Salt stress responses on protein profile in Vigna unguiculata L
Автор: Johnson M, Dooslin Mary D, Babu A
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 4 т.7, 2011 года.
Бесплатный доступ
The present study was aimed to elucidate the salt tolerant, salt inducible and salt sensitive protein of the Vigna unguiculataby Sodium Deodyl Sulphate - Poly Acrylamide Gel Electrophoresis. of Vigna unguiculataexposed to different environmental conditions exhibited a plethora of physio-chemical responses. The seedlings treated with various concentrations of Naat third day showed maximum of 85 bands, with nine active regions and their MW-Rf values ranged from 0.012 to 0.891. The seedlings treated with same experimental set up at fifth day showed maximum number of 63 bands with eight active regions and their MW-Rf values ranged from 0.108 to 0.891. On 5thday seedlings showed the isoperoxidase expression with various sizes of bands. The soluble protein showed a gradual increase during the initial stage and after fifth day there was gradual decrease in their content.
Salt stress, protein, sds- page, isoperoxidase, proline
Короткий адрес: https://sciup.org/14323555
IDR: 14323555
Текст научной статьи Salt stress responses on protein profile in Vigna unguiculata L
Environmental stress could be defined in plants as any change in growth within the plants natural habitat, which alters or disrupts its metabolic homeostasis. A few examples for plant metabolites involve in biotic/ abiotic stress response include compounds such as polyols, mannitol and sorbitol dimethyl sulfonicum compounds such as dimethy sulfoniopropionate, glycine, betains, sugar such as sucrose, trehalose and fructose; or amino acids such as proline and protein that serve as osmolytes and osmoprotectant to protect plants under extreme salt, drought and desiccation stresses. Salination is a widespread agricultural problem affecting 20% of the world's irrigated crop plants, and many other regions of the earth designated as arid and desert lands (Yamaguchi and Blumwold, 2005). Salt stress, drought and cold elicit a broad range of physiological and gene expression responses in plants (Zhu, 2002; Seki et al., 2003; Fujita et al., 2006; Yamaguchi- shinozaki and Shinozaki, 2006; Liu et al., 2007; Siddiqui et al., 2008). When plants are exposed to salinity in laboratory experiments, there is a rapid and temporary drop is growth rate followed by a gradual recovery to a new reduced rate of growth. The subsequent changes in growth rate and the underlying molecular or metabolic events are not so easily ascribed to water stress or to salt specific effects. As sessile organisms, plants are necessarily exposed to changing environmental conditions, often unfavorable. This situation has led them to develop evaluative strategies to recognize different environmental stresses and activate appropriate response.
An important adaptation found in many organisms subjected to water and other stresses is to accumulate certain organic compounds such as sucrose, amino acids (especially Proline) and several others that lower the osmotic potential and thus the water potential in cells without limiting enzyme function. Singh et al., (1985) and Yancey et al., (1982) observed the synthesis and accumulation of osmoprotective low molecular weight metabolites in plant responses to salts stress. Plant metabolic analysis has been used to examine the effects of gene deletions and transgenes (Roessner et al., 2000, 2001; Weckwerth et al., 2004), to elucidate the effects of physiological processes and physical stresses such as extremes of salinity or temperature (Johnson et al., 2003; Kaplan et al., 2004; Kim et al., 2007) and to study the genetics of metabolism (Keurentjes et al., 2006). Other applications include screening mutant plant populations, plant food sources and medicinal herbs or genetically manipulated (GM) crops to identify biomarkers relating to desirable or undesirable traits. Based on this background, the present investigation was initiated to extend the good work already done in our laboratory on other equally important species Vigna unguiculata L. The specific objectives of the investigation are to 1). Make a comparative study of the influence of NaCl on growth and development of Vigna unguiculata seedlings 2). Assess the relative changes of protein pattern, isoperoxidase profiles and proline content in different developmental stages 3). Elucidate the salt tolerant, salt sensitive and salt inducible proteins of Vigna unguiculata.
MATERIALS AND METHODS
and mortar and centrifuged at 12,000 rpm for 10 min and the supernatant was collected and used for iso enzyme (peroxidase) analysis (Smila et al., 2007). The Poly acrylamide gel electrophoresis was performed by Anbalagan (1999) method. The staining and fixation of the enzyme was performed by the Sadasivam and Manickam (1992) method. For the quantitative estimation of proline the standard procedure was followed (Bates et al., 1973).
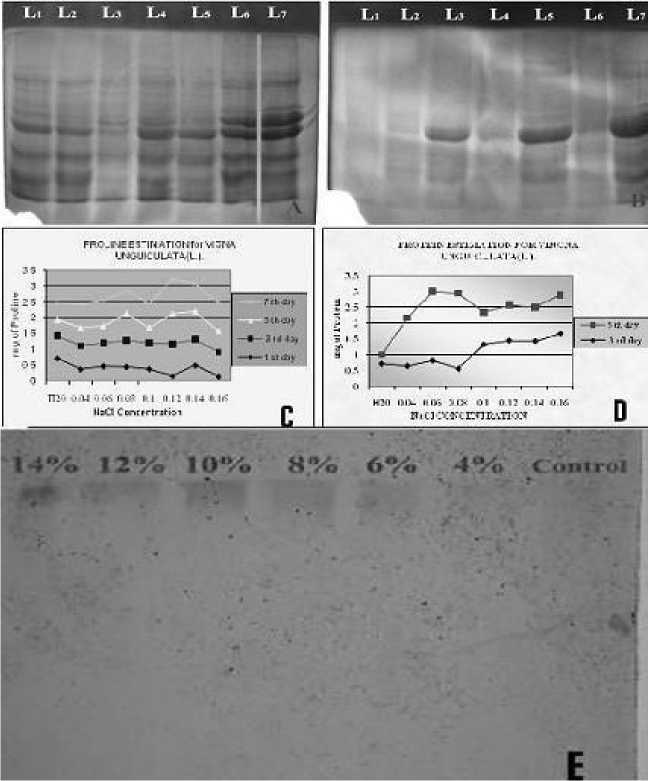
Figure 1 Salt Stress responses on Protein Profile in Vigna unguiculata L
-
A) SDS - PAGE Profile of 3rd day seedlings treated with different concentrations of NaCl B) SDS - PAGE Profile of 5th day seedlings treated with different concentrations of NaCl C) Graphical view of the protein level at various stress conditions
-
D) Graphical view of the proline level at various stress conditions
-
E) Isoperoxidase expression of salt treated and control seedlings
RESULTS
Seeds of V. unguigulata soaked for 24h, 48h and 72h in 1% Nacl, showed different percentage of seed germination and seedlings growth. The seeds soaked for more than 24 h showed very less percentage of germination and growth. The germinated seedlings were cultured on MS medium augmented with various percentage viz., 0, 2, 4, 6, 8, 10, 12 and 14 of salt (Nacl). The seedlings showed the maximum tolerance up to 10% of Nacl. The exomorphic character such as shoot length, fresh weight and dry weight were also affected differently at diverse concentrations. The Nacl treated plants showed that the reduction in height was progressively and consistently increased with increasing Nacl concentrations. The graphical view of the protein at different stage of the seedlings was shown in Fig 1C. The prolines of seedlings at different developmental stages were illustrated in Fig. 1 D.
Table 1 : SDS – PAGE banding pattern of Different concentrations of NaCl treated Vigna unguiculata L. seedlings on third day
|
MW - Rf |
Different Concentration of Nacl percentage |
||||||
|
Control (0%) |
4% |
6% |
8% |
10% |
12% |
14% |
|
|
0.012 |
+ |
- |
- |
- |
- |
- |
- |
|
0.192 |
- |
- |
- |
+ |
+ |
- |
- |
|
0.200 |
+ |
- |
- |
- |
- |
- |
- |
|
0.204 |
- |
- |
- |
- |
- |
+ |
+ |
|
0.253 |
- |
- |
- |
+ |
- |
- |
- |
|
0.265 |
+ |
- |
- |
- |
+ |
+ |
+ |
|
0.289 |
+ |
+ |
+ |
+ |
- |
+ |
+ |
|
0.304 |
- |
- |
- |
- |
+ |
- |
- |
|
0.313 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
0.326 |
- |
- |
+ |
- |
+ |
- |
- |
|
0.337 |
+ |
+ |
- |
- |
- |
+ |
+ |
|
0.385 |
+ |
- |
- |
- |
- |
- |
- |
|
0.409 |
- |
- |
- |
- |
- |
+ |
+ |
|
0.434 |
+ |
- |
- |
- |
- |
- |
- |
|
0.445 |
- |
- |
+ |
+ |
+ |
+ |
+ |
|
0.457 |
- |
+ |
- |
- |
- |
- |
- |
|
0.481 |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
0.518 |
+ |
+ |
+ |
- |
- |
- |
- |
|
0.542 |
- |
- |
- |
+ |
+ |
+ |
+ |
|
0.602 |
- |
- |
- |
- |
+ |
+ |
+ |
|
0.638 |
- |
- |
- |
- |
- |
+ |
+ |
|
0.650 |
+ |
- |
- |
- |
- |
- |
- |
|
0.662 |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
0.746 |
- |
- |
- |
- |
- |
+ |
+ |
|
0.783 |
- |
- |
- |
+ |
+ |
+ |
+ |
|
0.795 |
+ |
+ |
- |
- |
- |
- |
- |
|
0.831 |
- |
- |
- |
- |
+ |
- |
- |
|
0.843 |
- |
- |
- |
+ |
- |
- |
- |
|
0.867 |
+ |
- |
- |
+ |
+ |
+ |
- |
|
0.891 |
+ |
+ |
+ |
- |
- |
- |
- |
Proteome analyses have identified many proteins that are inducible by salt stress including the metabolism related genes. The relative positions of the protein bands as revealed by SDS- PAGE of the V. unguiculata seedlings under different stress conditions are shown in Fig 1A and 1B. The seedlings treated with various concentrations of Nacl at third day showed maximum of 85 bands, with nine active regions and their MW-Rf values ranged from 0.012 to 0.891. The seedlings treated with same experimental set up at fifth day showed maximum number of 63 bands with eight active regions and their MW-Rf values ranged from 0.108 to 0.891. Fig 1A and B illustrated the banding profile of salt stressed and control seedling of Vigna unguiculata. On the 3rd day the isoperoxidase expression was not obtained. On 5th day seedlings showed the isoperoxidase expression with various sizes of bands (Fig. 1 E). Maximum size of band was observed in the seedlings treated with 8 percentage of Nacl. The isoperoxidase expression was observed in the seedlings treated with 6 percentages of Nacl and above.
Table 2: SDS – PAGE banding pattern of Different concentrations of NaCl treated Vigna unguiculata
L. seedlings on fifth day
|
MW - Rf |
Different Concentration of Nacl percentage |
||||||
|
0% |
4% |
6% |
8% |
10% |
12% |
14% |
|
|
0.108 |
- |
- |
- |
+ |
- |
- |
- |
|
0.152 |
+ |
- |
- |
- |
- |
- |
- |
|
0.163 |
- |
+ |
- |
- |
- |
- |
- |
|
0.173 |
+ |
+ |
+ |
- |
- |
- |
- |
|
0.195 |
+ |
- |
+ |
- |
- |
- |
- |
|
0.217 |
- |
- |
+ |
- |
- |
- |
- |
|
0.239 |
+ |
+ |
+ |
- |
- |
- |
+ |
|
0.260 |
- |
- |
- |
- |
+ |
+ |
+ |
|
0.271 |
- |
+ |
- |
+ |
- |
- |
- |
|
0.282 |
- |
- |
+ |
- |
- |
- |
- |
|
0.304 |
- |
- |
- |
+ |
- |
- |
- |
|
0.326 |
- |
- |
- |
- |
- |
- |
+ |
|
0.358 |
- |
- |
- |
- |
+ |
+ |
- |
|
0.380 |
+ |
+ |
- |
- |
- |
- |
- |
|
0.413 |
- |
- |
+ |
+ |
+ |
+ |
+ |
|
0.434 |
+ |
- |
- |
- |
- |
- |
- |
|
0.456 |
- |
- |
- |
- |
- |
+ |
+ |
|
0.467 |
- |
+ |
+ |
+ |
+ |
- |
- |
|
0.489 |
+ |
- |
- |
- |
- |
- |
- |
|
0.500 |
- |
- |
- |
- |
+ |
- |
- |
|
0.505 |
- |
- |
- |
- |
+ |
+ |
- |
|
0.565 |
- |
- |
- |
- |
+ |
+ |
+ |
|
0.597 |
+ |
+ |
+ |
- |
- |
- |
- |
|
0.619 |
- |
- |
+ |
+ |
- |
- |
- |
|
0.706 |
+ |
- |
- |
- |
- |
- |
- |
|
0.765 |
+ |
+ |
- |
- |
- |
- |
- |
|
0.717 |
- |
+ |
- |
- |
- |
- |
- |
|
0.760 |
- |
- |
+ |
- |
- |
- |
- |
|
0.782 |
- |
- |
- |
+ |
- |
- |
- |
|
0.836 |
+ |
- |
- |
- |
- |
- |
- |
|
0.869 |
- |
+ |
+ |
+ |
+ |
- |
- |
|
0.891 |
- |
- |
- |
+ |
+ |
+ |
+ |
The protein profiles were classified in to three categories based on their expression viz., salt tolerant protein, salt inducible protein and salt sensitive protein. The present study elicited following salt inducible protein viz., MW- Rf 0.192, 0.204, 0.253, 0.301, 0.325, 0.406, 0.445, 0.457,
0.542, 0.602. 0.662. 0.746, 0.783, 0.831, 0.843 and 0.867 on the third day seedling of Vigna unguicualta . The following salt inducible proteins MW – Rf 0.108, 0.163, 0.217, 0.260, 0.271, 0.282, 0.304, 0.326, 0.358, 0.413, 0.456, 0.467, 0.500, 0.505, 0.565, 0.619, 0.717, 0.760, 0.782, 0.869 and 0.891 were identified on the 5th day salt stressed seedling of Vigna unguiculata . The following proteins MW – Rf. 0.173, 0.195, 0.239, 0.265, 0.289, 0.313, 0.337, 0.380, 0.518, 0.597, 0.765, 0.795 and 0.891 were expressed in the control and salt stressed seedling, they are called as salt tolerant proteins. The following proteins MW-Rf. 0.012, 0.152, 0.200, 0.385, 0.433, 0.489, 0.650, 0.706 and 0.836 failed to express in the salt treated seedling, they are called salt sensitive proteins.
DISSUSSION
Growth of any organ is associated with an additional synthesis of protein which are building blocks of protoplasm and are again the resultant on inter- mediatory metabolism. The soluble protein showed a gradual increase during the initial stage and after fifth day there was gradual decrease in their content. This may be due to reduction in chlorophyll contents. The leaves were turned pale yellowish green. This is a symptom of chlorosis. The chlorophylls in photosynthetic pigments established a complex with protein and when chlorophyll was decreased, there was decrease in protein content also.
Proline generally alleviated the inhibitory effect of salinity on the studied parameters. This alleviation was generally associated with K+/Na+ ratio of shoots and roots. Hegde and Joshi (1974) found that K+ / Na+ ratio was higher in salt tolerance than sensitive cultivars and recommended it as a suitable selection criterion for salt tolerance. It is clear from Fig 1 D that proline is very high at every period of salt stress. Our result depicting proline accumulation is in agreement with other reports (Routely, 1966). The greater accumulation of proline in these plants, presumably render them drought tolerant. A possible reason for this increased level of proline during the salt stress could be an alteration in the activities of the enzymes involved in the biosynthesis and degradation of proline. Proline is thought to play a multifunctional role in the defense mechanisms. It acts as a mediator of osmotic adjustment, a stabilizer of subcellular structure, a scavenger of free radicals, an energy salt and stress related signal (Nanjo et al., 2003).
A strong correlation between the accumulation of proline tolerance of drought stress has been demonstrated by over expression of the ∆’ pyrroline- 5- carboxylatesynthesis gene p5cs or by anti-sense suppression of the proline dehydrogenase (proDH) gene in various plants. Seedlings of Vigna unguiculata exposed to different environmental conditions exhibited a plethora of physio-chemical responses. Salt stress causes the production of reactive oxygen radicals or species (ROS). Mechanisms of ROS detoxification exist in all plants and can be categorized as enzymatic (SOD, APX, POX, GR, etc) and non-enzymatic (AA, flavanones, anthocyanins, etc) (Shao et al., 2008). The results of the present study also confirm the Shao et al., observation. Results of the present study also showed the isoperxidase content acceleration in accordance with the salt stress. In conclusion, the data presented here revealed that salinity induced changes in protein and isoperoxidase profiles in seeds and seedling of Vigna unguiculata. It is necessary to make further study on the structural and functional roles of these salt tress responsive polypeptide to enhance our understanding of the salt stress responses in Vigna unguiculata .
Список литературы Salt stress responses on protein profile in Vigna unguiculata L
- Anbalagan, K. (1999). An introduction to electrophoresis, Electrophoresis Institute Yercaud, Tamil Nadu, India. pp 105.
- Bates, L.S., Waldren, R.P. and Tear, L.D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil, 39,205-207.
- Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, and Shinozaki, K. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology, 9 (4), 436-442.
- Hegde, B. A. and Joshi, G. V. (1974). Pattern of photosynthesis in a saline indica var. of rice Kalarata. The Proceedings of the symposium on use of radiations and radioisotopes in studies on plant productivity, Pantnagar, 755 -763.
- Johnson, H.E. Broadhurst, D. Goodacre, R. and Smith, A. R. (2003). Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry, 62, 919-928.
- Kim, J.K., Bamba, T., Harada, K., Fukusaki, E. and Kobayashi, A. (2007). Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J Exp. Bot., 58, 415-424.
- Kaplan, F., Kopka, J., Haskell, D.W., Zhao, W., Schiller, K.C., Gatzke, N., Sung, D.Y. and Guy, C.L. (2004). Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol., 136, 4159-4168.
- Keurentjes, J.J.B., Fu, J.Y., De Vos, C.H.R., Lommen, A., Hall, R.D., Bino, R.J., Van der Plas L.H.W., Jansen, R.C., Vreugdenhil, D. and Koornneef, M. (2006). The genetics of plant metabolism. Nat. Genet., 38, 842-849.
- Liu, J. X., Srivastava, R., Che, P. and Howell, S. H. (2007). An Endoplasmic Reticulum Stress Response in Arabidopsis is Mediated by Proteolytic Processing and Nuclear Relocation of a Membrane-Associated Transcription Factor, bZIP28. The Plant Cell, 19, 4111-4119.
- Lowry, O. H., Rosebrough, N. J., Farr, A. L. and Randal R. J. (1951). Protein measurement with folin phenol reagent. J. Biol. Chem., 193, 265 -275.
- Nanjo, T., Fujita, M., Seki, M., Kato, T., Tabata, S. and Shinozaki,. (2003). Toxicity of free proline revealed in an Arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant and Cell Physiology, 44, 541-548.
- Roessner, U., Luedemann, A.,Brust, D., Fiehn, O., Linke, T., Willmitzer, L. and Fernie, A. R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell, 13, 11-29.
- Routley, D.G. (1966). Proline Accumulation in Wilted Ladino Clover Leaves. Crop Sci., 6, 358-361.
- Sadasivam, S. and Manickam, A. (1992). Biochemical methods for Agricultural Science. Wiley Eastern Ltd. and Tamil Nadu Agricultural University, Coimbatore, India.
- Seki, M., Kamei, A., Yamaguchi-Shinozaki, K. and Shinozaki, K. (2003). Molecular responses to drought, salinity and frost: common and different paths for plant protection. Current Opinion in Biotechnology, 14 (2), 194-199.
- Shao H.B., Chu L.Y., Lu Z.H., Kang C.M. (2008). Primary oxidant scavenging and redox signaling in higher plants. Int J Biol Sci.; 4, 8-14
- Smila, K.H., Johnson, M. and Rajasekarapandian, M. (2007). Studies on varietal difference, tissue specificity and developmental variation of esterase and peroxidase isozymes in pearl millet [Pennisetum glaucum (L.) R. Br.]. Indian Journal of Biotechnology, 6, 91-99.
- Siddiqui Z., Khan M.A., Kim B.G., Huang, J.S. and Kwon, T.R. (2008). Physiological responses of Brassica napus genotypes to combined drought and salt stress. Plant Stress, 2 (1), 78-83.
- Singh, N.K., Handa, A.K., Hasegaura, P.M. and Bressan, R.A. (1985). Proteins associated with adaptation of cultured tobacco cells to NaCl. Plant Physiology, 79, 126-37.
- Weckwerth, W., Wenzel, K. and Fiehn, O. (2004). Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics, 4, 78-83
- Yamaguchi, T. and Blumwald, E. (2005). Developing salt-tolerant crop plants: challengesand opportunities. Trends in Plant Science, 10, 615-620.
- Yamaguchi-Shinozaki, K. and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stress. Annual Review of Plant Biology, 57, 781-803.
- Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D. and Somero, G. N. (1982). Living with water stress: evolution of osmolyte systems. Science, 217, 1214-1222.
- Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annual Review of Plant Biology, 53, 247-273.


