Разнообразие культивируемых бактерий-деструкторов монохлорированных бифенилов в почвах охраняемого ландшафта
Автор: Королев Н.А., Кирьянова Т.Д., Егорова Д.О.
Журнал: Вестник Пермского университета. Серия: Биология @vestnik-psu-bio
Рубрика: Микробиология
Статья в выпуске: 3, 2024 года.
Бесплатный доступ
В работе использованы аэробные бактериальные штаммы из рабочей коллекции лаборатории микробиологии техногенных экосистем «ИЭГМ УрО РАН», выделенные ранее из почв охраняемого ландшафта ООПТ Осинская лесная дача. Наибольший деструктивный потенциал в отношении бифенила и его хлорированных производных выявлен у 16 штаммов. На основании анализа нуклеотидной последовательности гена 16S рРНК установлена филогенетическая принадлежность данных штаммов. Показано, что доля представителей рода Achromobacter составила 56.25%, Rhodococcus - 18.75%, Pseudomonas - 12.5%, Delftia и Stenotrophomonas - по 6.25%. Штаммы родов Achromobacter и Rhodococcus осуществляли деструкцию 2-хлор-, 3-хлор- и 4-хлорированных бифенилов с эффективностью 13-100% и 43-100% соответственно. Эффективность деструкции монохлорбифенилов для представителей рода Pseudomonas составляла выше 80%, за исключением 69%-го разложения 4-хлорбифенила штаммом Pseudomonas sp. Osa 27. Штамм Stenotrophomonas sp. Osa 13 наиболее активно (71%) разлагал 4-хлорбифенил, но проявлял наименьшую активность (34%) в отношении 3-хлорированного бифенила. Штамм Delftia sp. Osa 20 осуществлял полную деструкцию 3-хлор- и 4-хлорбифенилов, но не осуществлял трансформацию 2-хлорбифенила. Таким образом, аэробные бактериальные культуры, выделенные из почв охраняемого ландшафта, обладают способностью окислять сложные ароматические соединения, опасные для окружающей среды.
Монохлорированные бифенилы, achromobacter, rhodococcus, pseudomonas, stenotrophomonas, delftia, биодеструкция
Короткий адрес: https://sciup.org/147246119
IDR: 147246119 | УДК: 579.222/579.8 | DOI: 10.17072/1994-9952-2024-3-285-299
Текст научной статьи Разнообразие культивируемых бактерий-деструкторов монохлорированных бифенилов в почвах охраняемого ландшафта
Полихлорированные бифенилы (ПХБ) уже на протяжении нескольких десятилетий являются экологической проблемой глобального масштаба. Подтверждение этому – Международное соглашение от 2001 г. (Стокгольмская конвенция), в котором указана необходимость избавления от ПХБ как в процессах производства, так и в местах складирования и в природных резервуарах [Final act ..., 2001]. Проникновение ПХБ в организмы живых существ в незначительных количествах приводит к ряду негативных последствий, проявляющихся в нарушении целостности и передачи наследственной информации, нарушении работы основных органов и систем. ПХБ обладают высокой липофильностью, за счет чего проникают в жировые ткани и переходят по цепям питания на верхние трофические уровни [Adams et al., 2016; Müller et al., 2017; Warenik-Bany et al., 2019; Reddy et al., 2019; Devi, 2020]. В большинстве работ основное внимание уделяется воздействию на животных и человека высоко хлорированных бифенилов [Adams et al., 2016; Müller et al., 2017; Warenik-Bany et al., 2019; Reddy et al., 2019; Devi, 2020; Negret-Bolagay et al., 2021]. Однако низко хлорированные конгенеры, содержащие от 1 до 3 атомов хлора в молекуле, также могут оказывать воздействие, при этом их содержание в окружающей среде обусловлено не только проникновением в природу из промышленных смесей, но и образованием в результате анаэробной деструкции высоко хлорированных бифенилов.
В настоящее время ПХБ выявлены на таких территориях, где никогда не было их производства и они не использовались для промышленных целей [Трегер, 2013; Zhang et al., 2014; Zhu et al., 2020; Negret-Bolagay et al., 2021]. Способность к трансграничному переносу за счет высокой сорбционной составляющей привела к проникновению ПХБ в новые, территориально удаленные друг от друга, биотопы [Трегер, 2013]. Присутствие опасного загрязнителя вызывает изменения в составе ценозов, и в первую очередь, в составе почвенных сообществ [Negreet-Bolagay et al., 2021]. Преимущество в выживании получают организмы, обладающие устойчивостью к негативному воздействию поллютанта, либо способные использовать данный поллютант как источник углерода и /или энергии. Основной группой организмов, наиболее быстро адаптирующихся к новым загрязнителям, являются аэробные бактерии [Negreet-Bolagay et al., 2021]. Выявление бактерий, осуществляющих разложение/трансформацию ПХБ, происходит с использованием модельных соединений, а именно незамещенного бифенила (встречающегося в природных источниках) и монохлорированных бифенилов (входящих в список ПХБ, но обладающих более низким потенциалом опасности, чем высоко хлорированные бифенилы) [Kim, Picardal, 2000; Park et al., 2001; Hatamian-Zarmi et al., 2009].
Способность к разложению ПХБ, в том числе и монохлорированных бифенилов, выявлена у штаммов родов Achromobacter, Alcaligenes, Aquamicrobium, Arthrobacter, Bacillus, Brevibacillus, Brevibacterium, Castellaniella, Ceriporia, Chitinophaga, Comamonas, Cupriavidus, Enterobacter, Hydrogenophaga, Janibacter, Janthinobacterium, Luteibacter, Mesorhizobium, Ochrobactrum, Paenibacillus, Pandoraea, Phanerochaete, Pseudomonas, Rhodococcus, Shigella, Sphingobium, Sphingomonas, Stenotrophomonas, Subtercola, Talaromyces и Williamsia [Hou et al., 2000; Pieper, Seeger, 2008; Cao et al., 2011; Ponce et al., 2011; Colbert et al., 2013; Somaraja et al., 2013; Liang et al., 2014; Nam et al., 2014; Ilori et al., 2015; Hu et al., 2015; Atago et al., 2016; Shuai et al., 2016; Kour et al., 2019]. География выделения штаммов-деструкторов ПХБ обширная и охватывает все континенты. Однако преимущественными резервуарами для выявления бактерий с деградативной активностью в отношении ПХБ являются территории, длительное время загрязненные высокими концентрациями данных поллютантов [Masai et al., 1995; Furukawa, 2000; Sakai et al., 2003; Jia et al., 2008; Xu et al., 2011; Bako et al., 2021].
Цель настоящего исследования – изучение возможности развития штаммов-деструкторов хлорбифенилов в почвах, не подверженных загрязнению ПХБ.
Материалы и методы исследования
Бактериальные штаммы
Для изучения биодеградативной активности к бифенилу и хлорбифенилам были отобраны 54 аэробных бактериальных штамма, ранее выделенные из почв территории ООПТ – охраняемого ландшафта регионального значения Осинская лесная дача в кварталах 11 и 32, и хранящиеся в режиме криоконсервации (15% глицерин, -80ºС (Evosafe-seriesTM VF620-86, SNIJDERS Scientific, Holland)) в лаборатории микробиологии техногенных экосистем «ИЭГМ УрО РАН» [Бузмаков, Гатина, 2009; Егорова и др., 2017].
Реактивы, среды
В работе использовали аналитически чистые химические реактивы: бифенил (>98%), 2-хлорбифенил (2ХБ) (>98%), 3-хлорбифенил (3ХБ) (>98%), 4-хлорбифенил (4ХБ) (>98%),4-хлорбензойная кислота (4ХБК) (>98%), 3-хлорбензойная кислота (3ХБК) (>98%), 2-хлорбензойная кислота (2ХБК) (>98%) фирмы Sigma-Aldrich (Steinheim, Germany).
Среда LB состава (г/л): дрожжевой экстракт – 5.0, триптон – 10.0, хлорид натрия – 10.0.
Среда К1 состава (г/л): K 2 HPO 4 ×3H 2 O - 3.2, NaH 2 PO 4 ×2H 2 O - 0.4, (NH 4 ) 2 SO 4 - 0.5, MgSO 4 ×7H 2 O - 0.15, Ca(NO 3 ) 2 - 0.01.
Для получения плотной питательной среды вносили агар-агар в концентрации 10 г/л.
Периодическое культивирование на бифениле
Штаммы после криоконсервации восстанавливали на плотной среде LB, и культивировали в термостате суховоздушном ТС-1/80 СПУ (Санкт-Петербург, Россия) при 28ºС, 7 сут. Методом посева с плотной среды в жидкую, культуры далее помещали в колбы Эрленмейера объемом 250 мл, содержащих 100 мл минеральной среды К1. В качестве источника углерода в колбы вносили бифенил до конечной концентрации 1.0 г/л. Культивирование проводили на термостатируемой круговой качалке (Environmental Shaker-Incubator ES 20/60, BioSan, Латвия) при 120 об/мин и 28˚С в течение 5 сут. Измерение оптической плотности культуры производили на спектрофотометре BioSpec-mini (Shimadzu, Япония), при длине волны 600 нм (ОП 600 ).
Деструкция монохлорированных бифенилов
Разложение монохлорированных бифенилов осуществляли в экспериментах с отмытыми клетками. Бактериальную культуру, предварительно выращенную в минеральной среде К1 с бифенилом в качестве источника углерода до ОП 600 =1.0 о.е., центрифугировали при 9 660 g в течение 3 мин. на центрифуге MiniSpin (Eppendorf, Germany). Клетки бактериальной культуры, ресуспендированные в среде К1, концентрировали до ОП 600 =2.0 о.е. и помещали по 1 мл во флаконы с завинчивающимися крышками. Монохлорбифенилы вносили в виде ацетонового раствора до конечной концентрации 50 мг/л. Инкубацию производили на термостатируемой круговой качалке (Environmental Shaker-Incubator ES 20/60, BioSan, Латвия) при 120 об/мин и +28˚С. Концентрацию монохлорбифенилов оценивали через 24 ч. инкубации.
Анализ концентрации монохлорбифенилов
Количественный анализ монохлорированных бифенилов проводили в условиях ГХ-МС: газовый хроматограф Agilent 6890N с масс-селективным детектором и кварцевой капиллярной колонкой НР-5MS (длина 30 м, диаметр 0.25 мм) (Agilent Technology, США). При программировании температуры согласно [Hernandez et al., 1997]. Расчет содержания хлорбифенила в каждом исследуемом образце проводили методом внутренней нормализации. На основании полученных расчетных площадей пиков оценивали содержание хлорбифенила после процесса биодеструкции.
Эффективность деструкции рассчитывали по формуле
D (%) = 100 – 2С24, где С24 – концентрация монохлорбифенила в образце через 24 ч. инкубации, 2 – коэффициент пересчета.
Анализ концентрации монохлорбензойных кислот
Наличие хлорбензойных кислот определяли методом ВЭЖХ. Для анализа культуральную жидкость очищали от бактериальных клеток центрифугированием (9 660 g, 3 мин., центрифуга miniSpin
(Eppendorf, Германия)). Наличие в надосадочной жидкости хлорбензойных кислот определяли на хроматографе LC-20A (Shimadzu, Япония) с колонкой Discovery C18 (150 х 4.6 мм или 250 х 4.6 мм) (Supelco, Sigma-Aldrich, США) и УФ-детектором при 205 нм, а также на хроматографе LicArt (Лабконцепт, Россия) с колонкой Inspire C18 (5мкМ, 250 х 4.6 мм) (Dicma Technologies Inc., Китай) и детектором UV-62 при 205 нм. Анализ проводили в системе ацетонитрил-0.1%-ный Н 3 РО 4 (70:30). Качественную идентификацию производили на основе сравнения времени удержания вещества в экспериментальном образце и времени удержания контрольных соединений (монохлорированных бензойных кислот). Количественную оценку производили методом внутренней нормализации на основании пересчета площадей пиков опытных и контрольных образцов.
Статистический анализ
Все эксперименты проводили в трехкратной повторности. Полученные данные обрабатывали с использованием стандартных пакетов компьютерных программ Microsoft Excel и Statistica 6.0.
Амплификация гена 16S pРНК
Лизис клеток штаммов для получения матрицы ДНК производили в гидроксиде натрия (100 мкл, 0.1М) с последовательным нагреванием до +98ºС и охлаждением до -20ºС. Амплификацию генов 16S рРНК на матрице ДНК бактерий осуществляли на приборе С1000 Touch (Bio-Rad, США) с универсальными бактериальными праймерами 27F и 1492R.
Определение нуклеотидной последовательности гена 16S pРНК
Определение нуклеотидных последовательностей гена 16S рРНК проводили с применением набора реактивов «GenSeq-100» (Синтол, Россия) на автоматическом секвенаторе Нанофор 05 (Синтол, Россия) согласно рекомендациям производителя. Анализ полученных последовательностей осуществляли с использованием программ Sequence Scanner v. 2.0, MEGA Х . Поиск гомологичных последовательностей осуществляли по международной базе данных EzBioCloud . Для построения филогенетических деревьев использовали метод «neighbor-joining» программы MEGA Х. Эволюционные расстояния рассчитывали с использованием метода «p-distance». Статистическую достоверность ветвления («bootstrap»-анализ) оценивали на основе 1000 альтернативных деревьев.
Результаты и их обсуждение
Из лабораторной коллекции были отобраны аэробные бактериальные штаммы, ранее выделенные из почв ООПТ Осинская лесная дача. В результате культивирования в минеральной среде К1 с бифенилом, как единственным источником углерода, установлено, что 32 штамма способны использовать бифенил как ростовой субстрат. Оптическая плотность жидких культур данных штаммов достигала значений 0.5– 1.2 о.е. при длине волны 600 нм. В результате дальнейшего скрининга, было установлено, что способностью к разложению монохлорированных бифенилов обладают 16 штаммов.
Анализ гена 16S рРНК показал, что штаммы-деструкторы принадлежат к родам Achromobacter , Rho-dococcus , Pseudomonas , Delftia и Stenotrophomonas (рис. 1–5). Интересно отметить, что основную долю (56.25%) среди отобранных штаммов-деструкторов монохлорбифенилов составляют штаммы рода Achromobacter (рис. 1). В литературе описано несколько представителей данного рода, способных осуществлять разложение хлорзамещенных бифенилов. На примере штаммов Achromobacter sp. B-218, Achromobacter sp. BP3 и Achromobacter sp. 3YC3 показана хромосомная и плазмидная локализация генов деструкции бифенила/ПХБ [Witzig et al., 2006; Hong et al., 2009; Ilori et al., 2015]. Наиболее подробно особенности деструкции хлорбифенилов описаны для штамма Achromobacter xylosoxidans IR08, который способен утилизировать 4,4’-диХБ без накопления токсичных промежуточных продуктов [Ilori et al., 2008a].
Штаммы рода Rhodococcus составили 18.75% от общего числа штаммов-деструкторов монохлорбифенилов, выделенных из почв ООПТ Осинская лесная дача (рис. 2), а штаммы рода Pseudomonas – 12.5% (рис. 3).
Полученные ранее данные, описанные в литературе, позволяли предположить, что представители родов Rhodococcus и Pseudomonas, среди выявленных штаммов-деструкторов, должны быть доминирующими [Masai et al., 1995; Furukawa, 2000; Park et al., 2001; Pieper, Seeger, 2008; Hatamian-Zarmi et al., 2009; Nam et al., 2014; Atago et al. 2016; Shuai et al., 2016; Егорова и др., 2017, 2018; Bhattacharya, Khare, 2017; Devi, 2020; Bako et al., 2021]. Однако полученные результаты отличаются от ожидаемых. Доминирующую позицию занимают представители рода Achromobacter. В литературе представлены единичные сообщения, о выделении штаммов данного рода, способных разлагать сложные органические соединения, в том числе хлорированные бифенилы, из незагрязненных почв [Flavia et al., 2018; Tarlachkov et al. 2020;
Hara, Takatsuka, 2022]. Большинство известных деструкторов ПХБ рода Achromobacter изолированы из почв с различной химической нагрузкой [Witzig et al., 2006; Hong et al., 2009; Ilori et al., 2015]. Вероятно, данное различие может быть обусловлено уникальностью территории, на которой производился отбор образцов почв. Данная территория не была подвержена негативному воздействию ПХБ и находится в зоне охраняемого ландшафта, что, по-видимому, способствует развитию уникальных бактериоценозов.
__Osa б riOsa?
"LosalS
Osnl6
_ Osa24
Osal
J ---------------------------------Osa3
I——Osa4
La 网切卧 "LMG 3431 (HG324051)
Osa26
A. kerstersiil^^G 3 441 (HG324052)
A. span 竝 s LMG 5911 (АҮГ70848)
A. d&leyi^-IG 3458 (HG324053)
A. piechaudii^S^C 102461 (BCTKO1000022)
---A. marplaiensis B2 (EU150134)
--------A. mucicolens LMG 26685 (HE613446)
j-A agilis LMG 3411 (HG324050)
I—J. animicus LMG 26690 (HE613448)
14 insua\is LMG 26845 (HF586506)
U. aegrifiicie^ LMG 26852 (HF5B65 07)
p■ A. ruhlandiiLMG 1866 (CADIJL010000070)
—A. insolitus DSM 23807 (CP019325)
-----------------------A. alo^era^AVA-]. (LC094463)
-----------------------------------J. aestuarii KS-M25 (MH65175 。)
且 xybsoxidcim NBR.C 15126 (CP00695 8)
-I J. denitrificans DSM 30026 (¥14907)
J. ruhlandii ATCC 15749 (AB010840)
L A. pulmonis LMG 26696 (HE79855 2)
_A 口砲勿 LMG 2685 7 (HF5 865 08)
A. dol&m LMG 26&40' (HF5&6509)
—A. v&tensilvae LMG 303 78 (LT9765 03)
0.0010
Рис. 1. Филогенетическое дерево, построенное для представителей рода Achromobacter на основании анализа нуклеотидной последовательности гена 16S рРНК.
Штаммы, исследованные в настоящей работе, выделены жирным шрифтом. Анализ включал 31 нуклеотидную последовательность. Эволюционный анализ проводился в MEGA X
[Phylogenetic tree constructed for representatives of the genus Achromobacter based on analysis of the nucleotide sequence of the 16S rRNA gene.
The strains studied in this work are highlighted in bold. The analysis included 31 nucleotide sequences. Evolutionary analysis was carried out in MEGA X]
「 R f;fcerens!sFXJ9.536 (OP167981)
I_ R. ox\ : bertzo7iivorans S2-17 (KY765341)
尺 M 例 ms 飮 sNBRC 14363 (BCXBO1000074)
艮 j 第庁 DSM 44719 (FNTLOWOOOO1) 员 koreefisis DSM 44498 [TNSVO1000005) ______________兄 h-onopohus XEAU-X1L.12 (KF887492)
--- --------Л tukisamuensis JCM 11308 (jgL1102290) -------------Л. maanshayiemis DSXI44675 (jgi. 11022 8 6)
- --------尺 tiaqmgeusis Z1 (MH205096)
,—R.pedocdaVCn (KT301938)
—--------Л. са/ісіііриүе/isis \IBRL 3 5 3 (JN164649)
,——旦“位 Ktm-20 (MF405107)
"I------------------- 兄痴位。必 UC33 〔 КТ301939)
-------------R gannanensis Ml (KX887333) --------------------灵 speiaei C9-5 (MK605286)
-----------------尺切#賞械 DL 2 ML 杨 6?321) -------------A. ч-r^Isimie^is NBRC 1006 0 ; (BA\W01000105) --------------R. ^iatomas DSM 44892 (jgi. 1102288)
. ! Л qingshengii JCM 15477 (LRRJO1000O16)
i-------- 兄 baikonure^is GTC 1041 (AB071951) ------------注母七忘打血 sNBRC 14531 (ВС\таО1 00002 3)
Osa 39
OsaS
-------Osa 11 冗任^筋叩。加 NBRC 15567 (BCRM01O00055)
! --------冗 cercidiphylli ҮШ 65003 (EU325542)
'--------Л cerastii C5 (FR714842)
----------------------------------^ psychrotolerans СМА.^ 1533 (КҮЗ 17932)
R. trifolii^ (FR714S43)
氏 corynebacterioides DSM 20151 (AF430066)
0.0050 1
Рис. 2. Филогенетическое дерево, построенное для представителей рода Rhodococcus на основании анализа нуклеотидной последовательности гена 16S рРНК.
Штаммы, исследованные в настоящей работе, выделены жирным шрифтом. Анализ включал 30 нуклеотидных последовательностей. Эволюционный анализ проводился в MEGA X
[Phylogenetic tree constructed for representatives of the genus Rhodococcus based on analysis of the nucleotide sequence of the 16S rRNA gene.
The strains studied in this work are highlighted in bold. The analysis included 30 nucleotide sequences. Evolutionary analysis was carried out in MEGA X]
P. farris SWRI79 (GCA 019145235.1 21)
L P" piscicola P50 (LR797558)
_ P. nunensis In5 (GQ254719)
」 R 加 m 威位 NBRC 103147 (BDAF01000092) 1P. silesiensis АЗ (KX276 592)
Bc^i^FBF102 (LOHF0100003 3)
「 P.coronqfaciens CFBP 2216 (LJRTO1000005)
】 F 齿历 (DOAB1069)
■ P. avellanae BPIC 631 (AKBS0100B74)
7 尸• саш^Ь 加白 CFBP 2341 (AJ492827)
-P. caricapapaxa^ ATCC 33615 (D84010) P. 府迎 CFBP 3225 (JYHE01000183) -P, amygdali CFBP 3206 (JYHBO1000005) P tremae CFBP 6111 (AJ492826)
-P, 11G3 価血 ATCC 13522 (AB021402) г P. 5y 内拜飲@ KCTC 12500 (KI657453)
P. congelansDSM 14939 (FNJHO1000022) 尸.血如勿勿依 0 JCM 2400 (AB021378) P. cerasi 8 (LT222319)
Osa 27
П Osa 28
P sapomphilaDSM.9151 (FNTJO1000001)
sesami SI-P133 (EU912472) ----尸. c^rnicola K1S02-6 (QYUR01000006) -----P. ghrc 妙 KMM 9500 (LC011944)
0 温口
Рис. 3. Филогенетическое дерево, построенное для представителей рода Pseudomonas на основании анализа нуклеотидной последовательности гена 16S рРНК.
Штаммы, исследованные в настоящей работе, выделены жирным шрифтом. Анализ включал 25 нуклеотидных последовательностей. Эволюционный анализ проводился в MEGA X
[Phylogenetic tree constructed for representatives of the genus Pseudomonas based on analysis of the nucleotide sequence of the 16S rRNA gene.
The strains studied in this work are highlighted in bold. The analysis included 25 nucleotide sequences. Evolutionary analysis was carried out in MEGA X]
Выявлены единичные представители родов Delftia и Stenotrophomonas (на долю каждого приходится 6.25%) среди отобранных штаммов-деструкторов монохлорированных бифенилов (рис. 4, 5). В литературе описан штамм Stenotrophomonas maltophila GS-103, проявляющий активность в отношении 2-хлорбифенила [Somaraja et al., 2013]. Штаммы рода Delftia , осуществляющие разложение монохлорбифенилов, в настоящей работе выявлены впервые.
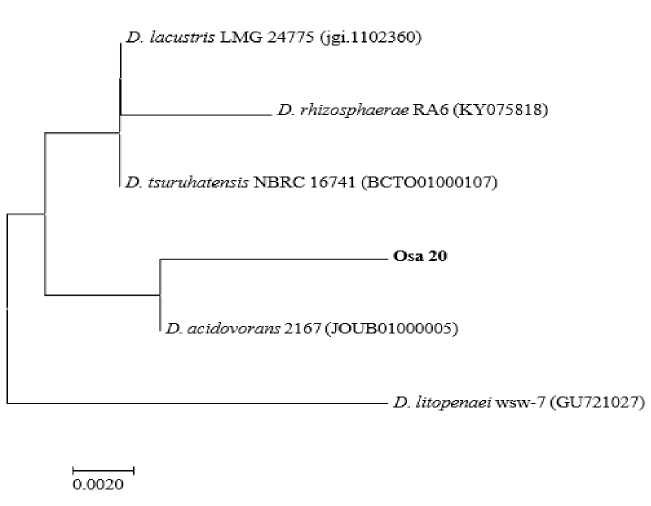
Рис. 4. Филогенетическое дерево, построенное для представителей рода Delftia на основании анализа нуклеотидной последовательности гена 16S рРНК.
Штаммы, исследованные в настоящей работе, выделены жирным шрифтом. Анализ включал 6 нуклеотидных последовательностей. Эволюционный анализ проводился в MEGA X
[Phylogenetic tree constructed for representatives of the genus Delftia based on analysis of the nucleotide sequence of the 16S rRNA gene.
The strains studied in this work are highlighted in bold. The analysis included 6 nucleotide sequences. Evolutionary analysis was carried out in MEGA X]
---------------Osa 13
.S 此/ raeDSM 18941 (LDJJO1000011)
£ nitritireducens DSM 12575 (LDJGO1000040)
S. pictomm JCM 9942 (BAZIO1000187)
----------------- ------------S humi DSM 1 &929 (Ы>Л01000044)
S acidaminiphila JCM 13310 (LDJOO1000053)
S mori DY006 (ON514073)
--------------------------------S koreensis DSM 17805 (LDJHO1000020)
0.0050
Рис. 5. Филогенетическое дерево, построенное для представителей рода Stenotrophomonas на основании анализа нуклеотидной последовательности гена 16S рРНК.
Штаммы, исследованные в настоящей работе, выделены жирным шрифтом. Анализ включал 9 нуклеотидных последовательностей. Эволюционный анализ проводился в MEGA X
[Phylogenetic tree constructed for representatives of the genus Stenotrophomonas based on analysis of the nucleotide sequence of the 16S rRNA gene.
The strains studied in this work are highlighted in bold. The analysis included 9 nucleotide sequences. Evolutionary analysis was carried out in MEGA X]
С использованием метода ГХ-МС установлена остаточная концентрация монохлорированных бифенилов в экспериментах по биодеградации с отмытыми клетками (табл. 1). Расчет эффективности деструкции показал, что для 2-хлорбифенила данный показатель составил 22.9–100%, для 3-хлорбифенила – 13.34–100%, а для 4-хлорбифенила – 8.74–100%. Выявлено два штамма Achromobacter sp. Osa 3 и Achromobacter sp. Osa 24, которые разлагают все три конгенера хлорбифенила на 100%. Следует отметить, что штамм Delftia sp. Osa 20 осуществляет 100%-ную деструкцию 3- и 4-хлорбифенилов, при этом не проявляет активности в отношении 2-хлорбифенила. Высокие показатели деструкции характерны для представителей родов Pseudomonas и Rhodococcus . Штаммы Rhodococcus ruber P25 и Rhodococcus wrati-slaviensis КТ112-7 осуществляют 100%-ную биоконверсию 2- и 4-хлорбифенилов за 24 ч. [Плотникова и др., 2012; Егорова и др., 2018]. Pseudomonas sp. CB-3 разлагает 100% 4-хлорбифенила в начальной концентрации 50 мг/л за 12 ч., а штамм Burkholderia xenovorans LB400 (идентифицированный в ранних работах как Pseudomonas ) осуществляет трансформацию данного субстрата на 98% за 96 ч. [Bhattacharya, Khare, 2017; Xing et al., 2020]. Штаммы, изолированные с территории Нигерии, Ralstonia sp. SA-3 и Ralstonia sp. SA-4 осуществляют разложение всех конгенеров монохлорированных бифенилов на 88–99% за 6–10 ч. при начальной концентрации 100 ppm [Adebusoye et al., 2008]. В работе Ilori с соавторами [2008] показано, что штамм Achromobacter xylosoxidans IR08 осуществляет полную биодеструкцию 2-хлорбифенила, 3-хлорбифенила и 4-хлорбифенила при начальной концентрации 0.27 ммоль/л за 96 ч. [Ilori et al., 2008b]. Таким образом, исследованные в настоящей работе бактериальные культуры не уступают по своей биодеградативной активности в отношении монохлорированных бифенилов известным штаммам, выделенным из загрязненных биотопов.
Таблица 1
Концентрация (мг/л) монохлорированных бифенилов через 24 ч. биодеструкции [Concentration (mg/l) of monochlorinated biphenyls after 24 hours of biodegradation]
|
Штамм |
2-ХБ |
3-ХБ |
4-ХБ |
|
Osa 1 |
23.75±0.01 |
26.2±0.02 |
45.63±0.04 |
|
Osa 3 |
0 |
0 |
0 |
|
Osa 4 |
38.55±0.02 |
19.52±0.08 |
1.98±0.02 |
|
Osa 6 |
9.18±0.02 |
6.93±0.04 |
19.55±0.05 |
|
Osa 7 |
0 |
43.33±0.03 |
0 |
|
Osa 8 |
19.45±0.05 |
0 |
18.51±0.02 |
|
Osa 11 |
13.94±0.03 |
0 |
18.23±0.03 |
|
Osa 13 |
24.32±0.05 |
33.09±0.02 |
14.26±0.01 |
|
Osa 16 |
15.95±0.04 |
30.78±0.02 |
41.52±0.02 |
|
Osa 20 |
50±0.1 |
0 |
0 |
|
Osa 24 |
0 |
0 |
0 |
|
Osa 25 |
0 |
22.51±0.04 |
27.68±0.05 |
|
Osa 26 |
15.61±0.02 |
19.31±0.04 |
36.67±0.03 |
|
Osa 27 |
6.83±0.01 |
6.24±0.03 |
15.32±0.02 |
|
Osa 28 |
3.81±0.01 |
5.15±0.05 |
5.07±0.02 |
|
Osa 29 |
28.71±0.02 |
27.71±0.03 |
16.15±0.03 |
Анализ основных метаболитов бактериальной трансформации монохлорированных бифенилов с применением метода ВЭЖХ показал, что, за исключением разложения 2-хлорбифенила штаммами Rhodo-coccus sp. Osa 11 и Delftia sp. Osa 20, в культуральной среде присутствуют монохлорированные бензойные кислоты (табл. 2).
Интересно отметить, что для штаммов Achromobacter sp. Osa 3 и Achromobacter sp. Osa 24, полностью разлагающих монохлорбифенилы, через 24 ч. инкубации в среде зафиксировано незначительное количество 2-хлорбензойной (0.678–0.889 мг/л) и 3-хлорбензойной кислот (0.014–0.041 мг/л), тогда как 4-хлорбензойная кислота накапливалась в большем количестве (4.081–4.204 мг/л). Можно предположить, что штаммы Osa 3 и Osa 24 осуществляют дальнейшую трансформацию образующихся в качестве метаболитов 2- и 3-хлорбензойных кислот.
Штамм Rhodococcus sp. Osa 11 осуществлял деструкцию 2-хлорбифенила на 72.12%, однако в его культуральной среде не обнаружены вероятные метаболиты. Полученный результат свидетельствует о разложении 2-хлорбифенила штаммом Osa 11 до соединений основного обмена клетки.
Анализ образующихся метаболитов позволяет предположить, что процесс деструкции монохлориро-ванных бифенилов штаммами, выделенными из почв ООПТ Осинская лесная дача, происходит по классическому бифенильному пути с окислением под действием бифенил диоксигеназы незамещенного кольца в молекуле хлорбифенила [Egorova et al., 2020].
Таблица 2
Концентрация (мг/л) монохлорированных бензойных кислот через 24 ч. биодеструкции [Concentration (mg/l) of monochlorinated benzoic acids after 24 hours of biodegradation]
|
Штамм |
2-ХБК |
3-ХБК |
4-ХБК |
|
Osa 1 |
0.108±0.001 |
0.164±0.002 |
0 |
|
Osa 3 |
0.889±0.001 |
0.014±0.002 |
4.081±0.004 |
|
Osa 4 |
0.238±0.002 |
0.174±0.004 |
2.112±0.002 |
|
Osa 6 |
0.554±0.002 |
0.039±0.001 |
1.315±0.002 |
|
Osa 7 |
0.420±0.002 |
3.391±0.003 |
3.758±0.002 |
|
Osa 8 |
0.215±0.003 |
0.093±0.001 |
2.054±0.001 |
|
Osa 11 |
0 |
2.304±0.004 |
0.789±0.001 |
|
Osa 13 |
0.472±0.002 |
0.123±0.002 |
1.979±0.003 |
|
Osa 16 |
0.079±0.001 |
0.104±0.002 |
3.667±0.002 |
|
Osa 20 |
0 |
1.346±0.004 |
1.863±0.002 |
|
Osa 24 |
0.678±0.02 |
0.041±0.001 |
4.204±0.004 |
|
Osa 25 |
0.365±0.001 |
0.192±0.002 |
1.892±0.003 |
|
Osa 26 |
0.695±0.002 |
5.369±0.004 |
3.766±0.003 |
|
Osa 27 |
0.011±0.001 |
1.430±0.003 |
3.637±0.003 |
|
Osa 28 |
0.117±0.001 |
1.016±0.003 |
2.067±0.002 |
|
Osa 29 |
0.285±0.002 |
2.748±0.002 |
4.563±0.003 |
Заключение
В результате проведенных исследований установлено, что в почвах ООПТ Осинская лесная дача присутствуют аэробные бактерии, обладающие биодеградативным потенциалом в отношении монохлориро-ванных бифенилов. Анализ филогенетического разнообразия показал, что выделенные штаммы-деструкторы принадлежат к родам Achromobacter, Rhodococcus , Pseudomonas , Stenotrophomonas, Delftia . Эффективность деструкции монохлорированных бифенилов бактериальными культурами варьировала от 8.74% до 100%. Наибольшей биодеградативной активностью характеризовались штаммы Achromobacter sp. Osa 3 и Achromobacter sp. Osa 24, осуществляющие 100%-ную деструкцию всех конгенеров монохлорбифенилов. Образование в качестве основных метаболитов монохлорированных бензойных кислот свидетельствует о том, что трансформация хлорбифенилов выделенными штаммами происходит по классическому бифенильному пути. Таким образом, выделенные из незагрязнённых ПХБ почв бактерии, обладают высоким биодеградативным потенциалом в отношении монохлорированных бифенилов.
Список литературы Разнообразие культивируемых бактерий-деструкторов монохлорированных бифенилов в почвах охраняемого ландшафта
- Бузмаков С.А., Гатина Е.Л. Зонирование особо охраняемой природной территории «Осинская лесная дача» // Географический вестник. 2009. № 1. С. 51-55. https://cyberleninka.ru/article/n/zonirovanie-osobo-ohranyaemoy-prirodnoy-territorii-osinskaya-lesnaya-dacha (дата обращения: 21.06.2024).
- Егорова Д.О. и др. Биоремедиация почвы, длительное время загрязненной дихлордифенилтрихлор-этаном, с использованием аэробного штамма Rhodococcus wratislaviensis CH628 // Почвоведение. 2017. № 10. С. 1262-1269. https://doi.org/10.7868/S0032180X1710001X.
- Егорова Д.О. и др. Особенности разложения хлорированных бифенилов штаммом Rhodococcus wratislaviensis КТ112-7 в условиях засоления // Прикладная биохимия и микробиология. 2018. Т. 54, № 3. С. 253-263. https://doi.org/10.7868/S0555109918030042.
- Плотникова Е.Г. др. Особенности разложения 4-хлорбифенила и 4-хлорбензойной кислоты штаммом Rhodococcus ruber P25 // Микробиология. 2012. Т. 81, № 2. С. 159-170. https://doi.org/10.1134/S0026261712020117.
- Трегер Ю. СОЗ - стойкие и очень опасные // The Chemical Journal. 2013. № 1. P. 30-34. https://tcj.ru/journal/jan-fev-2013.
- Adams C.I.M. et al. Toxicological effects of polychlorinated biphenyls (PCBs) on freshwater turtles in the United States // Chemosphere. 2016. Vol. 154. P. 148-154. https://doi.org/10.1016/j.chemosphere.2016.03.102.
- Adebusoye S.A. et al. Characterization of multiple novel aerobic polychlorinated biphenyl (PCB)-utilizing bacterial strains indigenous to contaminated tropical African soils // Biodegradation. 2008. Vol. 19, № 1. P. 145159. https://doi.org/10.1007/s10532-007-9122-x.
- Atago Y. et al. Identification of novel extracellular protein for PCB/biphenyl metabolism in Rhodococcus jostii RHA1 // Bioscience, Biotechnology, and Biochemistry. 2016. Vol. 80, № 5. P. 1012-1019. https://doi.org/10.1080/09168451.2015.1127134.
- Bako C.M. et al. Biodegradation of PCB congeners by Paraburkholderia xenovorans LB400 in presence and absence of sediment during lab bioreactor experiments // Environmental Pollution. 2021. Vol. 271. Article 116364. https://doi.org/10.1016/j.envpol.2020.116364.
- Bhattacharya A., Khare S.K. Biodegradation of 4-chlorobiphenyl by using induced cells and cell extract of Burkholderia xenovorans // Bioremediation Journal. 2017. Vol. 21. P. 109-118. https://doi.org/10.1080/10889868.2017.1282940.
- Cao Y.M. et al. Analysis of PCBs degradation abilities of biphenyl dioxygenase derived from Enterobacter sp. LY402 by molecular simulation // New Biotechnology. 2011. Vol. 29, № 1. P. 90-98. https://doi.org/10.1016/j.nbt.2011.08.005.
- Colbert C.L. et al. Structural characterization of Pandoraea pnomenusa B-356 biphenyl dioxygenase reveals features of potent polychlorinated biphenyl-degrading enzymes // PLoS One. 2013. Vol. 8, № 1. Article e52550. https://doi.org/10.1371/journal.pone.0052550.
- Devi N.L. Persistent Organic Pollutants (POPs): Environmental risks, toxicological effects, and bioremediation for Environmental Safety and Challenges for Future Research // Bioremediation of Industrial Waste for Environmental Safety / G. Saxena, R. Bharagava, eds. Singapore: Springer, 2020. Р. 53-76. https://doi.org/10.1007/978-981-13-1891-7_4.
- Egorova D.O. et al. Biodegradability of hydroxylated derivatives of commercial polychlorobiphenyls mixtures by Rhodococcus-strains // Journal of Hazardous Materials. 2020. Vol. 400. Article 123328. https://doi.org/10.1016/j.jhazmat.2020.123328.
- Final act of the Conference of Plenipotentiaries on the Stockholm, 22-23 May // UNEP / POPS/CONF/4. United Nations Environment Programme. Geneva, 2001. 44 p.
- Flavia A. et al. Degradation of atrazine by Pseudomonas sp. and Achromobacter sp. isolated from Brazilian agricultural soil // International Biodeterioration and Biodegradation. 2018. Vol. 130. P. 17-22. https://doi.org/10.1016/j.ibiod.2018.03.011.
- Furukawa K. Biochemical and genetic bases of microbial degradation of polychlorinated biphenyls (PCBs) // The Journal of General and Applied Microbiology. 2000. Vol. 46, № 6. P. 283-296. https://doi.org/10.2323/jgam.46.283.
- Hara T., Takatsuka Y. Aerobic polychlorinated biphenyl-degrading bacteria isolated from the Tohoku region of Japan are not regionally endemic // Canadian Journal of Microbiology. 2022. Vol. 68, № 3. P. 191-202. doi: 10.1139/cjm-2021-0056.
- Hatamian-Zarmi A. et al. Extensive biodegradation of highly chlorinated biphenyl and Aroclor 1242 by Pseudomonas aeruginosa TMU56 isolated from contaminated soils // International Biodeterioration and Biodegradation. 2009. Vol. 63, № 6. P. 788-794. https://doi.org/10.1016/j.ibiod.2009.06.009.
- Hernandez B.S. et al. Terpene-utilizing isolates and their relevance to enhanced biotransformation of polychlorinated biphenyls in soil // Biodegradation. 1997. Vol. 8, № 3. P. 153-158. https://doi.org/10.1023/A:1008255218432.
- Hong Q. et al. Isolation of a biphenyl-degrading bacterium, Achromobacter sp. BP3, and cloning of the bph gene cluster // International Biodeterioration and Biodegradation. 2009. Vol. 63, № 4. P. 365-370. https://doi.org/10.1016/j.ibiod.2008.10.009.
- Hou L.H. et al. Phylogenetic characterization of several para- and meta-PCB dechlorinating Clostridium species: 16s rDNA sequence analyses // Letters in Applied Microbiology. 2000. Vol. 30, № 3. P. 238-243. https://doi.org/10.1046/j.1472-765x.2000.00709.x.
- Hu J. et al. Sphingobium fuliginis HC3: a novel and robust isolated biphenyl-and polychlorinated biphen-yls-degrading bacterium without dead-end intermediates accumulation // PloS one. 2015. Vol. 10, № 4. Article e0122740. https://doi.org/10.1371/journal.pone.0122740.
- Ilori M.O. et al. Aerobic mineralization of 4,4'-dichlorobiphqnyl and 4-chlorobenzoic acid by a novel natural bacterial strain that grows poorly on benzoate and biphenyl // World J. Microbiol. Biotechnol. 2008a. Vol. 24. P. 1259-1265. https://doi.org/10.1007\s11274-007-9597-y
- Ilori M.O. et al. Degradation and mineralization of 2-chloro-, 3-chloro-and 4-chlorobiphenylby a newly characterized natural bacterial strain isolated from an electrical transformer fluid-contaminated soil // Journal of Environmental Sciences. 2008b. Vol. 20, № 10. P. 1250-1257. https://doi.org/10.1016/s1001-0742(08)62217-2.
- Ilori M.O. et al. Catabolic plasmid specifying polychlorinated biphenyl degradation in Cupriavidus sp. strain SK-4: Mobilization and expression in a pseudomonad // Journal of Basic Microbiology. 2015. Vol. 55, № 3. P. 338-345. https://doi.org/10.1002/jobm.201200807.
- Jia L.Y. et al. Isolation and characterization of comprehensive polychlorinated biphenyl degrading bacterium, Enterobacter sp. LY402 // J. Microbiol. Biotechnol. 2008. Vol. 18, № 5. P. 952-957. PMID: 18633297.
- Kim S., Picardal F.W. A novel bacterium that utilizes monochlorobiphenyls and 4-chlorobenzoate as growth substrates // FEMS Microbiology Letters. 2000. Vol. 185, № 2. P. 225-229. https://doi.org/10.1111/j.1574-6968.2000.tb09066.x.
- Kour D. et al. Gene manipulation and regulation of catabolic genes for biodegradation of biphenyl compounds // In New and Future Developments in Microbial Biotechnology and Bioengineering. 2019. P. 1-23. https://doi.org/10.1016/B978-0-444-63503-7.00001-2.
- Liang Y. et al. Potential for polychlorinated biphenyl biodegradation in sediments from Indiana Harbor and Ship Canal // International Biodeterioration and Biodegradation. 2014. Vol. 89. P. 50-57. https://doi.org/10.1016/j.ibiod.2014.01.005.
- Masai E. et al. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1 // Appl. Environ. Microbiol. 1995. Vol. 61, № 6. P. 2079-2085. https://doi.org/10.1128/aem.61.6.2079-2085.1995.
- Müller M.H.B. et al. Organochlorine pesticides (OCPs) and polychlorinated biphenyls (PCBs) in human breast milk and associated health risks to nursing infants in Northern Tanzania // Environmental research. 2017. Vol. 154. P. 425-434. https://doi.org/10.1016/j.envres.2017.01.031.
- Nam I.H. et al. Biodegradation of biphenyl and 2-chlorobiphenyl by a Pseudomonas sp. KM-04 isolated from PCBs-contaminated coal mine soil /// Bulletin of Environmental Contamination and Toxicology. 2014. Vol. 93, № 1. P. 89-94. https://doi.org/10.1007/s00128-014-1286-6.
- Negret-Bolagay D. et al. Persistent organic pollutants: the trade-off between potential risks and sustainable remediation methods // Journal of environmental Management. 2021. Vol. 300. Article 113737. https://doi.org/10.1016/j.jenvman.2021.113737.
- Park S.H. et al. Adaptive and cross-protective responses of Pseudomonas sp. DJ-12 to several aro-matics and other stress shocks // Current Microbiology. 2001. Vol. 43, № 3. P. 176-181. https://doi.org/10.1007/s002840010283.
- Pieper D.H., Seeger M. Bacterial metabolism of polychlorinated biphenyls // Journal of Molecular Microbiology and Biotechnology. 2008. Vol. 15, № 2-3. P. 121-138. https://doi.org/10.1159/000121325.
- Ponce B.L. et al. Antioxidant compounds improved PCB-degradation by Burkholderia xenovorans strain LB400 // Enzyme and Microbial Technology. 2011. Vol. 49, № 6-7. P. 509-516. https://doi.org/10.1016/j.enzmictec.2011.04.021.
- Reddy A.V.B. et al. Polychlorinated biphenyls (PCBs) in the environment: recent updates on sampling, pretreatment, cleanup technologies and their analysis // Chemical Engineering Journal. 2019. Vol. 358. P. 1186-1207. https://doi.org/10.1016/j.cej.2018.09.205.
- Sakai M. et al. 2-Hydroxypenta-2, 4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1 // Appl. Environ. Microbiol. 2003. Vol. 69, № 1. P. 427433. https://doi.org/10.1128/AEM.69.L427-433.2003.
- Shuai J. et al. Regional analysis of potential polychlorinated biphenyl degrading bacterial strains from China // Brazilian Journal of Microbiology. 2016. Vol. 47, № 3. P. 536-541. https://doi.org/10.1016/j.bjm.2014.12.001.
- Somaraja P.K. et al. Molecular characterization of 2-chlorobiphenyl degrading Stenotrophomonas maltophilia GS-103 // Bulletin of Environmental Contamination and Toxicology. 2013. Vol. 91, № 2. P. 148-153. https://doi.org/10.1007/s00128-013-1044-1.
- Tarlachkov S.V. et al. Draft genome sequence of glyphosate-degrading Achromobacter insolitus strain Kg 19 (VKM B-3295), isolated from agricultural soil // Microbiology Resource Announcements. 2020. Vol. 9, № 17. Article e00284-20. doi: 10.1128/MRA.00284-20.
- Warenik-Bany M. et al. Impact of environmental pollution on PCDD/F and PCB bioaccumulation in game animals // Environmental Pollution. 2019. Vol. 255. Article 113159 https://doi.org/10.1016/j.envpol.2019.113159.
- Witzig R. et al. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: links between benzene biodegradation and genes similar to those encoding isopropylbenzene dioxy-genases // Appl. Environ. Microbiol. 2006. Vol. 72, № 5. P. 3504-3514. https://doi.org/10.1128/AEM.72.5.3504-3514.2006.
- Xing Z. et al. Degradation Mechanism of 4-Chlorobiphenyl by Consortium of Pseudomonas sp. Strain CB-3 and Comamonas sp. Strain CD-2 // Current Microbiology. 2020. Vol. 77. P. 15-23. https://doi.org/10.1007/s00284-019-01791-9A.
- Xu L. et al. Congener selectivity during polychlorinated biphenyls degradation by Enterobacter sp. LY402 // Current Microbiology. 2011. Vol. 62, № 3. P. 784-789. https://doi.org/10.1007/s00284-010-9792-1.
- Zhang P. et al. Distribution and transfer pattern of polychlorinated biphenyls (PCBs) among the selected environmental media of Ny-Alesund, the Arctic: as a case study // Marine Pollution Bulletin. 2014. Vol. 89, № 1-2. P. 267-275.
- Zhu L. et al. Degradation mechanism of biphenyl and 4,4'-dichlorobiphenyl cis-dihydroxylation by non-heme 2,3 dioxygenases BphA: A QM/MM approach // Chemosphere. 2020. Vol. 247. Article125844. https://doi.org/10.1016/j.chemosphere.2020.125844.


