Rhizosphere microbiome signalling and biotic stress tolerance in plants
Автор: Sarkar R., Sarkar M., Roychoudhury A.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 2 т.21, 2025 года.
Бесплатный доступ
In the age of ever-increasing population density, the escalating demand for food has to be met. Irrespective of whether individuals prefer animal-derived food or not, the pressure on agricultural systems continues to intensify as it provides the most sustainable means to feed the masses. The climatic variability of crops or dietary preferences is only a part of the problem. The major issue lies in the viability of plants and their resistance to abiotic and biotic stresses. Being a huge threat to agriculture, numerous efforts have been made to study and subdue the effects of biotic stress on plants. One such sustainable approach could be the utilisation of microbes inhabiting the rhizosphere. In this review, we thus shed some light on the practical data to discuss the rhizosphere microbiome and how it affects the plant, particularly in conferring resistance to biotic stresses via priming the host plant and promoting the induced systemic resistance (ISR), antagonistic or competitive interactions with pathogens, strengthening insect pest resistance and control and disease suppression. The prospects that this field holds when combined with superior techniques like genetic engineering are immense for the development of a viable and fully-fledged plan for attaining sustainable agriculture.
Agricultural sustainability, biotic stress, plant pathogenesis, plant immunity, plant microbe interactions, plant disease resistance, rhizosphere engineering, rhizosphere microbiome
Короткий адрес: https://sciup.org/143184721
IDR: 143184721
Текст обзорной статьи Rhizosphere microbiome signalling and biotic stress tolerance in plants
Abbreviations: PAMPs: Pathogen-Associated Molecular Patterns; ETI: Effector-triggered immunity; ETS: Fffector-triggered susceptibility; NB-LRR: Nucleotide binding -leucine rich repeat; PR: Pathogenesis-related; SA: Salicylic acid; PTI: PAMP-triggered immunity; SAR: Systemic acquired resistance; ROS: Reactive Oxygen Species; JA: Jasmonic acid
A plant is said to exhibit ‘stress’ if the presence of any adverse factors negatively impacts its growth, development or normal physiological functioning. his ‘stress’ that we speak of is not only limited to plants but is also a phenomenon observed in every other organism from bacteria to humans. Plants lack an essential quality, viz., motility compared to most other life forms. Plants are sessile and thus have incorporated different strategies throughout evolution to combat all sorts of stress that they are subjected to. Many of these involve complex cross-talks between various pathways to deal with only one stress at a time. he two types of stresses that plants are subjected to, can broadly be classified as ‘abiotic’ and ‘biotic’. Abiotic stresses are contributed by environmental factors and may be either physical or chemical in nature. A plethora of components fall under this category like temperature extremes, drought or water stress, salinity stress, anoxia, oxidative stress, nutrient imbalances, etc. Biotic stresses on the other hand, encompass plant diseases, infections and herbivory contributed by different plant pathogens (bacteria, viruses, fungi and nematodes), animals and even plants themselves, a phenomenon known as allelopathy, which has even been used to our advantage in agriculture (Cheng and Cheng, 2015). Plant responses to these stresses involve a vast range of physiological, biochemical and molecular mechanisms, involving direct alterations in gene expression, like up regulation of stress-specific genes and down-regulation of housekeeping genes as well as transcription factors, epigenetic regulations and finally metabolic changes, leading to stress tolerance. Plants can further be temporarily ‘acclimatized’ to the stress condition to combat stress better. his can then lead to ‘adaptation’, which is reflected as permanent changes to the plant genome, making the stress-survival strategy inherently genetic. A good example of both of these processes can be seen in Yorkshire Fog grass (Holcus lanatus), which has found a way to combat arsenic toxicity while growing in heavy arsenate-contaminated soil via the induction of phytochelatin produced by the enzyme phytochelatin synthase as well as reducing the uptake of arsenic by causing changes to metal transport mechanisms (Meharg et al., 1993). Biotic and abiotic stresses can also interfere with the stress responses in plants when subjected together, leading to a greater severity. Instances have been seen in presence of higher temperature, which facilitates the spread of pathogens or weakens the plant defence mechanisms against biotic stress (Suzuki et al., 2014). In the field, multiple stress conditions are not only possible but practically quite likely to occur. A better understanding of plant stress becomes imperative to employ better defences against plant pathogenesis and predation.
Impact of biotic stress on agriculture and the role of rhizobial microbiome
It is well known that plants suffer highly from biotic stress. he impact however falls not only on agriculture but also on horticulture as well as the industry connected to the ornamental plants. here have been numerous reviews bringing to our notice, the mammoth losses that have been plaguing us (Oerke, 2006; Sharma et al ., 2017; Savary et al ., 2019; Savary et al. , 2012). As a safeguard measure, various strategies have been availed like the use of chemical pesticides, genetically modified (GM) crops, changes in cropping practices, etc. hey however exert their share of unwanted effects (Rani et al ., 2021; Nicolopoulou-Stamati et al ., 2016; Séralini et al ., 2011; Kumar et al ., 2020) on the environment. One particular approach in this regard is to make use of the potent microbes inhabiting the rhizosphere region. hese organisms not only hold the potential to be used as bio-control agents, but also can have highly beneficial roles concerning the promotion of plant growth. Henceforth, we delve deeper into the details of biotic stress, the plant mechanisms to deal with it and analyzing the effects of the rhizospheric microbiome in combating such stress.
Biotic stress
Abiotic stresses can affect plants in a technically predictable fashion, but the intricacy that plant interactions with other biological systems bring about is unmatched. Different bacteria, viruses, fungi, etc. have different modes of action on host plant which tackles such biotic stresses through their ‘immune system’. Not only do biotic stresses directly interfere with plant metabolism and cause plant death via the production of harmful or damaging molecules, but also exert effect on normal physiological processes like global down regulation of photosynthetic genes (Bilgin et al., 2010) which wreaks havoc in the plant system. A deeper understanding of such mechanisms can help in finding novel solutions to create induced resistance in plants. In general, biotic stresses are caused by organisms belonging to one of the four categories: pathogens, pests, herbivores and weeds.
Pathogens are microorganisms like fungi, bacteria, viruses or nematodes which disrupt the normal physiological processes in plants and cause diseases like wilts, rots, blights, etc. Pathogens of bacterial origin include Agrobacterium tumefaciens, whose tumourcausing gene and the respective i-plasmid are now heavily used in genetic engineering for gene transformations (Gelvin, 2003), Xanthomonas campestris pathovars and Erwinia amylovora to name a few (Mansfield et al., 2012). Next on the list are pests, including major insects and mites that can act as pests and cause severe damage to plants. hese organisms like aphids, caterpillars, beetles, etc. directly feed on plant tissue causing physical damage and can also introduce certain toxins or chemicals which impair plant growth and development. An old but significant data that was found in the book ‘New Directions for Biosciences Research in Agriculture: High-Reward Opportunities.’ claimed that as early as in 1983, a whopping $1.3 billion was spent on pesticides to combat the danger caused by plant pests. Over the years, numerous efforts have been made to invest more in eco-friendly methods like biopesticides, and novel strategies like genetic engineering are on a steady rise. Similar in the line of biotic stress caused by the physical damage, are the herbivores. he strategy to deal with them is however quite simple and involves preventing these animals from coming in contact with plants. his has more to do with human interventions like installing nets, fences or scarecrows to prevent herbivory. Plants however do have innate mechanisms as well to deal with herbivory. Last, but not least, are the weeds which although are not necessarily directly harmful, can compete with the cultivated plants for resources like water, nutrients and sunlight. his can usually be countered by proper cropping practices and occasional weeding. Allelopathy is also a mechanism of antagonistic effects shown by a plant species against another; however, it is not very commonly seen to affect plants in agriculture.
Plant resistance to biotic stress he resistance in plants to biotic stress occurs through successive lines of defences. Similar to skin, mucous membranes, etc. in humans, plants also have the first line of defence in the form of physical barriers. his can include thick cuticles, specialized secretory trichomes, waxes and thorns to repel larger herbivore animals. Similarly, plants also produce certain chemical compounds that can serve the same purpose. Secondary metabolites are one such category of chemical substances that the plants produce when they are under stress. As opposed to primary metabolites produced for growth at all times, secondary metabolite production is time and situation-specific. hese compounds may be involved in deterring or preventing the feeding of the plant tissue, serve as precursors in physical defence systems as mentioned earlier or can prove to be toxic to the pathogens. hese can include compounds like cyanogenic glycosides, glucosinolates, alkaloids, phenolics, sterols and even plant hormones like salicylic acid and methyl jasmonate. heir rate, situation-specificity and quantity of production can serve as markers to define and differentiate between the genetic level of resistance and susceptibility of different plant species (Bennett and allshrove, 1994).
Plants have established two levels of pathogen recognition which ultimately trigger their defence responses. he pathogens can enter the plant via different pathways including those utilizing cell-wall degrading enzymes, searching for portals of entry like hydathodes, stomata or physical wounds. he basal or the non-specific defence occurs through the involvement of Pattern Recognition Receptors (PRRs) on plant cells which can recognize Pathogen-Associated Molecular Patterns (PAMPs) (Monaghan and Zipfel, 2012). his PAMP-triggered immunity (P I) is then carried out by a series of downstream signalling and corresponding gene expression. A similar scenario is also seen for stress, caused due to herbivory. Insects or herbivorous animals have certain Herbivore-Associated Elicitors (HAEs), Herbivore-Associated Molecular Patterns (HAMPs) or other general/specific effector molecules which are also recognized by related PRRs in the affected plant. he defence mechanism includes inhibiting and blocking or modification of the pest metabolic pathways. An additional and proficient technique used by the plant systems involves the emission of certain volatile compounds to attract the natural enemies of these pests as well as signalling the neighbouring plants of the presence of the pathogen so that they can be predisposed to tackle the imminent threat, working in a fashion, similar to how interferons work in a human system against a viral attack (Santamaria et al., 2013). he second level of plant immune system is effector-triggered immunity (E I) which can induce hypersensitive responses to contain the spread of infection by killing the locally affected cells by programmed cell death. hese responses are triggered by the products obtained from specific activation and expression of plant resistance (R) genes during the detection of particular elicitors called Avr proteins, which are virulence-associated factors coded by the genes of the pathogen (Spoel and Dong, 2012). his results in effector-triggered susceptibility (E S) and the pathway involves the use of polymorphic nucleotide binding-leucine rich repeat (NB-LRR) proteins. he different pathogen elicitors are recognized by these proteins and activate corresponding defence responses. It is usually effective against biotrophic or hemibiotrophic pathogens, but not efficient against necrotrophs.
he process of infection of a plant by its pathogen involves the entire life cycle of the pathogen in the plant from the point of entry into the plant system, till it leaves the host. Pathogens that require the living host tissue and derive a long-term relationship of deriving nutrients from host cells without killing them are termed as biotrophs. Necrotrophs are pathogens that require dead host cells, aggressively killing the host for its survival. hey are the most harmful type of plant pathogens.
Hemibiotrophs start as biotrophs and then slowly transit to a necrotrophic phase. A state of survival of the plant pathogen in either the soil or in a non-native host till the beginning of the next disease cycle is termed as overwintering. Horizontal resistance refers to plant resistance against different strains or pathovars of a pathogen. It is largely non-specific and the resistance is polygenic, involving the activation of multiple different genes. On the other hand, vertical resistance refers to plant resistance against particular strain(s) of a pathogen which is mainly via activation of one or few specific genes. hile vertical resistance is more important for specific resistance against a particular disease, it is also more susceptible to being ineffective as pathogens evolve to deem their functions ineffective. Systemic acquired resistance is a distinct signal transduction pathway that is triggered after the formation of a necrotic lesion following the events of E I or as the manifestation of a symptom of the disease. After the first attack of the pathogen, a priming process occurs preparing the plant system for a more rapid and robust response upon successive encounters with the same or different strains of the pathogen. It is broad-spectrum, long-term protection provided by the pathogenesis-related (PR) proteins and other defence-related molecules. SAR is also seen to depend on the global accumulation of salicylic acid (SA). Unambiguously speaking, systemic acquired resistance (SAR) serves as a sort of memory response to an already encountered pathogen upon subsequent attacks (Ryals et al., 1996).
he zig-zag model of plant immunity occurs in four phases:
Phase 1: PAMP-triggered immunity is activated after the first attack of a new pathogen to prevent colonization of the pathogen.
Phase 2: Pathogens devise new effectors which can interfere with P I and result in effector-triggered susceptibility (E S) in the plant host.
Phase 3: A specific effector is recognized by one NB-LRR protein (gene-for-gene concept of disease resistance), either directly or indirectly, resulting in effector-triggered immunity (E I). E I is a more accelerated and amplified form of the P I response, resulting in enhanced resistance against the disease and can often lead to hypersensitive reactions at the site of infection.
Phase 4: Evolving pathogens create new bypass mechanisms by developing novel effector genes or by modifying the existing ones to suppress E I. Further, natural selection results in new R gene-specific products in the plant, so that E I can be triggered once again, offering host immunity against the pathogen. his cyclic event of heightening and lowering amplitude of resistance against a specific pathogen continues subsequently (Jones and Dangl, 2006).
Signalling pathways involved in biotic stress
A prevalent signalling pathway observed in both types of immunity is the early activation of ion channels present on the membranes and the subsequent increment in cytosolic calcium levels acting as a secondary messenger. Relatively recent studies have also unmasked that Reactive Oxygen Species (ROS) which are harmful secondary by products of oxidative metabolism, have a huge role in downstream signalling in both biotic and abiotic stress-induced responses. Identification of novel ROS receptors and associated ROS signalling pathways prove to be promising avenues of research which can be used to increase plant resilience to stress (Gechey et al. , 2006; Suzuki et al. , 2012; Mittler et al. , 2022). ROS signalling is often seen to activate mitogen-activated protein kinase pathways to bring about final effector functions (Son et al. , 2011). Schematic of various signalling processes occurring in the rhizosphere is represented in Fig. 1.
Out of all the plant hormones, three stand out as key regulators of mechanisms regulating biotic stress. hey are salicylic acid (SA), jasmonic acid (JA) and the only gas hormone, ethylene (E ). he SA pathway initiates resistance responses against biotrophic and hemibiotrophic pathogens, while the JA and ethylene pathways are triggered in response to necrotrophic pathogens and chewing insects. SA, as previously mentioned, also has a role in inducing systemic acquired resistance (SAR), providing prolonged protection against a wide range of pathogens. here is also evidence of collaborative interactions between defence pathways, operating for the three plant hormones to work more effectively. Notably other hormones like abscisic acid (ABA), auxin, cytokinin, etc. have also proven to be relevant in certain aspects of plant resistance (Gimenez et al., 2018).
Anthropogenic interventions in dealing with biotic stress
Plants have developed in-built mechanisms using an intricate interplay of molecules which are native to the plant. However, plants are often not alone in this battle and are seen to interact with a variety of organisms in their environment. e have briefly mentioned how parasites, predators, allelopathy, etc. aid plants in combating biotic stress, but we have left out one key player, whose contribution has recently been seen to surface. his contribution is made by beneficial microorganisms encompassing rhizosphere region and endophytic microbes. Before we discuss this specific partnership, we will look into the techniques utilized by plant breeders, farmers or researchers to artificially deal with biotic stress. Various methods, including a highly effective remote-sensing approach, have been employed in managing biotic stress. Exploring vegetative spectral reflectance through research could provide valuable insights into comprehending the impact of pest or pathogen attacks on the physical, physiological and chemical processes within plants. he underlying assumption is that stress may disrupt photosynthesis, affect the physical structure at the tissue and canopy levels, and potentially modify the absorption of light energy, thereby altering the reflectance spectrum. Recent advances in communication and technology can help guide remote sensing in a nondestructive and non-invasive way (Prabhakar, 2011). Remote sensing can also potentially be paired with machine learning strategies, utilized for precision agriculture such as neural networks for classification and k-means for clustering among other techniques. his can further help to drastically reduce statistical assumptions and promote early detection of biotic stress in plants (Behmann et al. , 2015).
Advances in ‘-omics’ based studies and their combined integration as in metabolomics-proteomics have been very helpful in the discovery of new plant biotic stress resistance genes. Using these techniques in forward and reverse genetics-based approaches can help decipher novel gene functions. his information can then be utilised to create elite plant cultivars through the process of cisgenics and transgenics (Kushalappa and Gunnaiah, 2013).
he desirable traits or characteristics from two different plants (in this case disease and pest resistance) can also be transferred to another plant by conventional methods like cross-breeding. Over the past, this has transitioned from processes like hybridization, composite crossing, multiline breeding and backcrossing to faster and less expensive processes taking advantage of molecular genetics methods like mutation, marker-assisted selection (MAS), targeted induced local lesions in genome ( ILLING) and virus-induced gene silencing (VIGS). Among them, ILLING is expected to gain a lot of popularity for being a non-transgenic, yet powerful method and can be used to precisely introduce mutations in the host genome (Hussain, 2015). Breeding-related techniques however can still suffer from a narrow initial genetic pool due to lack of proper screening and due to the emergence of new pathogens and pests over time.
A traditional way to deal with pests has been the use of chemical pesticides. However, out of the twelve persistent organic pollutants (POPs) that were classified in the Stockholm Convention, nine of them were pesticides (Lallas, 2001). hese compounds resist photolysis, chemical and biological breakdown, and are responsible for acute intoxications when they reach the body from contaminated sources like air, water or directly from food. Further, even though pesticides are important in agriculture having a global market that exceeds $30 billion per year, an estimated 40% of all crops are lost directly due to pest damage according to a report in 2007. Integrated pest management (IPM) using biopesticides or bioinsecticides is an eco-friendly approach of dealing with the side effects of chemical pesticides. It can include the use of naturally derived biochemicals or the microorganisms themselves having a pesticidal nature. he microorganisms also need not be inherently pesticidal; beneficial microorganisms of other kinds also exist like endophytic organisms which can promote plant growth and consecutively a higher vigour and better resistance systems of the plant. he use of transgenic crops has also been very fruitful and has led to some heavily used technologies like the use of Bt crops which originated back in 1996. his technology utilizes the heterologous expression of genes encoding for Cry proteins, which can kill a variety of insect pests. Enzyme inhibitors, lectins, manipulation of plant endogenous defences, use of RNA-interference (RNAi) technology, and creation of herbicide-tolerant crops are some of the ways to deal with pest attacks.
ransgenic varieties created via heterologous overexpression, constitutive expression of inducible antimicrobial factors like hydrolytic enzymes, H2O2 generating enzymes, defensins and induced activation of multiple genes, leading to easier and early attack to pathogens as well as their disposal, have been employed to create disease-resistant plants (Ferry and Gatehouse 2010). he use of genetic engineering to create transgenics is however complex and most of these events that succeed in laboratories are often not seen to translate well in the agricultural field. his, together with relatively higher costs and a lack of knowledge of the farmers has plagued this field for a long time.
Various other techniques can also be potent if given more time and effort, like the biochar effect, where a type of charcoal produced by the pyrolysis of organic materials can be added to the soil to increase the fertility and water retention capabilities of the soil as well as promote pathogen suppression, microbial population alteration and induced systemic resistance (Elad et al. , 2011). he use of hydroponics to completely eradicate the possible pathogenesis by soil-borne pathogens as well as to promote a properly balanced nutrient supply (Sambo et al. , 2019) might be possible to be complemented with the use of nanoparticle-based processes like the compounds of titanium (n iO2) and zinc (nZnO) to ameliorate further environmental stresses and counter the harmful effects of xenobiotic components (Silva et al. , 2022).
Rhizosphere microbiome
In order to describe the rhizosphere microbiome, we have to first shed some light on what exactly is a rhizosphere. he term "rhizosphere" is derived from the Greek words "rhiza," meaning "root," and "sphaira," meaning "sphere" which is the region of the soil surrounding the plant roots. he rhizosphere is a complex and highly dynamic zone of the soil that encompasses an indefinite area surrounding the roots of the plants. It acts as an interphase or a stage where a variety of biological activities take place with the roots of the plant and the diverse microbial communities of the soil. his region can contain up to 1011 microbial cells per gram of root having more than 30,000 prokaryotic species, behaving as one of the greatest reservoirs of biological diversity. he humongous diversity of microorganisms in this region and the occurrence of different physical, chemical and biological events are all guided by the influence of the living roots of the plants. he mutualistic effect of the rhizosphere is that it benefits both the plant and the microorganisms inhabiting it. he microorganisms derive their nutrition and receive optimal conditions for their growth and in return aid in nutrient recycling of the soil, disease suppression, plant health and overall proper functioning of the ecosystem. hat should be noted though is that not just the beneficial microorganisms inhabit this region, but it is also an ideal place for the plant pathogens which can colonize the rhizosphere and break down the proper functioning of the beneficial microbes. Even the opportunistic human pathogenic bacteria can take shelter in this region and ultimately reach our human bodies mediated by the plant. he microbes inhabiting the rhizosphere can include bacteria, fungi, oomycetes, protozoans, algae, viruses, nematodes and primitive organisms like archaea (Mendes et al., 2013). Plant chemical exudates are the most important criteria for determining the type of microorganisms that will be more prevalent in the microbiome and correspondingly the type of interactions that it takes part in. he type of plant secretions is also further seen to be dependent on the plant development (Chaparro et al., 2014). Other factors influencing microbiome diversity include soil type, prevalent climate and of course anthropogenic activities. Just as the human gut microbiome affects our health and can help in not only proper digestion but also in pathogenesis prevention, the rhizosphere microbiome significantly affects the physiological development and vitality of plants. Some of the commonly studied microorganisms which have been under study for their highly beneficial role in plant growth and health are nitrogen-fixers, plant growth-promoting rhizobacteria (PGPR), biocontrol microorganisms, mycoparasitic and mycorrhizal fungi and protozoans. On the other end of the spectrum lie pathogens like pathogenic fungi, oomycetes, nematodes and bacteria (Igiehon and Babalola, 2018).
he lack of appropriate tools to get an insight into the microbial community had kept this field of research in the dark for quite a while. his is mostly due to the fact that most of the microbes under in vitro conditions were non-culturable. However, with the emergence of nextgeneration sequencing (NGS) technologies which can provide genetic sequence information in record time, the process of gathering information has taken pace. NGS utilizes the versatility of culture-independent technique studies called metagenomics (Kunin et al., 2008). It is the study of the collection of genomes of microorganisms that are present in a sample collected from any environmental niche, for example, soil can provide valuable information like community structure and the possible complex interactions that can take place between the diverse microbial forms with each other and with the plant, making use of the information coded in their genes. A focused functional genomics approach can enable researchers to screen for specific functional activities of interest as well. Some classic techniques utilised has been (i) targeted amplification of the conserved regions of 16S rDNA and internal transcribed spacer (I S) to investigate the bacterial diversity and (ii) metatranscriptomics, by sequencing of cDNA obtained by reverse transcription of functional mRNA (Soni et al., 2017). A transition to metatranscriptomics, metaproteomics or metabolomics has helped to gather insights into the different functional molecules as opposed to information on just the diversity obtained from metagenomics-based studies. hen these methods are combined with the advanced combinatorial techniques of analytical chemistry, molecular biology and biophysics like gas chromatography-mass spectrometry (GC-MS), high performance liquid chromatography (HPLC)-MS, gel electrophoresis, nuclear magnetic resonance (NMR) spectroscopy, etc., they can be physically and accurately used as qualitative and quantitative measures to estimate the chemical composition at any point in the plant, rhizosphere or any other environmental niche (Zhang et al., 2012).
Rhizosphere signalling network
From plants to microbes
Different microbes present in the rhizosphere, are drawn towards plant roots by means of chemotaxis through phytochemicals secreted from the roots as part of root exudates. Microbes can detect and recognize these molecules, move towards the plant and associate with the root surface to form biofilms (O' Neal et al. , 2020; Kumar et al. , 2007, Feng et al. , 2018). It has been observed that usually root tips secrete polysaccharides (Dennis et al. , 2010), while oxidized compounds like amino acids and organic acids are secreted from elongation zones and meristem (Sharma et al. , 2020). For example, citric acid and fumaric acid released from cucumber and banana roots attracted Bacillus amyloliquefaciens SQR9 and Bacillus subtilis N11, respectively (Zhang et al. , 2013).
Plant root exudates contain phenolic compounds like flavonoids which act as chemotactic factors for bacteria. hese compounds induce expression of certain nodulation genes or nod factor genes. For example, flavonoids like 2-phenyl-1, 4-benzopyrone derivatives secreted from leguminous plants are recognized by nitrogen-fixing rhizospheric bacteria. Nod genes or nodulation factors consist of a chitin core made up of β-1,4-linked polymer of N-acetylglucosamine (GlcNAc) called lipo-chitooligosaccharides (LCOs). LCOs are different from chitin (a molecule associated with cell wall of fungi and exoskeletons of insects) with respect to the N-acyl moiety, consisting of a fatty acid chain varying in length, saturation and substitution groups. hese side chains are one of the factors conferring specificity between rhizobial strains and plant host targets (Oldroyd, 2013). LCOs have multiple plant growth related functions and one of them is in formation of root nodules. hese LCOs bind to plant receptors known as LysM receptor belonging to the lysine motif containing receptor-like kinase family (Liang et al., 2014). he LysM receptor has an extracellular LysM receptor-like kinase domain that binds to its cognate nod factor. his binding leads to a set of signalling events including accumulation of cytokinin and calcium spiking, initiation of root hair curling, development of an infection thread and subsequent rhizobial infection, leading to nodule formation (Oldroyd et al., 2011; Rose et al., 2012; van Zeijlet al., 2015). ithin the root nodules, bacteria fix atmospheric nitrogen and obtain photosynthetically prepared food from the host.
From microbes to plants
Microbe-associated molecular patterns (MAMPs) are microbial signalling molecules [such as peptidoglycan, chitin, flagellin, exopolysaccharides (EPS), hormones, volatile organic compounds (VOCs), antibiotics and extracellular enzymes] that are recognized by pattern recognition receptors (PRRs) present in host cell membranes. MAMPs have conserved chemical structures/patterns which are called signatures. his interaction between the two triggers a signalling cascade involving phytohormones such as SA, JA and E (Offor et al. , 2020). By this way, the plant can identify and interact with beneficial and pathogenic microbes. Interaction with beneficial microbes generally leads to colonization, whereas pathogenic microbes trigger plant defence responses.
he symbiosis between Mesorhizobium loti strain R7A with Lotus japonicu s initiates only after the firm binding of the EPS with the host receptors. In Lotus japonicus , two LysM receptors, viz., NFR1 and NFR5, are present, having high affinity for nod factors. Recognition of cognate nod factor by NFR1 leads to the expression of EPR3 in root hairs and epidermal cells of the susceptible zone. EPR3 is a LysM receptor kinase.
ild-type Mesorhizobium loti strain R7A (R7A) synthesizes and secretes an O-acetylated acidic EPS giving rise to mucoid colonies, while R7AexoU mutants of this strain (impaired in the exoU glucosyltransferase gene) secrete a penta-glycan (truncated EPS) and have a rough colony appearance. EPS expression is important for infection and nodule formation. From further studies, it was observed that EPR3 interacts with EPS and distinguishes between compatible and incompatible EPS and induces or inhibits infection and nodulation, respectively (Kawaharada et al. , 2015; Radutoiu et al. , 2003).
Microorganisms are able to drastically alter plant root system development, plant physiology, hormonal pathways and biomass production through the release of VOCs (volatile organic compounds). Other functions of VOCs include a direct source of nutrients, induce pathogen resistance and help in secondary metabolite production (Schulz-Bohm et al., 2017). Volatile compounds produced by diverse isolates of Fusarium oxysporum (a rhizosphere fungi) enhance shoot and root growth of Arabidopsis thaliana and tobacco by affecting auxin transport and signalling (Bital et al., 2015). VOCs emitted by the phytopathogen Alternaria alternata enhances photosynthesis, leading to the accumulation of high levels of cytokinins (CKs) and sugars, and early flowering in Arabidopsis (Sanchez-Lopez et al., 2016). Certain rhizobacteria like Pseudomonas fluorescens and Alcaligenes xylosoxidans metabolize α- pinene (a VOC) and utilize it as their sole carbon source (Kleinheinz et al., 1999).
Microbe-microbe interaction
Microbes harbor genes for the biosynthesis of different compounds that can alter the microbe–microbe interactions. For example, some compounds help them to interact with other microbes in its vicinity, while some are antibiotics or toxins which control the plenitude and diversity of other microbial groups by killing them.
Some microbes share primary metabolites. his allows microbes that are unable to produce said metabolite to thrive, especially in nutrient poor environment, leading to co-inhabitation of ecological niches (Frey-Klett et al. , 2011). However, this sharing of metabolites has a disadvantage. here could be a loss of the ability to produce certain primary metabolites which are shared. his makes the microbe metabolically obligated to its mutualistic partner. For example, certain endosymbiotic bacteria and gut microbes have significantly reduced genome sizes due to such metabolite sharing with their hosts. Burkholderia rhizoxinica is an intracellular symbiont of the phytopathogenic zygomycete Rhizopus microspores which causes blight in rice seedlings. he genome of B. rhizoxinica is smaller in size compared to free living Burkholderia species and harbors less transcriptional regulator genes (Lackner et al ., 2011).
Microbe to microbe signalling and communication within the root microbiome occurs through quorum sensing (QS). It is mediated by low-molecular-weight compounds called auto-inducers (AI). Important processes such as production of virulence factors or biofilm formation are regulated in a density dependent manner via QS. QS begins with the extracellular release of auto-inducers into the surrounding environment. In Gram-negative bacteria, two components of the S regulatory system are transcriptional activator protein (R protein) and the autoinducer molecule (AI). N-acyl homoserine lactones (AHLs) primarily function as AI in Gram-negative bacteria whereas for Gram-positive bacteria, QS is typically mediated by post-translationally modified oligopeptides (Visick and Fuqua, 2005). In a few strains of Gram-negative rhizobacteria, e.g., Burkholderia spp. and Stenotrophomonas maltophilia , communication occurs through diffusible-signal factors (DSFs) as the signaling compound. DSFs produced by the bacteria Burkholderia cenocepacia and Pseudomonas aeruginosa induces bacterial virulence, formation of biofilms and antibiotic tolerance in these important pathogens such as Xanthomonas campestris pv. campestris causing black rot in brassicas and Stenotrophomonas maltophilia in humans (Ryan et al ., 2015). Mixed biofilms can be produced by microbes that utilize similar or identical QS compounds. For example, mixed-species biofilm is formed by Streptococcus mutans , an important cariogenic bacterium and Streptococcus gordonii , an early colonizer of the tooth surface which leads to the formation of dental plaques. he LuxS/autoinducer-2(AI-2) system is involved in this mixed biofilm ( ang et al. , 2017).
Bacterial QS can also be detected by eukaryotes. Such instances are observed in Candida albicans, detecting farnesol and in Xanthomonas campestris, detecting cis-11-methyl-2-dodecenoic acid, both of which inhibits filamentation (Deng et al. , 2010).
erpenes and other VOCs are also involved in interactions between microbes and in between microbes and plants. hey can affect microbial gene expression and regulate different behaviours such as biofilm production, virulence and stress tolerance (Audrian et al. , 2015).
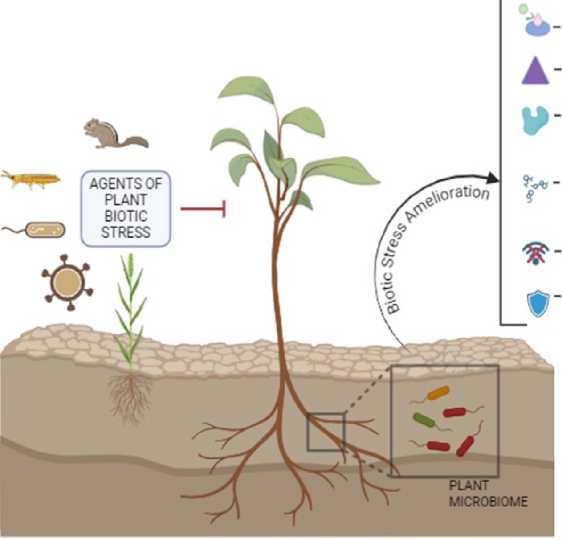
Competition with pathogens
Production of Siderophores
Production of cell* wall degrading enzymes Production of Antimicrobial compounds
Quorum quenching
ISR and SAR
Figure 1: Schematic highlighting the role of plant microbiome in mitigating biotic stress through various mechanisms.
Table 1. Different microbes inhabiting the rhizosphere and their modus operandi of remediating biotic stress
|
Microbe |
Functional attributes |
Application |
Reference |
|
Pseudomonas putida strain B10 |
Pyochelin and pyoverdine type of siderophore production |
Suppression of Fusarium wilt |
Haas & Défago 2005 |
|
Pseudomonas spp. GRC3 |
Chitinase production |
Control of F. oxysporum mediated root rot fungal diseases |
Lee et al. , 2009 |
|
Bacillus indicus M CC 5559 and Bacillus pumilus M CC 5560 |
Production of QSI, 4-phenylbutanoic acid |
Prevent biofilm production in many gram-negative and grampositive bacteria |
Nithya et al. , 2011 |
|
Halobacillus salinus |
Production of QSI |
Inhibits bioluminescence production by the gram-negative bacteria Vibrio harveyi |
easdale et al. , 2009 |
|
Bacillus nematocida B16 |
Extracellular alkaline serine protease Bace16 and neutral protease Bae16 production |
Inhibits the growth of nematode Panagrellus redivivus |
Niu et al. , 2010 |
|
Paecilomyces lilacinus strain 251 |
production of chitinases and proteases |
Control the root-knot nematode Meloidogyne incognita on tomato plants |
Kiewnick & Sikora 2006 |
|
Pseudomonas fluorescens CS417r |
rigger jasmonate and ethylene signalling pathways |
Induced systemic resistance against bacterial leaf pathogen P. syringae pv tomato |
Pieterse et al. , 1998 |
|
Rhizobium japonicum |
Synthesizing metabolites helping in antibiosis and inhibiton of spore germination |
Control of soybean root rot disease caused by Fusarium solani and Macrophomina phaseolina |
Al-Ani et al. , 2012 |
|
Klebsiella pneumoniae SnebYK |
Up regulation of genes PR1, PR2, PR5, and PDF1.2 and down regulation of PR3 |
Control of cyst nematode Heterodera glycines in soyabean |
Liu et al. , 2018 |
|
Trichoderma asperellum GDFS1009, Bacillus amyloliquefaciens 1841 |
induction of BLR-1/BLR-2, VELVE , and NADPH oxidases genes |
Enhance plant growth and protection against Fusarium graminearum |
Karuppiah et al. , 2019 |
|
Bacillus velezensis F21 |
Increase in expression ofplant defense related genes RKY, MYB, bZIP, AP2, and NAC, and activity of defense enzymes, such as CA , POD and SOD |
Control of wilt caused by Fusarium oxysporum f. sp. niveum in watermelon |
Jiang et al. , 2019 |
Contributions of rhizospheric microbiome towards plant health
Root and soil derived microbes provide essential host functions that contribute directly to plant fitness, productivity and resistance towards biotic and abiotic stresses. Such microbiome directly and indirectly impacts plant performance and productivity. his includes enhanced uptake of nutrients like potassium and nitrogen, production of phytohormones and resistance towards various abiotic and biotic stresses. Here, we discuss the various benefits of rhizosphere microbes on nutrient uptake, phytohormone and resistance against abiotic stresses.
Phosphate solubilization here are abundant reserves of phosphate in the soil. hat makes it a limiting resource is the fact that phosphates are not present in the form suitable for plant uptake. Plants can only uptake soluble forms of phosphate such as mono- and di-basic phosphates. Many PGPR can convert such inaccessible complex-structured phosphates in the soil, e.g., tricalcium phosphate and aluminium phosphate, into soluble forms via the process of solubilization and mineralization. Phosphate solubilization occurs with the help of organic acids (like acetic acid, lactic acid, isobutyric acid, oxalic acid, citric acid, succinic acid, gluconic acid and 2-ketogluconic acid) secreted by microbes through metabolism of sugars present in root exudates. hese acids also act as good chelators of divalent Ca2+ ions. Phosphate is solubilized by the species of Achromobacter, Agrobacterium, Azotobacter, Beijerinckia, Bacillus, Burkholderia, Erwinia, Flavobacterium, Microbacterium, Rhizobium, Pseudomonas, Serratia and fungi such as Aspergillus, Penicillium, Fusarium, Chaetomium and Cephalosporium (Sharma et al., 2013; Goswami et al., 2016; Dukare et al., 2020).
Potassium and other minor element uptake
Bacterial species such as Bacillus mucilaginosus, Bacillus edaphicus, Pseudomonas spp. Acidothiobacillus ferrooxidans, Bacillus circulans,
Paenibacillus spp., and Burkholderia spp. are well recognized K solubilizing bacteria (KSB). KSB solubilizes soil K mineral by different mechanisms, including: (i) lowering the pH, (ii) increasing chelation of the cations bound to K and (iii) acidolysis of the nearby area (Etesami et al. , 2017). Besides K, soil microbes also make bioavailability of other trace elements such as iron (Fe), zinc (Zn), sulphur (S) and Si to the plants (Adesemoye et al. , 2009). Microbially produced iron-chelating siderophores, gluconate or the derivatives of gluconic acids, e.g., 2-keto-gluconic acid, 5-keto-gluconic acid and other organic acids facilitate the mineralization of these minor elements (Dukare et al. , 2020).
Phytohormone production
Auxins
Auxin, i.e., indole-3-acetic acid (IAA), is a very important phytohormone that is involved in multiple developmental processes such as cell elongation, cell division, tissue differentiation and apical dominance. Among PGPR, Azospirrilum spp., Azotobacter spp., Aeromonas spp., Burkholderia spp., Enterobacter spp., Pseudomonas spp., and Rhizobium spp. produce IAA. Rhizobacteria-produced IAA enhances the plant root system by making it more branched, producing longer and broader branches and having more surface area. his in turn increases its ability to uptake nutrients from the soil. IAA is synthesized by microbes in two ways: L-tryptophan-dependent (three pathways are known) and -independent pathways. Plants secrete L-tryptophan in their root exudates which most PGPRs use to produce IAA via the L-tryptophan-dependent pathway. Some other microbes like Azospirillum brasilense use L-tryptophan independent pathway to produce most of the IAA, while the rest comes from using L-tryptophan (Ahmad et al ., 2008; Goswami et al ., 2016).
Cytokinins
Cytokinins are amino purines where N6 is substituted. Cytokinins help in the development of vascular bundles in embryos, leaf expansion, branching, production of more chlorophyll molecules, promote root growth, breaking seed dormancy during germination, and delay senescence. Microbes of the genera
Pseudomonas, Azospirillum, Bacillus, Proteus, Klebsiella, Escherichia and Xanthomonas can produce cytokinins. Isopentenyl transferase (encoded by ipt gene) is the enzyme, catalysing the rate-limiting step in the cytokinin biosynthesis pathway. It transfers the isopentenyl moiety from dimethylallyl diphosphate (DMAPP) to adenosine monophosphate (AMP) to initiate biosynthesis. Bacteria transfer isopentenyl moiety from 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate (HMBDP) to AMP during cytokinin production (Maheshwari et al. , 2015; ong et al. , 2015; Goswami et al. , 2016).
Gibberellins
Gibberellins (GAs) are a large family of phytohormones. Most GAs are found in higher plants with some in fungi. hey regulate multiple plant developmental processes such as cell division, stem elongation, seed germination, leaf expansion, flowering and senescence. GAs are produced in Azospirillum sp., Rhizobium sp., Bacillus sp., Acetobacter diazotrophicus and Herbaspirillum seropedicae . Rhizobacteria predominantly synthesize gibberellic acid (GA3) as the primary GA-like substance. In bacteria, the GA biosynthesis pathway begins from the geranyl–geranyl diphosphate (GGPP) as a starting compound and involves a sequence of reactions catalyzed by several cellular enzymes ( ong et al. , 2015; Salazar-Cerezo et al. , 2018; Goswami et al. , 2016).
Abscisic acid and ethylene
Abscisic acid (ABA) is another stress-induced hormone that is involved in plant root hydraulic conductivity, biochemical/physiological processes and regulating transcription factors of many stress-responsive genes. Plants build up ABA in stress conditions that elicit a response to cope with the adverse environmental conditions. ABA has a major role during drought stress in combating reactive oxygen species (ROS) that are generated and in stomatal closure. Aside from drought, ABA is also involved in mitigating salinity and cold stress. It is observed that plants exposed to harsh stress conditions have a higher accumulation of ethylene (E ). E causes several cellular and biochemical injuries in the plants exposed to adverse environmental conditions. ACC deaminase-producing rhizobacteria irreversibly convert the precursor of E hormone, ACC, into less harmful compounds such as α-ketobutyrate and ammonia during such conditions. his helps the plant gain increased resistance towards pathogens as well as conditions such as salinity stress (Qi et al., 2018; Dukare et al., 2020).
Contributions of rhizosphere microbiome in combating abiotic stress
Plants face various environmental stresses like extremes of temperature, drought, salinity, UV and pathogen attacks. Salinity, particularly, affects crop yield by impacting major processes like photosynthesis and respiration. his leads to the plant being nutritionally deficient and elevates its Na+ levels (Zahedi et al. , 2012). Reactive Oxygen Species (ROS) are generated under such stresses. hese ROS are highly reactive compounds which are known to harm biomolecules such as proteins, nucleic acids and lipids. In response to ROS generation, the plant triggers antioxidant enzymes like peroxidase (POX), superoxide dismutase (SOD), catalase (CA ), ascorbate peroxidase (APX) and glutathione reductase (GR) which can convert ROS into less harmful molecules such as water and oxygen. Application of plant growth-promoting rhizobacteria (PGPR) such as B. cereus AR156 can increase the activities of antioxidant enzymes in Lycopersicon esculentum (tomato) affected by salinity stress ( ang et al ., 2012). Rhizobacteria-produced exopolysaccharides (EPS) are capable of retaining moisture. hey increase the host tolerance towards stress by regulating metabolism and by accumulating stress-responsive osmolytes such as proline, amino acids and trehalose (called compatible solutes). hese are important in maintaining membrane permeability, enzyme integrity and protein function. Additionally, some microbes are known to produce volatile organic compounds (VOCs). VOCs produced by Bacillus subtilis GB03 downregulate the expression of the HK 1 (high-affinity K+ transporter 1) gene in Arabidopsis thaliana roots while upregulating it in shoots, which thereby reduces sodium accumulation and enhances tolerance towards stress (Goswami et al ., 2016; Dukare et al ., 2020).
Contributions of rhizosphere microbiome in combating biotic stress
So far, we have seen how rhizospheric microbes help in promoting plant growth through nutrient availability and hormone production along with the ability to tolerate various abiotic stresses. However, coming to our main focus of attention, we discuss how they can help a plant in warding off fungi, bacteria and nematode induced biotic stress and how they can prime the host immunity towards susceptibility to pathogens (Fig. 1).
Resistance against fungal phytopathogens
Siderophore-producing rhizobacteria compete with fungi for limited Fe nutrients and make them inaccessible. his process results in pathogen inhibition via hindering fungal germination, metabolism and in turn their virulence. Iron is an essential nutrient required by both plants and microbes. Fe+3 is present abundantly in the soil as insoluble oxides and hydroxides which are inaccessible to plants and microbes alike. Here, plants can release compounds capable of chelating iron, thus making them soluble. his solubilised iron is then reduced with the help of extracellular enzymes and the resulting reduced iron gets absorbed. Another way is in absorbing the chelated iron complex and reducing iron inside the plant. hese iron chelating compounds are called siderophores. Chemically they are low-molecular weight compounds, below 1 kDa, with functional groups (such as hydroximates and catechols) which can bind iron reversibly. Rhizospheric bacteria such as Pseudomonas usually produce siderophores. For example, Pseudomonas fluorescens and Pseudomonas aeruginosa produce pyochelin and pyoverdine type of siderophores. Ferripyoverdine, i.e., pyoverdine complexed with Fe3+ interacts with specific outermembrane receptors present in the producer microbe and plants. Fe3+ is absorbed, transported into the cytoplasm and then reduced to Fe2+. he advantage of producing siderophores is the fact that pathogens are unable to absorb the iron–siderophore complex. Due to the resulting Fe deficiency, the growth of pathogens is hindered (Payne, 1994; Haas & Défago, 2005).
Fusarium udum, Pythium sp., Phytophthora sp., and Rhizoctonia spp. are some soil-borne fungal pathogens responsible for reduced crop yield and quality. Bacterial species of the genera Bacillus, Paenibacillus, Pseudomonas, Azotobacter, Streptomyces,and Lysobacter have been recognised as beneficial for plants, acting as growth promoters, biofertilizers and biocontrol agents against these pathogens (Naing et al., 2015). hese bacteria are known to exhibit biocontrol activities through various mechanisms which include mycoparasitism via secretion of lytic enzymes (like chitinase, glucanase and cellulase), production of antibiotics, competitive exclusion of nutrients and space, formation of biofilms, ROS generation and induction of host defence (Saraf et al., 2014).
Chitin is a linear polymer of N-acetylglucosamine joined by β-1, 4-glycosidic bonds. It is one of the major structural polysaccharides found in fungal cell walls providing them structure and integrity. Degradation of chitin via cell wall degrading enzymes such as chitinases and glucanases is the primary mechanism of biocontrol (Manjula and Podile, 2005). Chitin is broken down into deacylated oligomers like chitosan, monomers like N-acetylglucosamine and the disaccharide chitobiose. his leads to the structural and cellular disintegration, lysis and eventually death of the fungal cells. For example, inhibition of Pythium aphanidermatum which causes damping-off of cucumber seedlings by Actinoplanes philippinensis, Microbispora rosea, Micromonospora chalsea and Streptomyces griseoloalbus by the production of β-1,3-glucanases, β-1,4-glucanases and β-1,6-glucanases (El- arabily 2006). hese enzymes are also involved in degradation of fungal cell walls of F. oxysporum and in controlling root rot fungal diseases by Pseudomonas spp. GRC3 (Lee et al. , 2009).
Bacteria belonging to genus Bacillus are known to produce a wide range of compounds having antibacterial and antifungal properties. Compounds such as subtilin, subtilosin A, asA and sublancin are synthesized from ribosomes. Other compounds like bacilysin, chlorotetain, mycobacillin, rhizocticins, bacillaene, difficidin and lipopeptides which belong to the surfactin, iturin and fengycin families are synthesized by non-ribosomal peptide synthetases (NRPSs) and/or polyketide synthases (PKS) (Leclere et al., 2005). Iturin for instance, inhibits pathogen growth by creating pores in the cell membrane which results in extensive leakage of cytoplasmic potassium ions. Besides, multiple compounds having antifungal activity include metabolites such as hydrogen cyanide, ammonia, alcohols, sulfides, ketones, aldehydes, cyclic lipopeptides, polyketides and phenylpyrrole (Fouzia et al., 2015).
Resistance against bacterial phytopathogens by quorum quenching
Cell-to-cell communication in bacteria known as quorum sensing (QS) involves the synthesis, release and detection of diffusible signalling molecules. hey are called autoinducers (AIs) since they can induce their own biosynthesis. N-acyl homoserine lactones (AHLs) are one of the well-studied QS signalling molecules. A complex is produced when AHLs (produced by LuxI genes) reaches a threshold level and binds to their cognate receptors known as LuxR. his complex is involved in regulating various processes like bioluminescence, secondary metabolite production, biofilm formation, sporulation and in inducing competence (Fuqua and Greenberg, 1998).
Quorum quenching (QQ) is the process of disrupting QS, either by degrading QS signals or by inhibiting the QS signalling pathway. he enzymes responsible for degradation of QS signalling molecules are called QQ enzymes, while chemicals that inhibit QS signalling pathways are called QS inhibitors (QSIs).
QQ enzymes fall in four catalytic classes: lactonases, amidases also known as amidohydrolases or acylases, reductases and cytochrome oxidases. Lactonases cleave and open the homoserine lactone ring, amidases break AHLs at the amide bond and release fatty acids and homoserine lactone reductases convert 3-oxo-substituted AHLs to their corresponding 3-hydroxyl-substituted forms and cytochrome oxidases oxidise the acyl chain in AHLs. Examples of such enzymes include AHL lactonase from Bacillus spp. (Dong et al. , 2002), AHL acylase from Ralstonia sp. (Lin et al., 2003) and oxido-reductase from Burkholderia sp. (Chan et al. , 2011).
In contrast to QQ enzymes, QSIs inhibit QS in three major pathways. Firstly, they can inhibit autoinducer production. AHL autoinducers are produced from two crucial precursors, S-adenosylmethionine (SAM) and an acylated acyl carrier protein (acyl-ACP). Both of these metabolites are essential for the survival of the bacteria and thus their production cannot be inhibited. Instead, the synthesis of AHLs from SAM and acyl-ACP can be inhibited (Schaefer et al., 1996).
he second method involves interfering with the exchange of autoinducers between cells. Most of the AHLs are capable of diffusing through the cell membrane, but AI-2 requires a specific transporter. In Escherichia coli , LsrK, a cytoplasmic enzyme is involved in phosphorylation of AI-2 post-transportation. In case AI-2 gets phosphorylated extracellularly, the negative charge on phospho-AI-2 does not allow its subsequent transport into the cell.
hirdly, QSIs can interfere with signal perception and transduction. hese molecules can prevent binding of AIs to their cognate receptors or modify the conformation of the receptor-signal complex. his in turn prevents subsequent dimerization of the receptor or interaction of the complex with appropriate DNA regions (such as gene promoters). As a result, the downstream expression of QS associated genes and their corresponding functions are prevented.
Such QSI molecules are produced by multiple bacterial genera such as Bacillus, Halobacillus, Alteromonas and Pseudomonas . Bacillus indicus M CC 5559 and Bacillus pumilus M CC 5560 cell extracts are capable of preventing biofilm production in many bacteria by hampering QS. 4-phenylbutanoic acid present in the cell extracts act as the compound that prevents biofilm production (Nithya et al. , 2011). Halobacillus salinus , a marine Gram-positive bacterium, is capable of inhibiting bioluminescence by the Gramnegative bacteria, Vibrio harveyi via secretion of QSIs ( easdale et al. , 2009).
Resistance against parasitic nematodes
Plant-parasitic nematodes (PPNs) are microscopic worms that migrate through soil searching for plants from whom they can derive water and nutrients via the vascular tissues. hese nematodes attack the underground parts of plants and cause serious yield loss. hat often happens is that damage by such PPNs remain unnoticed as aboveground symptoms are scarce plus there are no proper diagnosis for nematodes. PPNs can be classified into three major groups based on feeding habits: the migratory ectoparasites (e.g., Belonolaimus spp., Xiphenema spp. and Trichodorus spp.), the migratory endoparasites (e.g., Pratylenchus spp. and Radopholus spp.), and the sedentary endoparasites, [e.g., the root-knot nematodes (RKNs) (Meloidogyne spp.) and the cyst nematodes (CNs) (Heterodera spp. and Globodera spp.] (Li et al., 2015). Nematophagous fungi and bacteria are present within the rhizophere which are able to neutralize PPNs through various methods, often against specific developmental stages of their life cycles.
Nematophagous bacteria are a group of soil bacteria that can inhibit PPNs. Pasteuria is one such genus, which consists of multiple species of Gram-positive endospore forming nematophagous bacteria. Pasteuria penetrans is known to parasitize Meloidogyne spp. (rootknot nematodes), Pasteuria nishizawae against Globodera spp. and Heterodera spp. (cyst nematodes), Pasteuria thornei against Pratylenchus spp. (root-lesion nematodes), and Pasteuria usgae against Belonolaimus spp. (sting nematode) (Preston et al., 2003). hat P. penetrans does is to develop highly resistant endospores which can adhere to the cuticle of second-stage juveniles (J2) of nematodes. hen these endospores germinate, they form an infection peg penetrating the cuticle, reaches the pseudocoelom and grows into mycelial microcolonies. Due to the microcolonies growing and subsequently releasing its spores, the nematodes meet their demise. In female nematodes, growth of P. penetrans in the pseudocoelom reduces their fecundity (Li et al., 2015; Davies et al., 2011). Another example of nematophagous bacteria is the endospore forming bacteria, Bacillus nematocida (B16). It parasitises the nematode, Panagrellus redivivus. he bacteria produce VOCs like benzaldehyde and 2-heptanone to attract the nematode for consuming it. Once in the host alimentary tract, it secretes extracellular proteases which cause internal tissue damage and ultimately lead to host death. Proteases secreted include an alkaline serine protease Bace16 and a neutral protease Bae16 among others (Niu et al., 2010). Another classic example is Bacillus thuringiensis. his bacterium produces proteinaceous protoxin crystals (known as Cry proteins) in the nematode gut during its stationary growth phase, especially during sporulation. Under alkaline conditions, the crystals solubilize followed by proteolytic cleavage of the protoxin into active toxin which in turn binds to receptors. he toxin in turn gets inserted into the apical membrane to form pores. his results in osmotic imbalance, followed by lysis of internal organs and eventually death of the nematode (Schnepf et al., 1998).
Aside from nematophagous bacteria, there are nematophagous fungi which are also capable of combating PPN. here are four major groups of nematophagous fungi:
-
(i) Nematode-trapping fungi. hese fungi are from the order Orbiliales (Ascomycota) and utilize specialized trap like structures formed from their hyphae. raps include constricting rings and five different types of adhesive traps, i.e., sessile adhesive knobs, stalked adhesive knobs, adhesive nets, adhesive columns and non-constricting rings. hese saprophytic fungi become parasitic in the presence of their nematode prey. hey are classified into four genera: Arthrobotrys , Dactylellina , Drechslerella and Gamsylella (Jiang et al. , 2017).
-
(ii) Endoparasitic fungi. hey produce different kinds of spores, capable of infecting nematodes, namely encysting spores, adhesive conidia and ingesting conidia. hese parasitic fungi complete their entire vegetative life cycle within the nematode. Drechmeria coniospora is one such example. It produces large amounts of conidia. On maturing, they can adhere to its cuticle via an adhesive bud, form an infection vesicle and trophic hyphae within the nematode gut. ithin three days, more conidia are produced as the PPN meet their demise (Li et al. , 2015; Moosavi and Zare, 2012).
-
(iii) Egg and female-parasitic fungi. hey use appressoria, which is a specialized penetration peg or use lateral mycelial branches along with extracellular hydrolytic enzymes such as chitinases and proteases to infect nematode egg shells. his leads to the breakdown of egg shell layers and infection. Examples include Pochonia chlamydosporia, Paecilomyces lilacinus, Clonostachys rosea and Lecanicillium psalliotae (Li et al. , 2015; Moosavi and Zare, 2012).
-
(iv) oxin-producing fungi. hese produce toxins that immobilize nematodes before their hyphae penetrate through the nematode cuticle. he toxins produced belong to various different chemical groups including alkaloids, peptides, terpenoids, macrolides, oxygen heterocycle and benzo compounds. Examples include Paecilomyces lilacinus, Penicillium spinulosu, Nematoctonus robustus, Beauveria bassiana, Harposporium anguillulae and Lachnum papyraceum (Li et al. , 2014).
Resistance by priming host immunity
Root and soil-associated microbes prime host plant immunity by inducing systemic resistance (ISR). Microbes are known to produce various elicitor molecules that activate phytohormone signalling pathways such as jasmonic acid (JA), salicylic acid (SA) or ethylene (E ). hese phytohormones trigger ISR in the host. Several MAMPs such as flagellar proteins, chitin, LPSs, AHLs, cyclic lipopeptides, VOCs (e.g., 2, 3-butanediol, acetoin), siderophore and antibiotics are also capable of activating ISR. Microbiomes can reinforce the defensive capabilities of plants by interrupting the plantpathogen interactions, which subsequently confers plant resistance. he beauty of induced resistance is that it is not only expressed at the site of induction, but also in plant parts that are at a significant distance from the inducer, hence called systemic. Generally, induced resistance confers an enhanced level of resistance compared to the basal level of horizontal resistance, already present in the plant. Plants can also develop induced resistance as a result of any previous infection by a pathogen. he infected plant part produces compounds that prime the plant for any future infection by the same pathogen. his is termed as SAR.
Microbial proteins called PAMPs are identified by membrane bound receptors called PRRs. PAMPs are essential components of pathogens like bacterial flagellin or fungal chitin. Molecules such as cell wall or cuticular fragments, released by pathogens during invasion, are also detected by plants. hese molecules are called danger-associated molecular patterns (DAMPs). Recognition of these molecules induces the first line of defence known as PAMP-triggered immunity (P I), conferring basal horizontal resistance to the plant.
o prevent P I, pathogens have evolved a number of effector proteins which can disrupt P I, collectively known as avirulence proteins (Avr). In response to Avr proteins, plants evolved resistance (R) genes whose gene products are known to scavenge pathogen Avr proteins. his confers the plant effector triggered immunity (E I), which is the second line of defence (Boyd et al. , 2013; Dodds and Rathjen, 2010).
P I and E I triggers an induced resistance in tissues distal from the infection site. his induced resistance can be triggered by three ways. he first involves infection by a pathogenic microorganism and triggers the salicylic acid (SA) signalling pathway. he protein Nonexpresser of PR genes1 (NPR1) is expressed downstream to SA signalling. NPR1 remains in the cytoplasm of cells as an oligomer by the help of disulphide bonds. Following SA induction, NPR1 dissociates into the monomer and interacts with different transcription factors like RKY family of transcription factors which leads to the expression of SA responsive genes such as multiple pathogenesis-related (PR) genes. he second pathway involves wounding of plant tissues by herbivores, triggering the jasmonic acid (JA) signalling pathway. In uninduced cells, Jasmonate Zim-domain (JAZ) proteins bind to the transcription factor MYC2 preventing transcription of JA responsive genes. Jasmonic acid interacts with isoleucine to form jasmonoyl-isoleucine (JA-Ile). Another protein Coronatine Insensitive1 (COI1) binds to JA-Ile forming a complex. his complex on binding to JAZ, removes it from MYC2 and subsequently polyubiquitinates it. Ubiquitinated JAZ gets degraded by 26S proteosome. he freed MYC2 can then transcribe JA responsive genes. Both these signalling pathways lead to the expression of pathogenesis-related (PR) genes. PR proteins have antimicrobial properties by which they can combat the pathogens. he third pathway involves resistance induced by non-pathogenic root-associated rhizobacteria. Rhizobacteria do not lead to the production of PR proteins or other antimicrobial compounds. ranscriptome analysis showed that rhizobacteria induced Arabidopsis root had almost similar transcript levels as uninduced roots. he difference was observed post pathogen infection. Different strains of Pseudomonas spp. were used to confer resistance to Arabidopsis thaliana. hen the induced plants were subjected to infection by leaf pathogen P. syringae pv. tomato, they showed a much faster and stronger response compared to uninduced plants (Pieterse et al., 2014; Choudhary et al., 2007).
Implementation of rhizosphere microbes in agriculture raditional agriculture is dependent upon the use of agrochemicals such as pesticides, synthetic fertilizers, herbicides along with irrigation with untreated wastewater. Over the years, this has led to various challenges owing to injudicious use of agrochemicals like reduced soil fertility, fluctuations in climate, environmental damage and expansion in urban areas. In addition, there has been development of pesticideresistant pests because of which there is sufficient yield loss per year. Hence, to keep up with the food demand for the growing population, different methods need to be developed which can increase crop productivity and at the same time should not adversely affect the environment. his is necessary to ensure food security for the future generations (Saeed et al., 2021).
One promising strategy is to utilize beneficial microbes, such as rhizobacteria, as biofertilizers. Not only do they reduce our dependence on traditional agrochemicals, but they also provide the plant with benefits which can indirectly help in increasing crop yield. As discussed in earlier sections, plant growthpromoting rhizobacteria (PGPR) are beneficial bacteria present within the rhizobacteria. Some of them help in nitrogen fixation, i.e., converting atmospheric nitrogen (N 2 ) into ammonium (NH4+) which the plants can take up and utilize. hey are also responsible for the production of bioactive compounds such as phytohormones, inhibiting the growth of plant pathogens such as bacteria, fungi and nematodes via secretion of antimicrobials or by simply competing with them for limited resources, tolerance to abiotic stresses such as drought, salinity and cold, and by inducing resistance in plants via SAR and ISR. Some examples include bacteria of the genera Azospirillum, Azotobacter, Acetobacter, Gluconacetobacter, Azoarcus, Bacillus, Paenibacillus, Burkholderia, Herbaspirillum, Clostridium, Klebsiella, Enterobacter, Citrobacter and Pseudomonas.
Besides bacteria, there are fungi of the genera Aspergillus, Penicillium and Trichoderma which are involved in solubilizing phosphates. hey are able to solubilize insoluble mineral phosphates mostly via organic acid formation and convert P into soluble forms that plants can readily utilize. Bacillus spp. and Pseudomonas spp. are also able to perform the job, though better results are obtained when using them along with P-solubilizing fungi and AMF. Moreover, these microbes play a critical role in solubilizing micronutrients like iron (Fe), manganese (Mn) and zinc (Zn) and making them available to the plants for uptake, which are often deficient in soil due to intensive farming practices. Overall, rhizospheric microbes have significant potential for improving agricultural sustainability and productivity (Altomare and ringovska, 2011).
In one study, maize was cultivated on two different kinds of soil: calcareous calcisol soil of Sirdarya, Uzbekistan and loamy sand of Muencheberg, Germany. Maize plants were inoculated with three PGPR strains, Pseudomonas alcaligenes PsA15, Bacillus polymyxa BcP26 and Mycobacterium phlei MbP18. he calsicol soil was deficient in nutrients as well as alkaline. As a result, the untreated maize plants were unable to grow optimally. hen the three strains were added, the plant exhibited enhanced growth and development. If the results are compared with loamy soil, it did not show any sufficient changes on PGPR application. his was probably because loamy soil is rich in nutrients and hence did not show any effects of PGPR application. It was evident from this experiment that the efficiency of bacterial inoculants in promoting plant growth was affected in turn by edaphic factors such as nutrient content, pH and moisture content (Egamberdiyeva, 2007).
In an experiment performed by hrane et al. (2000), the activity of the biocontrol agent Pseudomonas fluorescens DR54 was tested against Pythium ultimum, a fungal plant pathogen affecting the roots and causing root rot disease. Both the organisms were inoculated on sugar beet seeds and a microcosmic study was performed. he result was a clear improvement of plant germination and emergence as observed after a period of seven days when compared to control samples without P. fluorescens. he pathogenic fungus showed greatly reduced mycelial density, showed a reduction in the formation of oospores and showed sub-optimal intracellular activities. Metabolite study also revealed the presence of a cyclic lipopeptide called viscosinamide (source being Pseudomonas fluorescens DR54) which was present on the locations where encysted zoospores of the fungus appeared. he encysted zoospores would naturally show reduction/loss of motility towards the surface of the roots, as a result of which the pathogen would be prevented from colonizing and proliferating inside the host plant. hus, the presence of the biocontrol agent in the rhizosphere would resist Pythium infection in the plant by hampering the mycelial growth as well as the zoospore motility ( hrane et al., 2000).
he inoculation of Pseudomonas libanensis R1 alone or in combination with Claroideoglomus claroideum BEG210 could alleviate the deleterious effects of salinity stress (SS), heavy metal stress (MS) or both (SS + MS) in soil. hey perform it by improving plant growth, chlorophyll content and physiological status (electrolyte leakage, proline and malondialdehyde content), thereby enhancing multiple stress (SS + MS) tolerance in Helianthus annuus . Application of P. libanensis alone or in combination with C. claroideum also reduced the deleterious effects of multiple stresses by decreasing Ni and Na+ uptake under SS + MS. he findings conclusively suggested that inoculation of plant beneficial bacteria (PBB), arbuscular mycorrhizal fungi (AMF) or their combination has significant potential to improve the growth of plant under salinity and heavy metal stress (Ma et al ., 2019).
Another species of Pseudomonas called Pseudomonas stutzeri E25, and Stenotrophomonas maltophilia CR71, were seen to independently promote the growth of Lycopersicon esculentum cv Saladette, a cultivar of tomato, when grown under greenhouse conditions. he experimental plant exhibited enhanced features such as improved shoot and root length together with a net increase in the plant biomass (calculated as fresh weight). his was the result of various VOCs released by these bacterial strains including acetoin and dimethyl disulphide (DMDS). he role of acetoin in plant growth and disease resistance have already been studied before (Ryu et al., 2003; Rudrappa et al., 2010). DMDS is also another plant growth promoter which serves as reserves of reduced sulphur, which can be beneficial for wild-type plants growing in the sulfur deficient soil. Furthermore, when pure DMDS was analysed, it was also seen to inhibit the mycelial growth of the foliar pathogen, Botrytis cinerea, thereby also acting as a potent biocontrol agent. hus, it could be predicted that the strains E25 and CR71, when applied in a consortium, hold true potential in plant agricultural scenarios (Rojas-Solís et al., 2018).
omato plants are also susceptible to wilt disease by the causative agent, Fusarium oxysporum f.sp. lycopersici (Fol). o treat this, the 34 strain of Trichoderma asperellum was assayed in four sets of increasing iron concentrations [1, 10, 100 and 1000 μM supplied in the form of chelated ED A/Fe (III)] in the nutrient solution where both the pathogen and the plant were present. Among the four sets, a reduction in pathogen load in the plant shoots was only significant where 34 was supplied with 10 μM Fe. It was thus hypothesized by the researchers that iron competition is a major player which helps 34 to exert its biocontrol activity against the Fusarium pathogen. Iron concentrations exceeding 10 μM suppressed the 34 siderophore synthesis and thereby the competition for iron with Fol was greatly inhibited. 34 was also seen to alleviate Fe cytotoxicity in plants subjected to high iron concentrations (100 and 1000 μM). he control plants (without 34 in the medium) seemed to have significantly reduced height and dry weight, owing to the toxic effect of high levels of iron. It was observed that 34 could promote plant height at even the optimal Fe concentration of 10 μM. he positive effects of 34 were thus much evident in combating both biotic and abiotic stresses in the experimental tomato plant (Segarra et al. , 2010).
Reactive Oxygen Species (ROS) is produced by plants to initiate a hypersensitive response against invading plant pathogens like obligate parasites. However, accumulation of high levels of ROS can prove to be harmful for the plant itself causing severe oxidative damage (Sharma et al., 2012). hus, plants are equipped with a plethora of ROS scavenging enzymes, one of which is superoxide dismutase (SOD). Here, conclusive evidence of the working of a microbial consortium was observed, comprising of three microbes namely Pseudomonas sp., Trichoderma sp. and Rhizobium sp.. which was able to stimulate enhanced SOD activity in plants undergoing oxidative stress. his consortium was also seen to beneficially suppress the collar rot disease of chickpea plants (Cicer arietinum) which is mediated by the pathogen, Sclerotium rolfsii. Maximum activity of phenylalanine ammonia lyase (PAL) as well as the highest accumulation of toxic phenolic compounds was seen in the experimental chickpea plants under the activity of the triple microbial consortium. Compared to individually applied microbes, the consortium was also seen to prevent variations in lignin deposition in chickpea plant under the attack of the S. rolfsii pathogen. hus, the augmented/synergistic response of all these activities as seen in the consortium of the three microbes indicate that such a treatment can be used to enhance the host defence responses against an invading pathogen (Singh et al., 2013).
able 1 represents the different microbes inhabiting the rhizosphere and their modus operandi of remediating biotic stress.
Conclusion and future perspectives ill this point, we have considered all the mechanisms by virtue of which the rhizosphere microbiome can benefit plants. e have also realized that their uses are not just limited to amelioration of biotic stress, but they also have a serious role in other avenues which can beneficially affect plant growth. ‘Change’ or rather ‘improvement’ in this case, is the only constant. A series of possible approaches usually mentioned under the umbrella term of ‘soil-microbiome engineering’ that have been newly put into practise or those that warrant attention and further research are mentioned below:
A lot of experiments and models have employed the use single species of microbes. o deal with specific stresses, the soil can be treated with a microbial consortium containing a collection of different microbes working synergistically. his requires ample research into the bio-compatibility of microorganisms ensuring conditions and strains that can co-establish in the microbiome without competition. Successful implementation of close to ideal conditions have yielded synergism in biocontrol activity against plant pathogens by not only bacterial strains but also by heterogenous populations of bacteria and fungi (Yendyo et al., 2017; Jain et al., 2013). hrough further research, synthetic microbial communities (SynComs) have been formulated utilizing powerful -omics based approaches with good understanding of plant-microbe interactions (Pradhan et al., 2022). Utilization of dry laboratory experiments involving interpretation of dynamics of interactions along with other statistical data can ease the process of finding beneficial microbe combinations.
Certain customizations and formulations can greatly enhance plant resistance against a series of pathogens when carried out with soil amendments and proper nutrient management. Improved shelf life and a higher efficiency of microbes need to be obtained through genetic engineering (a few cases discussed earlier), as these hold the secret for better viability and translatability of in-vitro -tested microbes to the field conditions where the environmental factors like temperature, pH, water availability, soil-type etc. still greatly affect the outcome of the process. his process can be part of a bigger picture where we utilize rhizosphere engineering to not only target microbes, but also make amendments and improvements in plants for their root exudate manipulation or enhanced natural defences (Oger et al. , 1997).
A serious flaw with genetic engineering could be the possible horizontal gene transfer (HG ) of essential genes (a phenomenon observed in the spreading of antibiotic resistance) from the beneficial microbes to the pathogens, leading to a complete reversal of the outcome. here need to be novel ways through which we can counteract this process, like via utilizing synthetic fatty acids (Getino et al. , 2015) or using CRISPR-mediated interference to HG (Marraffini and Sontheimer, 2008). However, the latter method has received some criticism not only due to certain limitations, but also for the inherent cost of the techniques (Gophna et al. , 2015).
As more and more data are being generated, there lies the need for efficient bioinformatics-based tools to handle the ‘Big Data’ of sequencing information.
e also must look forward to increase the practicability of using rhizospheric microbiome. his will include steps to educate the farmers, agronomists and other stakeholders and call for proper schemes for easier adoption of these practices. he economic viability is also a major concern as the cost of these methods would be reflected through a rise in the price of crops in the market, and thus would be needed to be kept in check via rational laws of the land.
Dealing with all these concerns and possibly integrating this method with other prevalent methods of ameliorating biotic stress in plants will provide a holistic approach to achieve the target of agricultural sustainability and consecutively, environmental sustainability.
CONFLICTS OF INTEREST he authors declare that they have no potential conflicts of interest.


