving fungal cultivability for natural products discovery. Frontires in Microbiology, 2021, 12: 706044 (doi: 10.3389/fmicb.2021.706044).
- Liu H., Jia Y. Ergot alkaloids: synthetic approaches to lysergic acid and clavine alkaloids. Natural Product Reports, 2017, 34(4): 411-432 (doi: 10.1039/c6np00110f).
- Gerhards N., Matuschek M., Wallwey C., Li S.M. Genome mining of ascomycetous fungi reveals their genetic potential for ergot alkaloid production. Archives Microbiology, 2015, 197(5): 701-713 (doi: 10.1007/s00203-015-1105-4).
- Chan J.D., Agbedanu P.N., Grab T., Zamanian M., Dosa P.I., Day T.A., Marchant J.S. Ergot Alkaloids (re)generate new leads as antiparasitics. PLoS Neglected Tropical Diseases, 2015, 9(9): e0004063 (doi: 10.1371/journal.pntd.0004063).
- Smakosz A., Kurzyna W., Rudko M., D^sal M. The usage of ergot (Clavicepspurpurea (Fr.) Tul.) in obstetrics and gynecology: a historical perspective. Toxins, 2021, 13(7): 492 (doi: 10.3390/toxins13070492).
- Kralova M., Bergougnoux V., Frebort I., CRISPR/Cas9 genome editing in ergot fungus Claviceps purpurea. Journal of Biotechnology, 2021, 325: 341-354 (doi: 10.1016/jjbiotec.2020.09.028).
- Kralova M., Frebortova J., Pencik A., Frebort I. Overexpression of Trp-related genes in Claviceps purpurea leading to increased ergot alkaloid production. New Biotechnology, 2021, 61: 69-79 (doi: 10.1016/j.nbt.2020.11.003).
- Wong G., Lim L.R., Tan Y.Q., Go M.K., Bell D.J., Freemont P.S., Yew W.S. Reconstituting the complete biosynthesis of D-lysergic acid in yeast. Nature Communication, 2022, 13(1): 712 (doi: 10.1038/s41467-022-28386-6).
- Lieberman A., Kupersmith M., Estey E., Goldstein M. Treatment of Parkinson's disease with bromocriptine. New England Journal of Medicine, 1976, 295(25): 1400-1404 (doi: 10.1056/NEJM197612162952504).
- Winblad B., Fioravanti M., Dolezal T., Logina I., Milanov I.G., Popescu D.C., Solomon A. Therapeutic use of nicergoline. Clinical Drug Investigation, 2008, 28(9): 533-552 (doi: 10.2165/00044011-200828090-00001).
- Tandowsky R.M. Clinical evaluation of combined hydrogenated ergot alkaloids (hydergine) in arterial hypertension: with special reference to their action in central manifestations. Circulation, 1954, 9(1): 48-56 (doi: 10.1161/01.cir.9.1.48).
- Johnson J.W., Ellis M.J., Piquette Z.A., MacNair C., Carfrae L., Bhando T., Ritchie N.E., Saliba P., Brown E.D., Magolan J. Antibacterial activity of metergoline analogues: revisiting the ergot alkaloid scaffold for antibiotic discovery. ACS Medicinal Chemistry Letters, 2022, 13(2): 284291 (doi: 10.1021/acsmedchemlett. 1c00648).
- Фитопрепараты ВИЛАР: научно-справочное издание /Под ред. Т.А. Сокольской. М., 2009.
- Савина Т.А., Савин П.С., Бобылева Р.И. Экзогенная биорегуляция развития Oaviceps purpurea (Fr.) Tul. (обзор). Вопросы биологической, медицинской и фармацевтической химии, 2018, 21(12): 28-34 (doi: 10.29296/25877313-2018-12-06).
- Бобылева Р.И., Савин П.С. Физиологические и технологические аспекты биосинтеза эр-гоалкалоидов в сапрофитной культуре Oaviceps purpurea (Fr.) Tulasne (обзор). Вопросы биологической, медицинской и фармацевтической химии, 2019, 22(10): 30-36 (doi: 10.29296/25877313-2019-10-05).
- Бобылева Р.И., Савин П.С. Изучение морфологических и физиолого-биохимических особенностей штамма Claviceps purpurea (Fries) Tulasne bkmf-2641d в сапрофитной культуре. Вопросы биологической, медицинской и фармацевтической химии, 2021, 24(12): 57-62 (doi: 10.29296/25877313-2021-12-09).
- Franzmann C., Schröder J., Münzing K., Wolf K., Lindhauer M.G., Humpf H. Distribution of ergot alkaloids and ricinoleic acid in different milling fractions. Mycotoxin Research, 2011, 27(1): 13-21 (doi: 10.1007/s12550-010-0070-7).
- Menzies J.G., Turkington T.K. An overview of the ergot (Claviceps purpurea) issue in western Canada: challenges and solutions. Canadian Journal of Plant Pathology, 2015, 37(1): 40-51 (doi: 10.1080/07060661.2014.986527).
- Klotz J.L. Activities and effects of ergot alkaloids on livestock physiology and production. Toxins, 2015, 7(8): 2801-2821 (doi: 10.3390/toxins7082801).
- Klotz J.L., Nicol A.M. Ergovaline, an endophytic alkaloid. 1. Animal physiology and metabolism. Animal Production Science, 2016, 56: 1761-1774 (doi: 10.1071/AN14962).
- Reddy P., Hemsworth J., Guthridge K.M., Vinh A., Vassiliadis S., Ezernieks V., Spangenberg G.C., Rochfort S.J. Ergot alkaloid mycotoxins: physiological effects, metabolism and distribution of the residual toxin in mice. Scientific Reports, 2020, 10(1): 9714 (doi: 10.1038/s41598-020-66358-2).
- Bauermeister A., Aguiar F., Marques L., Malta J., Barros F., Callejon D., Lopes N. In vitro metabolism evaluation of the ergot alkaloid dihydroergotamine: application of microsomal and biomimetic oxidative model. Planta Medica, 2016, 82(15): 1368-1373 (doi:1 0.1055/s-0042-111732).
- Lünne F., Niehaus E.-M., Lipinski S., Kunigkeit J., Kalinina S. A., Humpf H.-U. Identification of the polyketide synthase PKS7 responsible for the production of lecanoric acid and ethyl lecano-rate in Claviceps purpurea. Fungal Genetics and Biology, 2020, 145: 103481 (doi: 10.1016/j.fgb.2020.103481).
- Flieger M., Stodûlkovâ E., Wyka S.A., Cerny J., Grobarova V., Pichova K., Novak P., Man P., Kuzma M., Cvak L., Broders K.D., Kolarik M. Ergochromes: heretofore neglected side of ergot toxicity. Toxins (Basel), 2019, 11(8): 439 (doi: 10.3390/toxins11080439).
- Uhlig S., Botha C.J., Vrelstad T., Rolen E., Miles C.O. Indole-diterpenes and ergot alkaloids in Cynodon dactylon (Bermuda grass) infected with Claviceps cynodontis from an outbreak of tremors in cattle. Journal of Agricultural and Food Chemistry, 2009, 57(23): 11112-11119 (doi: 10.1021/jf902208w).
- Kozak L., Szilagyi Z., Vagy B., Kakuk A., Tyth L., Molnar I., Pycsi I. Inactivation of the indole-diterpene biosynthetic gene cluster of Claviceps paspali by Agrobacterium-mediated gene replacement. Applied Microbiology and Biotechnology, 2018, 102(7): 3255-3266 (doi: 10.1007/s00253-018-8807-x).
- Kozak L., Szilagyi Z., Töth L., Pöcsi I., Molnar I. Functional characterization of the idtF and idtP genes in the Claviceps paspali indole diterpene biosynthetic gene cluster. Folia Microbiologica, 2020, 65(3): 605-613 (doi: 10.1007/s12223-020-00777-6).
- Dopstadt J., Neubauer L., Tudzynski P., Humpf H.-U. The epipolythiodiketopiperazine gene cluster in Claviceps purpurea: dysfunctional cytochrome P450 enzyme prevents formation of the previously unknown clapurines. PLoS ONE, 2016, 11(7): e0158945 (doi: 10.1371/journal.pone.0158945).
- Florea S., Panaccione D.G., Schardl C.L. Ergot alkaloids of the family Clavicipitaceae. Phytopathology, 2017, 107(5): 504-518 (doi: 10.1094/PHYT0-12-16-0435-RVW).
- Schwake-Anduschus C., Lorenz N., Lahrssen-Wiederholt M., Lauche A., Dänicke S. German monitoring 2012-2014: ergot of Claviceps purpurea and ergot alkaloids (EA) in feedingstuffs and their toxicological relevance for animal feeding. Journal für Verbraucherschutz und Lebensmittelsicherheit, 2020, 15: 321-329 (doi: 10.1007/s00003-020-01298-7).
- Tente E. Investigations into the molecular interactions between Claviceps purpurea, the causal agent of ergot, and cereal hosts. Doctoral thesis, University of Cambridge, 2020 (doi: 10.17863/CAM.64578).
- Wallwey C., Li S. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Natural Product Reports, 2011, 28(3): 496-510 (doi: 10.1039/c0np00060d).
- Robinson S.L., Panaccione D.G. Diversification of ergot alkaloids in natural and modified fungi. Toxins, 2015, 7: 201-218 (doi: 10.3390/toxins7010201).
- Young C.A., Schardl C.L., Panaccione D.G., Florea S., Takach J.E., Charlton N.D., Moore N., Webb J.S., Jaromczyk J. Genetics, genomics and evolution of ergot alkaloid diversity. Toxins (Basel), 2015, 7(4): 1273-1302 (doi: 10.3390/toxins7041273).
- Chen J., Han M., Gong T., Yang J., Zhu P. Recent progress in ergot alkaloid research. RSC Advances, 2017, 7(44): 27384-27396 (doi: 10.1039/C7RA03152A).
- Tasker N.R., Wipf P. Biosynthesis, total synthesis, and biological profiles of ergot alkaloids. Alkaloids: Chemistry and Biology, 2021, 85: 1-112 (doi: 10.1016/bs.alkal.2020.08.001).
- Hinsch J., Tudzynski P. Claviceps: the Ergot fungus. In: Molecular biology of food and water borne mycotoxigenic and mycotic fungi, 1st edn /R. Paterson, N.Lima (eds.). CRC Press, Boca Raton, 2015: 229-250.
- Miedaner T., Geiger H.H. Biology, genetics, and management of ergot (Claviceps spp.) in rye, sorghum, and pearl millet. Toxins (Basel), 2015, 7(3): 659-678 (doi: 10.3390/toxins7030659).
- Mantle P. Comparative ergot alkaloid elaboration by selected plectenchymatic mycelia of Claviceps purpurea through sequential cycles of axenic culture and plant parasitism. Biology (Basel), 2020, 9(3): 41 (doi: 10.3390/biology9030041).
- Wingfield B.D., Liu M., Nguyen H.D.T.; Lane F.A., Morgan S.W., De Vos L., Wilken P.M., Duong T.A., Aylward J.C., Martin P.A., Dadej K., De Beer Z.W., Findlay W., Havenga M.,
- Kolarik M., Menzies J.G., Naidoo K., Pochopski O., Shoukouhi P., Santana Q.C., Seifert K.A., Soal N., Steenkamp E.T., Tatham C.T., van der Nest M.A., Wingfield M.J. Nine draft genome sequences of Claviceps purpurea s.lat., including C. arundinis, C. humidiphila, and C. cf. spartinae, pseudomolecules for the pitch canker pathogen Fusarium circinatum, draft genome of Davidsoniella eucalypti, Grosmannia galeiformis, Quambalaria eucalypti, and Teratosphaeria destructans. IMA Fungus, 2018, 9(2): 401-418 (doi: 10.5598/imafungus.2018.09.02.10).
- Wyka S., Mondo S., Liu M., Nalam V., Broders K. A large accessory genome and high recombination rates may influence global distribution and broad host range of the fungal plant pathogen Claviceps purpurea. PLoS ONE, 2022, 17(2): e0263496 (doi: 10.1371/journal.pone.0263496).
- Wyka S.A., Mondo S.J., Liu M., Dettman J., Nalam V., Broders K.D. Whole-genome comparisons of ergot fungi reveals the divergence and evolution of species within the genus Claviceps are the result of varying mechanisms driving genome evolution and host range expansion. Genome Biology and Evolution, 2021, 13(2): evaa267 (doi: 10.1093/gbe/evaa267).
- Liu M., Findlay W., Dettman J., Wyka S.A., Broders K., Shoukouhi P., Dadej K., Kola Hk M., Basnyat A., Menzies J.G. Mining indole alkaloid synthesis gene clusters from genomes of 53 Claviceps strains revealed redundant gene copies and an approximate evolutionary hourglass model. Toxins, 2021, 13(11): 799 (doi: 10.3390/toxins13110799).
- Hicks C., Witte T.E., Sproule A., Lee T., Shoukouhi P., Popovic Z., Menzies J.G., Boddy C.N., Liu M., Overy D.P. Evolution of the ergot alkaloid biosynthetic gene cluster results in divergent mycotoxin profiles in Claviceps purpurea Sclerotia. Toxins, 2021, 13(12): 861 (doi: 10.3390/toxins13120861).
- Schardl C.L., Young C.A., Hesse U., Amyotte S.G., Andreeva K., Calie P.J., Fleetwood D.J., Haws D.C., Moore N., Oeser B., Panaccione D.G., Schweri K.K., Voisey C.R., Farman M.L., Jaromczyk J.W., Roe B.A., O'Sullivan D.M., Scott B., Tudzynski P., An Z., Arnaoudova E.G., Bullock C.T., Charlton N.D., Chen L., Cox M., Dinkins R.D., Florea S., Glenn A.E., Gordon A., Güldener U., Harris D.R., Hollin W., Jaromczyk J., Johnson R.D., Khan A.K., Leistner E., Leuchtmann A., Li C., Liu J., Liu J., Liu M., Mace W., Machado C., Nagabhyru P., Pan J., Schmid J., Sugawara K., Steiner U., Takach J.E., Tanaka E., Webb J.S., Wilson E.V., Wiseman J.L., Yoshida R., Zeng Z. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Oavicipitaceae reveals dynamics of alkaloid loci. PLoS Genetetics, 2013, 9(2): e1003323 (doi: 10.1371/journal.pgen.1003323).
- Oeser B., Kind S., Schurack S., Schmutzer T., Tudzynski P., Hinsch J. Cross-talk of the bio-trophic pathogen Claviceps purpurea and its host Secale cereale. BMC Genomics, 2017, 18: 273 (doi: 10.1186/s12864-017-3619-4).
- Oberti H., Abreo E., Reyno R., Feijoo M., Murchio S., Dalla-Rizza M., Rokas A. New draft genome sequence of the ergot disease fungus Claviceps paspali. Microbiology Resource Announcements, 2020, 9(29): e00498-20 (doi: 10.1128/MRA.00498-20).
- Tente E., Ereful N., Rodriguez A.C., Grant P., O'Sullivan D.M., Boyd L.A., Gordon A. Reprogramming of the wheat transcriptome in response to infection with Claviceps purpurea, the causal agent of ergot. BMC Plant Biology, 2021, 21: 316 (doi: 10.1186/s12870-021-03086-3).
- Pichova K., Pazoutova S., Kostovcik M., Chudickova M., Stodülkova E., Novak P., Flieger M., van der Linde E., Kolarik M. Evolutionary history of ergot with a new infrageneric classification (Hypocreales: Clavicipitaceae: Claviceps). Molecular Phylogenetics and Evolution, 2018, 123: 73-87 (doi: 10.1016/j.ympev.2018.02.013).
- Bouchenak-Khelladi Y., Verboom G.A., Savolainen V., Hodkinson T.R. Biogeography of the grasses Poaceae: a phylogenetic approach toreveal evolutionary history in geographical space and geologicaltime. Botanical Journal of the Linnean Society, 2010, 162(4): 543-557 (doi: 10.1111/j.1095-8339.2010.01041.x).
- Soreng R.J., Peterson P.M., Romaschenko K., Davidse G., Teisher J.K., Clark L.G., Barber P., Gillespie L.J., Zuloaga F.O. A worldwide phylogenetic classification of the Poaceae gramineae II: an update and a comparison of two 2015 classifications. Journal of Systematics and Evolution, 2017, 55(4): 259-290 (doi: 10.1111/jse.12150).
- Pazoutova S., Pesicova K., Chudickova M., Sratka P., Kolarik M. Delimitation of cryptic species inside Claviceps purpurea. Fungal Biology, 2015, 119(1): 7-26 (doi: 10.1016/j.funbio.2014.10.003).
- Liu M., Overy D.P., Cayouette J., Shoukouhi P., Hicks C., Bisson K., Sproule A., Wyka S.A., Broders K., Popovic Z., Menzies J.G. Four phylogenetic species of ergot from Canada and their characteristics in morphology, alkaloid production, and pathogenicity. Mycologia, 2020, 112(5): 974-988 (doi: 10.1080/00275514.2020.1797372).
- Oberti H., Dalla R.M., Reyno R., Murchio S., Altier N., Abreo E. Diversity of Claviceps paspali reveals unknown lineages and unique alkaloid genotypes. Mycologia, 2020, 112(2): 230-243 (doi: 10.1080/00275514.2019.1694827).
- Liu M., Tanaka E., Kolarik M. Neotypification of Claviceps humidiphila and recognition of C. bavariensis sp. nov. Mycotaxon, 2022, 137(1): 73-87 (doi: 10.5248/137.73).
- Wyka S., Broders K. Population biology and comparative genomics of Claviceps purpurea and other defensive mutualists in the Hypocreales. In: Рoster thesis of 2017 APS Annual Meeting, San Antonio, USA, 2017: 126 (doi: 10.13140/RG.2.2.26063.38567). '
- Liu M., Shoukouhi P., Bisson K.R., Wyka S.A., Broders K.D., Menzies J.G. Sympatric divergence of the ergot fungus, Claviceps purpurea, populations infecting agricultural and nonagri-cultural grasses in North America. Ecology and Evolution, 2021, 11(1): 273-293 (doi: 10.1002/ece3.7028).
- Cheng Q., Frost K., Dung J.K.S. Population genetic structure of Claviceps purpurea in cool-season grass seed crops of Oregon. Phytopathology, 2020, 110(11): 1773-1778 (doi: 10.1094/phyto-01-20-0005-r).
- Dung J.K.S., Duringer J.M., Kaur N., Scott J.C., Frost K.E., Walenta D.L., Alderman S.C., Craig A.M., Hamm P.B. Molecular and alkaloid characterization of Claviceps purpurea sensu lato from grass seed production areas of the U.S. Pacific Northwest. Phytopathology, 2021, 111(5): 831-841 (doi: 10.1094/PHYT0-07-20-0289-R).
- Pazoutova S., Olsovska J., Linka M., Kolinska R., Flieger M. Chemoraces and habitat specialization of Claviceps purpurea populations. Applied and Environmental Microbiology, 2000, 66(12): 5419-5425 (doi: 10.1128/AEM.66.12.5419-5425.2000).
- Douhan G.W., Smith M.E., Huyrn K.L., Westbrook A., Beerli P., Fisher A.J. Multigene analysis suggests ecological speciation in the fungal pathogen Claviceps purpurea. Molecular Ecology, 2008, 17(9): 2276-2286 (doi: 10.1111/j.1365-294X.2008.03753.x).
- Jungehülsing U., Tudzynski P. Analysis of genetic diversity in Claviceps purpurea by RAPD markers. Mycological Research, 1997, 101(1): 1-6 (doi: 10.1017/S0953756296001657).
- Slack J.M.W., Holland P.W.H., Graham C.F. The zootype and the phylotypic stage. Nature, 1993, 361(6412): 490-492 (doi: 10.1038/361490a0).
- Duboule D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development Supplement, 1994, 1994: 135-142 (doi: 10.1242/dev.1994.Supplement.135).
- Prud'homme B., Gompel N. Genomic hourglass. Nature, 2010, 468(7325): 768-769 (doi: 10.1038/468768a).
- Galis F., van Dooren T.J., Metz J.A. Conservation of the segmented germband stage: Robustness or pleiotropy? Trends in Genetics, 2002, 18(10): 504-509 (doi: 10.1016/S0168-9525(02)02739-7).
- Panaccione D.G. Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiology Letters, 2005, 251(1): 9-17 (doi: 10.1016/j.femsle.2005.07.039).
- Gilmore B.S., Alderman S.C., Knaus B.J., Bassil N.V., Martin R.C., Dombrowski J.E., Dung J.K.S. Simple sequence repeat markers that identify Claviceps species and strains. Fungal Biology and Biotechnology, 2016, 3(1): 1-13 (doi: 10.1186/s40694-016-0019-5).
- Shoukouhi P., Hicks C., Menzies J.G., Popovic Z., Chen W., Seifert K.A., Assabgui R., Liu M. Phylogeny of Canadian ergot fungi and a detection assay by real-time polymerase chain reaction. Mycologia, 2019, 111(3): 493-505 (doi: 10.1080/00275514.2019.1581018).
- Uhlig S., Rangel-Huerta O.D., Divon H.H., Rolen E., Pauchon K., Sumarah M.W., Vrelstad T., Renaud J.B. Unraveling the ergot alkaloid and indole diterpenoid metabolome in the claviceps purpurea species complex using LC-HRMS/MS diagnostic fragmentation filtering. Journal of Agricultural and Food Chemistry, 2021, 69(25): 7137-7148 (doi: 10.1021/acs.jafc.1c01973).
- Hulvova H., Galuszka P., Frebortova J., Frebort I. Parasitic fungus Claviceps as a source for biotechnological production of ergot alkaloids. Biotechnology Advances, 2013, 31(1): 79-89 (doi: 10.1016/j.biotechadv.2012.01.005).
- Yao Y., Wang W., Shi W., Yan R., Zhang J., Wei G., Liu L., Che Y., An C., Gao S., Overproduction of medicinal ergot alkaloids based on a fungal platform. Metabolic Engineering, 2022, 69: 198-208 (doi: 10.1016/j.ymben.2021.12.002).
- Hendrickson J.B., Wang J. A new synthesis of lysergic acid. Organic Letters, 2004, 6(1): 3-5 (doi: 10.1021/ol0354369).
- Umezaki S., Yokoshima S., Fukuyama T. Total synthesis of lysergic acid. Organic Letters, 2013, 15(16): 4230-4233 (doi: 10.1021/ol4019562).
- Majeska Cudejkova M., Vojta P., Valik J., Galuszka P. Quantitative and qualitative transcriptome analysis of four industrial strains of Claviceps purpurea with respect to ergot alkaloid production. New Biotechnology, 2016, 33(5 Pt B): 743-754 (doi: 10.1016/j.nbt.2016.01.006).
- Фонин В.С., Сидякина Т.М., Шаин С.С., Озерская С.М., Павлова Е.Ф. Изучение условий хранения промышленных штаммов паразитарной культуры спорыньи. Прикладная биохимия и микробиология, 1996, 32(4): 406-410.
- Tonolo A., Scotti T., Vero-Barcellona L. Morphological observations on different species of Claviceps Tul. grown in submerged culture. Scientific Reports of the Istituto Superiore di Sanita, 1961, 1: 404-422.
- Mantle P.G., Tonolo A. Relationship between the morphology of Claviceps purpurea and the production of alkaloids. Transactions of the British Mycological Society, 1968, 51(3-4): 499-505 (doi: 10.1016/S0007-1536(68)80017-8).
- Mantle P.G. Development of alkaloid production in vitro by a strain of Claviceps purpurea from Spartina townsendii. Transactions of the British Mycological Society, 1969, 52(3): 381-392 (doi: 10.1016/S0007-1536(69)80122-1).
- Strnadov6 K. UV-Mutanten bei Claviceps purpurea. Planta Medica, 1964, 12(4): 521-527 (doi: 10.1055/S-0028-1100208).
- Kren V., Pazoutova S., Sedmera P., Rylko V., Rehacek Z. High-production mutant Claviceps purpurea 59 accumulating secoclavines. FEMS Microbiology Letters, 1986, 37(1): 31-34 (doi: 10.1111/j.1574-6968.1986.tb01761.x).
- Smit R., Tudzynski P. Efficient transformation of Claviceps purpurea using pyrimidine auxotrophic mutants: cloning of the OMP decarboxylase gene. Molecular Genetics and Genomics, 1992, 234(2): 297-305 (doi: 10.1007/BF00283850).
- Strnadova K. A method of preparation and application of nitrous acid as a mutagen in Claviceps purpurea. Folia Microbiologic, 1976, 21(6): 455-458 (doi: 10.1007/BF02876936).
- Brauer K.L., Robbers J.E. Induced parasexual processes in Claviceps sp. strain SD58. Applied and Environmental Microbiology, 1987, 53(1): 70-73 (doi: 10.1128/aem.53.1.70-73.1987).
- Davis K.A., Sampson J.K., Panaccione D.G. Genetic reprogramming of the ergot alkaloid pathway of Metarhizium brunneum. Applied and Environmental Microbiology, 2020, 86(19): e01251-20 (doi: 10.1128/AEM.01251-20).
- Yu L., Xiao M., Zhu Z., Wang Y., Zhou Z., Wang P., Zou G. Efficient genome editing in Claviceps purpurea using a CRISPR/Cas9 ribonucleoprotein method. Synthetic and Systems Biotechnology, 2022, 7(2): 664-670 (doi: 10.1016/j.synbio.2022.02.002).
- van Engelenburg F., Smit R., Goosen T, van den Broek H., Tudzynski P. Transformation of Claviceps purpurea using a bleomycin resistance gene. Applied Microbiology and Biotechnology, 1989, 30: 364-370 (doi: 10.1007/BF00296625).
- Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 1995, 11(1): 53-55 (doi: 10.1002/yea.320110107).
- Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borko-vich K.A., Dunlap J.C. A hight hroughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(27): 10352-10357 (doi: 10.1073/pnas.0601456103).
- Kouprina N., Larionov V. Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma, 2016, 125(4): 621-632 (doi: 10.1007/s00412-016-0588-3).
- Crews C. Analysis of ergot alkaloids. Toxins (Basel), 2015, 7(6): 2024-2050 (doi: 10.3390/toxins7062024).
- Schummer C., Brune L., Moris G. Development of a UHPLC-FLD method for the analysis of ergot alkaloids and application to different types of cereals from Luxembourg. Mycotoxin Research, 2018, 34(4): 279-287 (doi: 10.1007/s12550-018-0322-5).
- Kuner M., Kühn S., Haase H., Meyer K., Koch M. Cleaving ergot alkaloids by hydrazinolysis — a promising approach for a sum parameter screening method. Toxins (Basel), 2021, 13(5): 342 (doi: 10.3390/toxins13050342).
- Kodisch A., Oberforster M., Raditschnig A., Rodemann B., Tratwal A., Danielewicz J., Korbas M., Schmiedchen B., Eifler J., Gordillo A., Siekmann D., Fromme F.J., Wuppermann F.N., Wieser F., Zechner E., Niewi ska M., Miedaner T. Covariation of ergot severity and alkaloid content measured by HPLC and one ELISA Method in inoculated winter rye across three isolates and three European countries. Toxins, 2020, 12(11): 676 (doi: 10.3390/toxins12110676).
- Franzmann C., Wächter J., Dittmer N., Humpf H.-U. Ricinoleic acid as a marker for ergot impurities in rye and rye products. Journal of Agricultural and Food Chemistry, 2010, 58(7): 42234229 (doi: 10.1021/jf1006903).
- Appelt M., Ellner F.M. Investigations into the occurrence of alkaloids in ergot and single sclerotia from the 2007 and 2008 harvests. Mycotoxin Research, 2009, 25(2): 95-101 (doi: 10.1007/s12550-009-0014-2).
- Miedaner T., Dänicke S., Schmiedchen B., Wilde P., Wortmann H., Dhillon B., Geiger H., Mirdita V. Genetic variation for ergot ( Claviceps purpurea) resistance and alkaloid concentrations in cytoplasmic-male sterile winter rye under pollen isolation. Euphytica, 2010, 173: 299-306 (doi: 10.1007/s10681-009-0083-5).
- Aboling S., Drotleff A., Cappai M., Kamphues J. Contamination with ergot bodies (Claviceps purpurea sensu lato) of two horse pastures in Northern Germany. Mycotoxin Research 2016, 32(4): 207-219 (doi: 10.1007/s12550-016-0253-y).
- Uhlig S., Vikmren T., Ivanova L., Handeland K. Ergot alkaloids in Norwegian wild grasses: A mass spectrometric approach. Rapid Communications in Mass Spectrometry, 2007, 21(10): 16511660 (doi: 10.1002/rcm.3005).
- Dopstadt J., Vens-Cappell S., Neubauer L., Tudzynski P., Cramer B., Dreisewerd K., Humpf H.-U. Localization of ergot alkaloids in sclerotia of Claviceps purpurea by matrix-assisted laser desorp-tion/ionization mass spectrometry imaging. Analytical and Bioanalytical Chemistry, 2017, 409(5): 1221-1230 (doi: 10.1007/s00216-016-0047-2).
- Tudzynski P., Correia T., Keller U. Biotechnology and genetics of ergot alkaloids. Applied Microbiology and Biotechnology, 2001, 57(5-6): 593-605 (doi: 10.1007/s002530100801).
- Haarmann T., Machado C., Lübbe Y., Correia T., Schardl C.L., Panaccione D.G., Tudzynski P. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry, 2005, 66(11): 1312-1320 (doi: 10.1016/j.phytochem.2005.04.011).
- Ryan K.L., Akhmedov N.G., Panaccione D.G. Identification and structural elucidation of er-gotryptamine, a new ergot alkaloid produced by genetically modified Äspergillus nidulans and natural isolates of Epichloe species. Journal of Agricultural and Food Chemistry, 2015, 63(1): 6167 (doi: 10.1021/jf505718x).
- Steen C.R., Sampson J.K., Panaccione D.G. A Baeyer-Villiger monooxygenase gene involved in the synthesis of lysergic acid amides affects the interaction of the fungus Metarhizium brunneum with insects. Applied and Environmental Microbiology, 2021, 87(17): e0074821 (doi: 10.1128/AEM.00748-21).
- Lorenz N., Wilson E.V., Machado C., Schardl C.L., Tudzynski P. Comparison of ergot alkaloid biosynthesis gene clusters in Claviceps Species indicates loss of late pathway steps in evolution of C. fusiformis. Applied and Environmental Microbiology, 2007, 73(22): 7185-7191 (doi: 10.1128/AEM.01040-07).
- Coyle C.M., Panaccione D.G. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Applied and Environmental Microbiology, 2005, 71(6): 3112-3118 (doi: 10.1128/aem.71.6.3112-3118.2005).
- Coyle C.M., Cheng J.Z., O'Connor S.E., Panaccione D.G. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Applied and Environmental Microbiology, 2010, 76(12): 3898-3903 (doi: 10.1128/AEM.02914-09).
- Liu M., Panaccione D.G., Schardl C.L. Phylogenetic analyses reveal monophyletic origin of the ergot alkaloid gene dmaW in fungi. Evolutionary Bioinformatics, 2009, 5: 15-30 (doi: 10.4137/ebo.s2633).
- Goetz K.E., Coyle C.M., Cheng J.Z., O"Connor S.E., Panaccione D.G. Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Current Genetics, 2011, 57(3): 201-211 (doi: 10.1007/s00294-011-0336-4).
- Jones A.M., Steen C.R., Panaccione D.G. Independent evolution of a lysergic acid amide in Äspergillus species. Applied and Environmental Microbiology, 2021, 87(24): e01801-21 (doi: 10.1128/AEM.01801-21).
- Nielsen C.A., Folly C., Hatsch A., Molt A., Schröder H., O'Connor S.E., Naesby M. The important ergot alkaloid intermediate chanoclavine-I produced in the yeast Saccharomyces cerevisiae by the combined action of EasC and EasE from Aspergillus japonicus. Microbial Cell Factories, 2014, 13(1): 95 (doi: 10.1186/s12934-014-0095-2).
- An C., Zhu F., Yao Y., Zhang K., Wang W., Zhang J., Wei G., Xia Y., Gao Q., Gao S. Beyond the cyclopropyl ring formation: fungal Aj_EasH catalyzes asymmetric hydroxylation of ergot alkaloids. Applied Microbiology and Biotechnology, 2022, 106(8): 2981-2991 (doi: 10.1007/s00253-022-11892-4).
- Jakubczyk D., Cheng J.Z., O'Connor S.E. Biosynthesis of the ergot alkaloids. Natural Product Reports, 2014, 31(10): 1328-1338 (doi: 10.1039/C4NP00062E).
- Wallwey C., Heddergott C., Xie X., Brakhage A.A., Li S. Genome mining reveals the presence of a conserved gene cluster for the biosynthesis of ergot alkaloid precursors in the fungal family Arthro-dermataceae. Microbiology (Reading), 2012, 158(Pt 6): 1634-1644 (doi: 10.1099/mic.0.056796-0).
- Fabian S.J., Maust M.D., Panaccione D.G. Ergot alkaloid synthetic capacity of Penicillium cam-emberti. Applied and Environmental Microbiology, 2018, 84(191): e01583-18 (doi: 10.1128/AEM.01583-18).
- Pertz H., Eich E. Ergot alkaloids and their derivatives as ligands for serotoninergic, dopaminergic, and adrenergic receptors. In: Ergot: the genus Claviceps /V. Kren, L. Cvak (eds.). Amsterdam, Harwood Academic Publishers, 1999: 411-440.
- Mantegani S., Brambilla E., Varasi M. Ergoline derivatives: receptor affinity and selectivity. Farmaco, 1999, 54(5): 288-296 (doi: 10.1016/s0014-827x(99)00028-2).
- Negerd M., Uhlig S., Kauserud H., Andersen T., H0iland K., Vrelstad T. Links between Genetic Groups, Indole alkaloid profiles and ecology within the grass-parasitic Claviceps purpurea species complex. Toxins, 2015, 7(5): 1431-1456 (doi: 10.3390/toxins7051431).
- Uhlig S., Petersen D. Lactam ergot alkaloids (ergopeptams) as predominant alkaloids in sclerotia of Claviceps purpurea from Norwegian wild grasses. Toxicon, 2008, 52(1): 175-185 (doi: 10.1016/j.toxicon.2008.05.002).
- Cheng J.Z., Coyle C.M., Panaccione D.G., O'Connor S.E. Controlling a structural branch point in ergot alkaloid biosynthesis. Journal of the American Chemical Society, 2010, 132(37): 1283512837 (doi: 10.1021/ja105785p).
- Wei X., Wang W.G., Matsuda Y. Branching and converging pathways in fungal natural product biosynthesis. Fungal Biology and Biotechnology, 2022, 9: 6 (doi: 10.1186/s40694-022-00135-w).
- Jakubczyk D., Caputi L., Hatsch A., Nielsen C.A.F., Diefenbacher M., Klein J., Molt A., Schröder H., Cheng J.Z., Naesby M., O'Connor S.E. Discovery and reconstitution of the cycloclavine biosynthetic pathway-enzymatic formation of a cyclopropyl group. Angewandte Chemie International Edition, 2015, 54(17): 5117-5121 (doi: 10.1002/anie.201410002).
- Hütter R., DeMoss J.A. Organization of the tryptophan pathway: a phylogenetic study of the fungi. Journal of Bacteriology, 1967, 94(6): 1896-1907 (doi: 10.1128/JB.94.6.1896-1907.1967).
- Hütter R., Niederberger P., DeMoss J.A. Tryptophan biosynthetic genes in eukaryotic microorganisms. Annual Review of Microbiology, 1986, 40: 55-77 (doi: 10.1146/annurev.mi.40.100186.000415).
- Crawford I.P., Eberly L. Structure and regulation of the anthranilate synthase genes in Pseudomonas aeruginosa: I. Sequence of trpG encoding the glutamine amidotransferase subunit. Molecular Biology and Evolution, 1986, 3(5): 436-448 (doi: 10.1093/oxfordjournals.molbev.a040408).
- Metzger U., Schall C., Zocher G., Unsöld I., Stec E., Li S.M., Heide L., Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proceedings of the National Academy of Sciences, 2009, 106(34): 1430914314 (doi: 10.1073/pnas.0904897106).
- Ruijter G.J., Visser J. Carbon repression in Aspergilli. FEMS Microbiology Letters, 1997, 151(2): 103-114 (doi: 10.1111/j. 1574-6968.1997.tb12557).
- Arst H.N., Cove D.J. Nitrogen metabolite repression in Aspergillus nidulans. Molecular Genetics and Genomics, 1973, 126: 111-141 (doi: 10.1007/BF00330988).
- Bignell E., Negrete-Urtasun S., Calcagno A.M., Haynes K., Arst Jr H.N., Rogers T. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Molecular Microbiology, 2005, 55(4): 1072-1084 (doi: 10.1111/j.1365-2958.2004.04472.x).
- Kang S., Metzenberg R.L. Molecular analysis of nuc-1+, a gene controlling phosphorus acquisition in Neurospora crassa. Molecular and Cellular Biology, 1990, 10(11): 5839-5848 (doi: 10.1128/MCB.10.11.5839).
- Otsuka H., Quigley F.R., Gryger J., Anderson A., Floss H.G. In vivo and in vitro evidence for N-methylation as the pathway specific step in ergoline biosynthesis. Planta Medica, 1980, 40(10): 109-119 (doi: 10.1055/s-2008-1074947).
- Yao Y., An C., Evans D., Liu W., Wang W., Wei G., Ding N., Houk K.N., Gao S. Catalase involved in oxidative cyclization of the tetracyclic ergoline of fungal ergot alkaloids. Journal of the American Chemical Society, 2019, 141(44): 17517-17521 (doi: 10.1021/jacs.9b10217).
- Wallwey C., Matuschek M., Li S. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Archives of Microbiology, 2010, 192(2): 127-134 (doi: 10.1007/s00203-009-0536-1).
- Arnold S.L., Panaccione D.G. Biosynthesis of the pharmaceutical^ important fungal ergot alkaloid dihydrolysergic acid requires a specialized allele of cloA. Applied and Environmental Microbiology, 2017, 83(14): e00805-17 (doi: 10.1128/AEM.00805-17).
- Robinson S.L., Panaccione D.G. Heterologous expression of lysergic acid and novel ergot alkaloids in Aspergillus fumigatus. Applied and Environmental Microbiology, 2014, 80(20): 6465-6472 (doi: 10.1128/AEM.02137-14).
- Haarmann T., Ortel I., Tudzynski P., Keller U. Identification of the cytochrome P450 monoox-ygenase that bridges the clavine and ergoline alkaloid pathways. ChemBiochem, 2006, 7(4): 645652 (doi: 10.1002/cbic.200500487).
- Riederer B., Han M., Keller U. D-Lysergyl peptide synthetase from the ergot fungus Claviceps purpurea. Journal of Biological Chemistry, 1996, 271(44): 27524-27530 (doi: 10.1074/jbc.271.44.27524).
- Walzel B., Riederer B., Keller U. Mechanism of alkaloid cyclopeptide synthesis in the ergot fungus Claviceps purpurea. Chemistry & Biology, 1997, 4(3): 223-230 (doi: 10.1016/s1074-5521(97)90292-1).
- Haarmann T., Lorenz N., Tudzynski P. Use of a nonhomologous end joining deficient strain (Aku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis. Fungal Genetics and Biology, 2008, 45(1): 35-44 (doi: 10.1016/j.fgb.2007.04.008).
- Ortel I., Keller U. Combinatorial assembly of simple and complex D-lysergic acid alkaloid peptide classes in the ergot fungus Claviceps purpurea. Journal of Biological Chemistry, 2009, 284(11): 6650-6660 (doi: 10.1074/jbc.M807168200).
- Havemann J., Vogel D., Loll B., Keller U. Cyclolization of D-lysergic acid alkaloid peptides. Chemistry & Biology, 2014, 21(1): 146-155 (doi: 10.1016/j.chembiol.2013.11.008).
- Berry D., Mace W., Grage K., Wesche F., Gore S., Schardl C.L., Young C.A., Dijkwel P.P., Leuchtmann A., Bode H.B., Barry S. Efficient nonenzymatic cyclization and domain shuffling drive pyrrolopyrazine diversity from truncated variants of a fungal NRPS. Proceedings of the National Academy of Sciences, 2019, 116(51): 25614-25623 (doi: 10.1073/pnas.1913080116).
- Baunach M., Chowdhurry S., Stallforth P., Dittmann E. The landscape of recombination events that create nonribosomal peptide diversity. Molecular Biology and Evolution, 2021, 38(5): 21162130 (doi: 10.1093/molbev/msab015).
- Wang P., Choera T., Wiemann P., Pisithkul T., Amador-Noguez D., Keller N.P. TrpE feedback mutants reveal roadblocks and conduits toward increasing secondary metabolism in Aspergillus fumigatus. Fungal Genetics and Biology, 2016, 89: 102-113 (doi: 10.1016/j.fgb.2015.12.002).
- Ryan K.L., Moore C.T., Panaccione D.G. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins (Basel), 2013, 5(2): 445-455 (doi: 10.3390/toxins5020445).
- Tudzynski P., Scheffer J. Claviceps purpurea: molecular aspects of a unique pathogenic lifestyle. Molecular Plant Pathology, 2004, 5(5): 377-388 (doi: 10.1111/j.1364-3703.2004.00237.x).
- Kodisch A., Wilde P., Schmiedchen B., Fromme F.-J., Rodemann B., Tratwal A., Oberforster M., Schiemann A., Jmrgensen L., Miedaner T. Ergot infection in winter rye hybrids shows differential contribution of male and female genotypes and environment. Euphytica, 2020, 216(4): 65 (doi: 10.1007/s10681-020-02600-2).
- Pageau D., Wauthy J., Collin J. Evaluation of barley cultivars for resistance to ergot fungus, Claviceps purpurea (Fr.) Tul. Canadian Journal of Plant Science, 1994, 74(3): 663-665 (doi: 10.4141/cjps94-118).
- Mette M.F., Gils M., Longin C.F.H., Reif J.C. Hybrid breeding in wheat. In: Advances in wheat genetics: from genome to field /Y. Ogihara, S. Takumi, H. Handa (eds.). Springer, Tokyo, 2015: 225-232 (doi: 10.1007/978-4-431-55675-6_24).
- Platford R.G., Bernier C.C. Resistance to Claviceps purpurea in spring and durum wheat. Nature, 1970, 226(5247): 770 (doi: 10.1038/226770a0).
- Gordon A., McCartney C., Knox R.E., Ereful N., Hiebert C.W., Konkin D.J., Hsueh Ya.-C., Bhadauria V., Sgroi M., O'Sullivan D.M., Hadley C., Boyd L.A., Menzies J.G. Genetic and transcriptional dissection of resistance to Claviceps purpurea in the durum wheat cultivar Green-shank. Theoretical and Applied Genetics, 2020, 133: 1873-1886 (doi: 10.1007/s00122-020-03561-9).
- Thakur R.P., Rai K.N. Pearl millet ergot research: advances and implications. In: Sorghum and millets diseases /J.F. Leslie (ed.). Iowa State Press, Ames, IA, USA, 2003: 57-64 (doi: 10.1002/9780470384923.ch9).
- Pazoutova S., Frederickson D.E. Genetic diversity of Claviceps africana on sorghum and Hypar-rhenia. Plant Pathology, 2005, 54: 749-763 (doi: 10.1111/j.1365-3059.2005.01255.x).
- Haarmann T., Rolke Y., Giesbert S., Tudzynski P. Ergot: from witchcraft to biotechnology. Molecular Plant Pathology, 2009, 10(4): 563-577 (doi: 10.1111/j.1364-3703.2009.00548.x).
- Wäli P.P., Wäli P.R., Saikkonen K., Tuomi J. Is the pathogenic ergot fungus a conditional defensive mutualist for its host grass? PLoS ONE, 2013, 8(7): e69249 (doi: 10.1371/journal.pone.0069249).
- Menzies J.G., Klein-Gebbinck H.W., Gordon A., O'Sullivan D.M. Evaluation of Claviceps purpurea isolates on wheat reveals complex virulence and host susceptibility relationships. Canadian Journal of Plant Pathology, 2017, 39(3): 307-317 (doi: 10.1080/07060661.2017.1355334).
- Platford R.G., Bernier C.C. Reaction of cultivated cereals to Claviceps purpurea. Canadian Journal of Plant Science, 1976, 56: 51-58 (doi: 10.4141/cjps76-009).
- Cooke R.C., Mitchell D.T. Sclerotium size and germination in Claviceps purpurea. Transactions of the British Mycological Society, 1966, 49(1): 95-100 (doi: 10.1016/S0007-1536(66)80039-6).
- Likar M., Grandi M., Strajn B.J., Kos K., Celar F.A. Links between genetic groups, host specificity, and ergot-alkaloid profiles within Claviceps purpurea (Fr.) Tul. on Slovenian grasses. Plant Disease, 2018, 102(7): 1334-1340 (doi: 10.1094/pdis-08-17-1179-re).
- Pazoutova S., Cagas B., Kolinska R., Honzatko A. Host specialization of different 424 populations of ergot fungus (Claviceps purpurea). Czech Journal of Genetics and Plant Breeding, 2002, 38(2): 75-81.
- Pazoutova S. The evolutionary strategy of Claviceps. In: Clavicipitalean fungi: evolutionary biology, chemistry, biocontrol and cultural impacts /F. White, C.W. Bacon, N.L. Hywel-Jones (eds.). Marcel Dekker, New York, 2002: 329-354.
- Dung J.K.S., Alderman S.C., Walenta D.L., Hamm P.B. Spatial patterns of ergot and quantification of sclerotia in perennial ryegrass seed fields in eastern Oregon. Plant Disease, 2016, 100(6): 1110-1117 (doi: 10.1094/PDIS-08-14-0787-RE).
- Dung J.K., Scott J., Cheng Q., Alderman S.C., Kaur N., Walenta D.L., Frost K.E., Hamm P.B. Detection and quantification of airborne Claviceps purpurea sensu lato ascospores from hirst-type spore traps using real-time quantitative PCR. Plant Disease, 2018, 102(12): 2487-2493 (doi: 10.1094/PDIS-02-18-0310-RE).
- Butler M.D., Alderman S.C., Hammond P.C., Berry R.E. Association of insects and ergot (Claviceps purpurea) in Kentucky bluegrass seed production fields. Journal of Economic Entomology, 2001, 94(6): 1471-1476 (doi: 10.1603/0022-0493-94.6.1471).
- Kaur N., Cating R.A., Rondon S.I., Scott J.C., Alderman S.C., Walenta D.L., Frost K.E., Hamm P.B., Dung J.K.S. Dispersal potential of ergot spores by insects foraging in perennial ryegrass fields in the Columbia Basin of Oregon and Washington. Crop, Forage & Turfgrass Management, 2019, 5(1): 1-5 (doi: 10.2134/cftm2019.04.0020).
- Alderman S.C., Halse R.R., White J.F. A reevaluation of the host range and geographical distribution of Claviceps species in the United States. Plant Disease, 2004, 88(1): 63-81 (doi: 10.1094/PDIS.2004.88.1.63).
- Шаин С.С. Биологические основы производства сырья спорыньи (Claviceps purpurea (Fr.) Tul.) в биотехнологической системе гриб—растение ^бзор). Прикладная биохимия и микробиология, 1996, 32(3): 275-279.
- Hanosova H., Koprna R., Valik J., Knoppova L., Frébort I., Dzurova L., Galuszka P. Improving field production of ergot alkaloids by application of gametocide on rye host plants. New Biotechnology, 2015, 32(6): 739-746 (doi: 10.1016/j.nbt.2015.01.008).
- Menzies J.G. The reactions of Canadian spring wheat genotypes to inoculation with Claviceps purpurea, the causal agent of ergot. Canadian Journal of Plant Science, 2004, 84(4): 625-629 (doi: 10.4141/P03-086).
- Gordon A., Basler R., Bansept-Basler P., Fanstone V., Harinarayan L., Grant P.K., Birch-more R., Bayles R.A., Boyd L.A., O'Sullivan D.M. The identification of QTL controlling ergot sclerotia size in hexaploid wheat implicates a role for the Rht dwarfing alleles. Theoretical and Applied Genetics, 2015, 128: 2447-2460 (doi: 10.1007/s00122-015-2599-5).
- Platford R.G., Bernier C.C., Evans L.E. Chromosome location of genes conditioning resistance to Claviceps purpurea in spring and durum wheat. Canadian Journal of Genetics and Cytology, 1977, 19: 679-682 (doi: 10.1139/g77-074).
- Komolong B., Chakraborty S., Ryley M., Yates D. Ovary colonization by Claviceps africana is related to ergot resistance in male-sterile sorghum lines. Plant Pathology, 2003, 52(5): 620-627 (doi: 10.1046/j.1365-3059.2003.00886.x).
- Parh D.K., Jordan D.R., Aitken E.A., Mace E.S., Jun-ai P., McIntyre C.L., Godwin I.D. QTL analysis of ergot resistance in sorghum. Theoretical and Applied Genetics, 2008, 117(3): 369-382 (doi: 10.1007/s00122-008-0781-8).
- Tente E., Carrera E., Gordon A., Boyd L.A. The role of the wheat reduced height (Rht)-DELLA mutants and associated hormones in infection by Claviceps purpurea, the causal agent of ergot. Phytopathology, 2022, 112(4): 842-851 (doi: 10.1094/PHYT0-05-21-0189-R).
- Kind S., Schurack S., Hinsch J., Tudzynski P. Brachypodium distachyon as alternative model host system for the ergot fungus Claviceps purpurea. Molecular Plant Pathology, 2017, 19(4): 1005-1011 (doi: 10.1111/mpp.12563).
- Kind S., Hinsch J., Vrabka J., Hradilova M., Majeska-Cudejkova M., Tudzynski P., Galuszka P. Manipulation of cytokinin level in the ergot fungus Claviceps purpurea emphasizes its contribution to virulence. Current Genetics, 2018, 64(6): 1303-1319 (doi: 10.1007/s00294-018-0847-3).
- Mirdita V., Dhillon B., Geiger H., Miedaner T. Genetic variation for resistance to ergot (Claviceps purpurea [Fr.] Tul.) among full-sib families of five populations of winter rye (Secale cereale L.). Theoretical and Applied Genetics, 2008, 118(1): 85-90 (doi: 10.1007/s00122-008-0878-0).
- Mirdita V., Miedaner T. Resistance to ergot in self-incompatible germplasm resources of winter rye. Journal of Phytopathology, 2009, 157(6): 350-355 (doi: 10.1111/j.1439-0434.2008.01499.x).
- Mahmood K., Orabi J., Kristensen P.S., Sarup P., Jmrgensen L.N., Jahoor A. De novo tran-scriptome assembly, functional annotation, and expression profiling of rye (Secale cereale L.) hybrids inoculated with ergot (Claviceps purpurea). Scientific Reports, 2020, 10(1): 13475 (doi: 10.1038/s41598-020-70406-2).
- Oeser B., Heidrich P.M., Müller U., Tudzynski P., Tenberge K.B. Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genetics and Biology, 2002, 36(3): 176-186 (doi: 10.1016/S1087-1845(02)00020-8).
- Wang Z., Wan L., Zhang X., Xin Q., Song Y., Hong D., Sun Y., Yang G. Interaction between Brassica napus polygalacturonase inhibition proteins and Sclerotinia sclerotiorum polygalacturonase: implications for rapeseed resistance to fungal infection. Planta, 2021, 253(2): 34 (doi: 10.1007/s00425-020-03556-2).
- Volpi C., Raiola A., Janni M., Gordon A., O'Sullivan D.M., Favaron F., D'Ovidio R. Claviceps purpurea expressing polygalacturonases escaping PGIP inhibition fully infects PvPGIP2 wheat transgenic plants but its infection is delayed in wheat transgenic plants with increased level of pectin methyl esterification. Plant Physiology and Biochemistry, 2013, 73: 294-301 (doi: 10.1016/j.plaphy.2013.10.011).


