Segmentation of Pre-processed Medical Images: An Approach Based on Range Filter
Автор: Amir Rajaei, Elham Dallalzadeh, Lalitha Rangarajan
Журнал: International Journal of Image, Graphics and Signal Processing(IJIGSP) @ijigsp
Статья в выпуске: 9 vol.4, 2012 года.
Бесплатный доступ
Medical image segmentation is a frequent processing step. Medical images are suffering from unrelated article and strong speckle noise. In this paper, we propose an approach to remove special markings such as arrow symbols and printed text along with medical image segmentation using range filter. The special markings are extracted using Sobel edge detection technique and then the intensity values of the detected markings are substituted by the intensity values of their corresponding neighborhood pixels. Next, three different image enhancement techniques are utilized to remove strong speckle noise as well enhance the weak boundaries of medical images. Finally range filter is applied to segment the texture content of different modalities of medical image. Experiment is conducted on ImageCLEF2010 database. Results show the efficacy of our proposed approach which lead to have precise content based medical image classification and retrieval systems.
Pre-processing, Special markings, Sobel edge detection technique, Medical image texture segmentation, Image enhancement, Texture filter, Range filter
Короткий адрес: https://sciup.org/15012372
IDR: 15012372
Текст научной статьи Segmentation of Pre-processed Medical Images: An Approach Based on Range Filter
Published Online September 2012 in MECS
Computer Aided Diagnosis (CAD) provides an effective tool for analyzing the particular anatomical structure of a medical image to have better diagnosis and decision-making. The number of medical images has grown significantly in the recent years. Medical images are very important for clinical diagnosis, pathology localization, treatment planning, computer-integrated surgery, surgical planning, post-surgical assessment, abnormality detection and so on. It is a well- established fact that the common characteristics of medical images such as unknown noise, poor image contrast, inhomogeneity, weak boundaries and unrelated articles will affect the content of the medical images.
Hence, to overcome mentioned dilemma, the preprocessing of medical images is a vital step in image processing to have better image segmentation and feature extraction. The performance of pre-processed of medical images will consequently influence the classification and retrieval performance in ContentBased Medical Image Retrieval (CBMIR) systems. Preprocessing stage deals with image enhancement, noise and special marks removal. In segmentation stage several methods are exist for automatic and semiautomatic medical image segmentation. However, medical image segmentation may fail to segment well the anatomical structure, the region of interest. Unknown noise, poor image contrast, inhomogeneity, weak boundaries and special marking existing in medical images have made the segmentation process extremely difficult. Besides, it is an application dependent technique to enhance the features and to remove the noise and special markings that exist in medical images [3, 4].
Peng et al. [5] proposed a pre-processing method including cutting out background area and normalization for CT brain images. In their proposed approach, an ellipse structure is constructed based on skull contour and then the lean imaging angle is being corrected. Červinka and Provazník [6] proposed pre-processing method on CT images. In their proposed approach, the histogram of the intensity in CT images is downsampled. Therefore, the low contrast and blurring regions in CT images are enhanced. Markov Random Field model which consider the geometrical constraints of the processed image is used to improve the accuracy resulting from the down-sampling procedure. Median filtering, open morphological operation and contrast enhancement are used by Hussien et a. [7] as the preprocessing stages to reduce noise and image enhancement. Sinha [1] investigated the use of the Eigen image methodology with focus on the image preprocessing steps. An improved minutia preserving smoothing algorithm for pre-processing of MR image is given by Zu et al. [2] based on solving a non-linear diffusion equation.
The techniques such as thresholding [8], region growing [9], fast greedy algorithm [10], Fuzzy C-mean (FCM) clustering and statistical models [11], active contour model [12] and watershed segmentation [13,14] have been proposed for medical image segmentation. Felzenszwalb and Daniel [15] proposed Minimum Spanning Tree (MST) for segmentation of medical images. MST is one of the graph-based segmentation methods that it is computationally efficient for capturing perceptually the important aspects of image regions.
However, it is prone to cause the problem of over segmentation with some long and narrow redundant areas between two regions. Lu et al. [16] describe an improved segmentation algorithm based on Minimum Spanning Tree (MST). To overcome the problem of over segmentation, an adaptive neighbor mode is defined by adding links between non-neighbor pixels of an image. They explored their proposed segmented method on three different modalities of medical images such as MR, CT and X-ray.
Gradient Vector Flow (GVF) snake model is used in image segmentation. In [17], GVF snake is proposed to overcome the inability of tracking concavity at boundary. However, noise existing in medical images may cause the boundaries of medical images not to converge. Hence, in these complicated images GVF still fails to capture object contours. To improve GVF snake model, Chuag and Li [18] proposed a new downstream algorithm based on Extended Gradient Vector Flow (E-GVF) for segmentation of multi-objects. In [19], they focused on texture features as well as region growing algorithm to automate segmentation of the MR images. Co-occurrence texture features and semi-variogram texture features are extracted from the image and then the seeded region growing algorithm is run on these feature spaces. With a given region of interest, a seed point is automatically picked up based on three homogeneity criteria. Medical ultrasound images are poor in contrast and they have strong speckle noise. Hence, traditional image segmentation methods fail to segment the ultrasound images satisfactorily. In [20], the ultrasound images are segmented using texture feature and graph cut method. The texture feature parameters are obtained according to the Gary level Co-occurrence Matrix (GLCM).
In addition to the above discussed literature survey based on medical image pre-processing and image segmentation, it is observed that the medical images suffer from special markings and speckle noise. Therefore, this unrelated article will affect the precision of medical image segmentation. It is also observed that numbers of segmentation methods are applied on only specific anatomical structure such as the use of GVF for segmentation of kidney images. Moreover, the proposed methods are exploited on particular medical image modalities. Medical ImageCLEF2010, which we used in this paper, is a database including various anatomical structures belonging to different medical image modalities. These several of medical image modalities are carrying special markings such as printed text, arrow symbols as well as strong speckle noise. The above mentioned deficiency has motivated us to propose an approach to remove special markings and speckle noise existed in medical Image to have precise medical image segmentation. Furthermore, the well-known characteristics of medical images such as unknown noise, poor image contrast, inhomogenity, weak boundaries and special marks affect the anatomical structure resulting in poor segmentation. Besides, the poor characteristics of medical images are moderated to have accurate medical image segmentation. On the other hand, the precise medical image segmentation lead to have an accurate and efficient content based medical image classification and retrieval systems.
The rest of the paper is as follows. Pre-processing of medical images to remove special markings and speckle noise is discussed in section II. The proposed approach to segment medical images based on texture is explained in section III. Section IV gives the details of experimentation followed by the results. The paper is concluded in section V.
-
II. Medical image preprocessing
-
A. Special Marking Removal
Medical images have different content structures. However, the special markings alter the content structure of medical images. ImageCLEF2010 includes various anatomical structures. But different shape of arrow symbols and printed text located in these images degrade further image processing and analysis. Removal of special markings existing in medical images will increase the quality of image segmentation, feature extraction and consequently will improve the image retrieval performance. Hence, a pre-processing step is required to eliminate such special markings and retain pure anatomical structure of images. Furthermore, removal of special markings should not modify the content of images. In this direction, we propose an approach to remove the arrow symbols and printed texts from the texture content of images. Fig.1 shows a number of sample images belonging to different medical image modalities of ImageCLEF2010.
From the medical images illustrated in Fig.1, it is observed that the existing special markings have distinct gray-level values and strong boundaries compare to anatomical structures. Thus, the arrow symbols and printed texts can be extracted using edge detection technique as explained in section A.1. The extracted symbols and texts are eliminated from the texture content of images as details are given in section A.2.
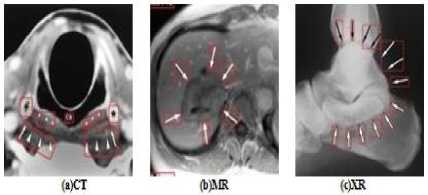
Figure1. (a) –(c) ImageCLEF2010, Different Medical Image Modalities
A.1. Extraction of Special Markings
Poor image contrast, inhomogenity and weak boundaries make the common edge detection techniques not able to extract the anatomical structures of medical images. However, the distinct gray-level values as well as the strong boundaries of symbols and texts make the edge detection techniques to extract these components to high level of accuracy. Hence, Sobel edge detector is utilized in this proposed approach to extract the special markings. In Sobel algorithm, the edges of an image can be found by approximating the gradient magnitude of the image. The image obtains the gradient as a result of convolving the image with the Sobel. The image edges are detected at those points where the gradient of image is maximum. Dilation as a morphological operation is then applied on the extracted edge image to have connected component. Fig.2 illustrates the applied Sobel edge detection technique on a MR image.
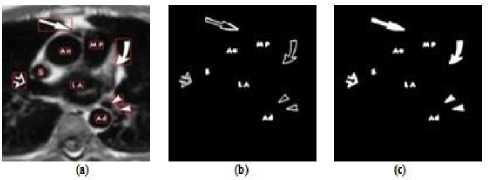
Figure2. (a) Original Image with Arrow Symbols and Printed Text which Shown in Red Box, (b) Extracted Special Markings, (c) Filled
Special Markings
A.2. Elimination of Special Markings
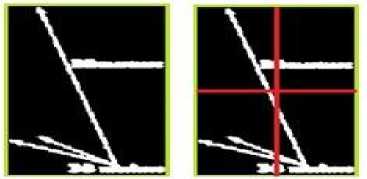
In this subsection, we propose an approach to eliminate the extracted arrow symbols and printed text in the texture content of an image. A bounding box is constructed surrounding each extracted symbols or text as shown in Fig.3(c). The bounding box is then divided into the four equal areas as illustrate in Fig.4(b). Every pixels located in the first quarter of the bounding box having the intensity value ‘255’ are considered. From the location of each obtained pixel, we start moving in both horizontal and vertical directions towards left and up. The movement continues until it crosses the border of the bounding box as shown in Fig.5(a).
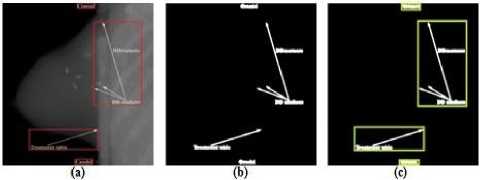
Figure3. (a) X-ray Image with Arrow Symbols and Printed Text, (b) Extracted Special Markings, (c) Special Marking are Surrounded with a Bounding Box
(a)
(b)
Figure4. (a) Extracted Special Markings, (b) Dividing Area
After traversing from each considered pixel to the border of the bounding box, the shortest of horizontal and vertical distances are found. Next, the pixel located on the other side of the computed shortest distance is supposed to be the candidate neighbor pixel for the considered pixel. Therefore, in the original image the intensity value of the corresponding considered pixel is replaced by the intensity value of the candidate neighbor pixel. Similarly, the discussed proposed approach is done for the second, third and fourth quarters. However, for each considered pixel located in the second quarter, we start moving in both horizontal and vertical direction towards right and up, Fig.5(b). In the third quarter, moving is started in both horizontal and vertical towards left and down, Fig.5(c). Finally, in the fourth quarter, moving is done by traversing in both horizontal and vertical directions towards right and down, Fig.5 (d). To summarize our proposed approach, the position of the candidate pixel with respect to each quarter is presented in Table 1. We illustrate the matrix representation of particular special marking, extraction and removal of that special marking in Appendix I. Fig.6 presents different medical image modalities that the special markings are removed.
T able 1. C andidate P ixel L ocation
|
Area |
Position of Candidate Pixel |
|
|
Shortest Distance in Horizontal Direction |
Shortest Distance in Vertical Direction |
|
|
First Quarter |
((X-shortestDintance-l), Y) |
(X.CY-shortestDistance-l)) |
|
Second Quarter |
((X-shottestDiiOncrl). Y) |
(X, (Y- shottstDistance-l)) |
|
Third Quarter |
((X-ihortesDistance-l), Y) |
(X. (Y-shotteitDistancH)) |
|
Fourth Quarter |
((X-ihoitestDistancrl).Y) |
(X. (Y-shortestDistancrl)) |
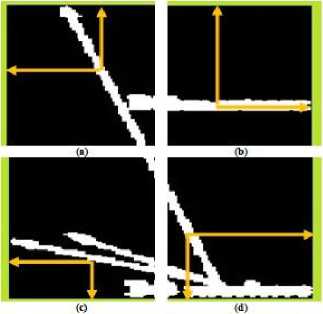
Figure5. (a)Traversing in I Quarter, (b) Traversing in II Quarter, (c)Traversing in III Quarter, (d) Traversing in IV Quarter
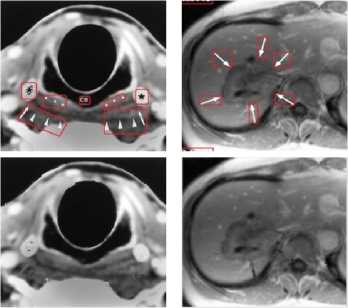
(I) CT (II) MR
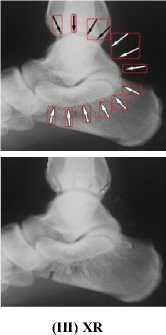
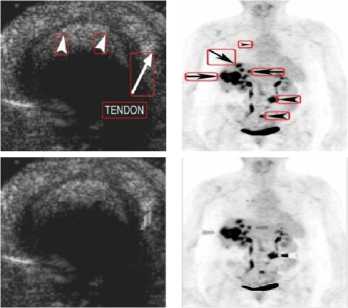
(IV) US
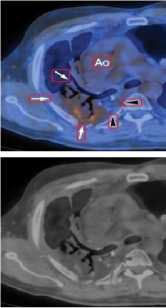
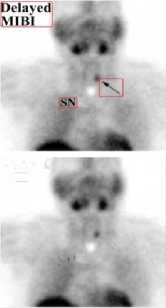
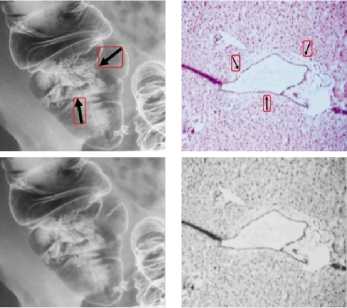
(VII) NM
(V) PET (VI) PET-CT
(VIII) XR-Angio
(IX) Micro
Figure6. Removal of Special Markings
-
B. Speckle Noise Removal
The contrast of medical images is very low and these images have strong speckle noise. We apply 2D adaptive noise-removal, median and 2D order-statistic filtering to remove the noise and enhance the weak boundaries of medical images. 2D adaptive noise removal is a low-pass filter on gray scale images. 2D adaptive noise removal uses a pixel wise adaptive wiener method based on the statistics estimated, mean and standard deviation, from a local neighborhood of size m by n for each pixel. Result of image enhancement using 2D adaptive noise removal is shown in Fig.7(b).
To remove ‘salt and pepper’ noise existing in medical images, we use median filtering. Median filtering is a nonlinear operation which is very effective to simultaneously reduce noise and preserve edges. Each output pixel contains the median value in p by q neighborhood around the corresponding pixel of the input image. Fig.7(c) shows removal of “salt and pepper” noise from a medical image using median filtering. The order statistic filter is then applied to enhance the regions of an anatomical structure. The 2D orderstatistic filtering replaces each pixel of an image by the nth order element in the sorted set of neighbors of size r by s specified by the nonzero elements in domain. Image edge enhancement using 2D order-statistics filtering is shown in Fig.7(d). Then, the speckle noise removal of medical images is done by the three discussed image enhancement functions as shown in Fig.7(e).
-
III. Segmentation
Image texture includes the local spatial pattern, scale and magnitude of brightness variations, smoothness or roughness of the image. The output image can be used as the basis for further image analysis such as image texture segmentation. Hence, in this paper we propose texture filtering to segment medical image texture. We propose to utilize the concept of range filtering for segmentation. Filtering is perhaps the most fundamental operation of image processing. The term filtering can be defined as the value of the filtered image at a given location. It is a function of the values of the input image in a small neighborhood of the same location. Filter operations can be used to sharpen or blur images, to selectively suppress image noise, to detect and enhance edges, or to alter the contrast of the image. The filters use the local statistical variations in an image to reveal elements of the image texture. Image texture segmentation using range filtering is discussed in subsection B.1.
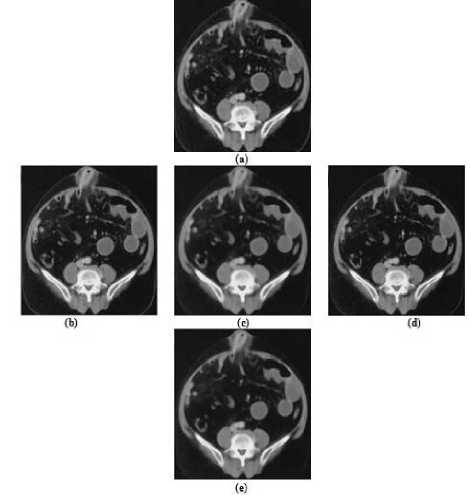
Figure7. (a) Original Image, (b) Applied Adaptive Noise Removal on an Image, (c) Applied Median Filtering on an Image, (d) Applied Order-statistic Filtering on an Image, (e) Pre-processed Image
-
A. Analyzing the Texture of an Image
Texture analysis refers to the characterization of regions in an image by their texture content. Texture analysis attempts to quantify intuitive qualities described by terms such as rough, smooth, silky or bumpy as a function of the spatial variation in pixel intensities. In this sense, the roughness or bumpiness refers to variations in the intensity values or gray levels.
Texture analysis is used in a variety of applications, including remote sensing, automated inspection and medical image processing. Texture analysis can be used to find the texture boundaries and texture segmentation. Texture analysis can be helpful when objects in an image are more characterized by their texture than by intensity and hence, traditional thresholding techniques cannot be used effectively. The texture analysis functions such as range filtering, standard deviation filtering and entropy filtering, filter an image using standard statistical measures. These statistics can characterize the texture of an image. They provide information about the local variability of the pixels intensity values in an image. In the areas with smooth texture, the range values in the neighborhood around a pixel will be small and similarity, the range values are large in the areas of rough texture.
The texture functions all operate in a similar way. They define a neighborhood around the pixel of interest calculate the statistic for that neighborhood and use the computed statistic value as the value of the pixel of interest in the output image. The example shown in Fig.8 illustrates how the range filtering function operates on a simple matrix. In this example, the value of element B (2, 4) is calculated from A (2, 4). Range filtering function use m by n pixels, in this example 3 × 3, neighborhood around the pixels.
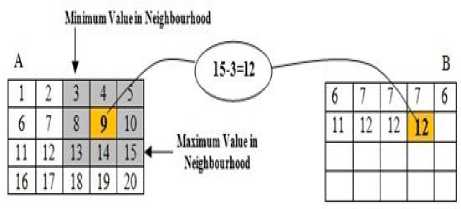
Figure8. Range Filtering Function
-
B. Medical Image Texture Segmentation
Medical image segmentation is a critical task for analyzing the structural content of the images. Surgical planning, early disease detection and 3D visualization can be provided for the physicians by proper image segmentation. Medical image texture segmentation can be widely applicable to evaluate an area of body that is not externally visible. The texture filter function can detect the texture regions of a medical image. It can be argued that there is a little variation in the gray level values of the background of medical images since the background is smooth. Hence in the foreground the surface contours of the anatomical structure exhibit more texture. Therefore, the foreground pixels of medical images have more variability and thus higher range values. Range filtering is one of the texture analysis methods that filter an image. Range filtering makes the edges and contours of the anatomical structures of medical images become more apparent. Consequently, the range filtering is explored in this paper for medical image texture segmentation as explained in the following subsection.
-
B.1. Range Filtering
A local sub range filtering uses the statistical sub range of the pixel intensities within the window. The range distance is often used in statistics as a measure of the sample variation. Edges are typically characterized by discontinuities in mean intensity. If the variations existing among the local intensity values are low then the local range distance is small. Similarly, the local range distance is large if a region has large discontinuities in intensity values. Hence, range filtering is able to detect pixel intensity values of the edges within a window. The output of the range filtering is the difference between maximum and minimum range values of the filtered window. The range values in the filtered window are multiplied by a constant value to provide strong edges. Local range filtering tend to have short calculation time as it operates on only a small number of input for each output pixel. Moreover, range filtering can have better segmentation through creating a structure element to extract the neighborhood for the local range of values. Hence a structuring element is created to extract the neighborhood for the local range of values. Fig.9 illustrates the range filtered medical image using the defined structure element.
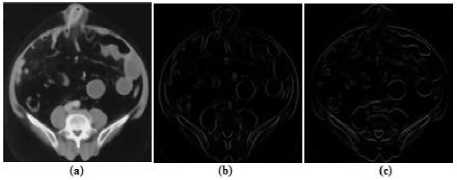
Figure9. (a) Pre-processed Image, (b) Range Filtered Medical Image using Horizontal Line Structure, (c) Range Filtered Image using Vertical Line Structure
-
IV. Experimentation
-
A. Image Dataset
In this paper, ImageCLEF2010 database is used. Experiments are conducted on different medical image modalities with various image sizes. The medical image modalities are nine in numbers namely, Compute Tomography (CT), Magnetic Resonance (MR), Ultrasound (US), Nuclear Medicine (NM), Xr-Angio, Positron Emission Tomography (PET), PET-CT, Micro and X-ray. Medical images include various anatomical structures and image orientation. However, the majority of images existing in this database carrying special marking such as arrow symbols with different shapes and sizes as well as printed texts along with speckle noise.
-
B. Experimental Setup
Medical images contain strong speckle noise. To remove noise and smooth the images, we have applied 2D adaptive noise removal using neighborhood of size 3 by 3. Moreover, medical images suffer from “salt and paper” noise. We remove the existing ‘salt and paper’ noise using median filtering. Each output pixel contains the median value of 9 by 9 neighborhoods surrounding the pixel of the input image. Further, it is well known that the content of medical images are inhomogeneous having weak boundaries. We use 2D order-statistic filtering by the 3rd order element in the sorted set of neighbors of size 3 by 3 in domain. Then, special markings in images are extracted by Sobel edge detection technique with the threshold value of 0.15. The extracted symbols and printed texts are dilated using diamond structuring element of size ‘3’. The pixel intensity values of the extracted special markings are replaced by the corresponding intensity values of their neighbor pixels as explained before.
In the segmentation step, we have exploited range filtering using ‘line’ structure element of size 1×2 and 2×1 in horizontal and vertical directions respectively. Next, we have applied sum operation on the images extracted using range filtering in both horizontal and vertical directions. Fig.10(a) shows the added image obtained from the medical images illustrated in Fig. 9(b) and Fig.9(c) respectively. In addition, to improve the extracted edges of the ranged filtered medical image as shown in Fig.10(a) we scale the image by multiplying with the fix value of ‘k’. In this experimentation, the value of ‘k’ is set to 20 empirically. Fig.10(b) presents the scaled range filtered medical image.
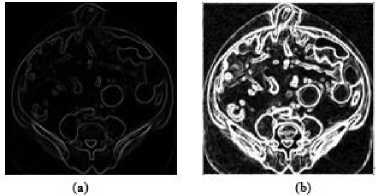
Figure10. (a) Range Filtered Medical Images after Filtering in Horizontal and Vertical Directions are Added, (b) Scaled Range
Filtered Medical Image
Next, we have used ‘clean’ morphological operation to remove all detected isolated pixels. Fig. 11 shows the segmentation of the anatomical structure of the medical image using our proposed texture segmentation method.
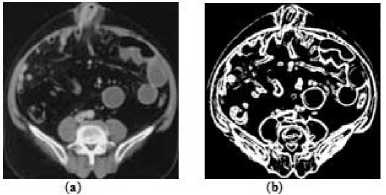
Figure11. (a) Original Image, (b) Medical Image Segmentation
-
C. Experimental Results
In this paper, experiments are conducted on nine different medical image modalities including different anatomical structures. Medical images are segmented after the pre-processing step. The segmented medical images using range filtering is as shown in Fig.12.
-
V. Conclusion
Pre-processing stage is an application dependent technique for enhancing the content of medical image based on removal of special markings and speckle noise. Removal of special markings and speckle noise existing in medical images will increase the quality of image segmentation. On the other hand, it will improve the accuracy and efficiency of content based medical image classification and retrieval systems. In this paper, special markings are extracted using Sobel edge detection technique. The extracted markings are then removed from the texture content of an image by substituting their pixel intensity value with the corresponding pixel intensity values of their neighborhood. Then, 2D adaptive noise removal, median and 2D order-statistic filtering are applied to remove strong speckle noise and enhance the weak boundaries of medical images. Finally, we segment various anatomical structures of different medical image modalities. The range filtering is exploited for segmentation based on texture. Results show the efficiency, simplicity and robustness of our proposed approach.
Acknowledgment
The authors would like to thank TM Lehman, Department of medical informatics, RWTH Achen, Germany, for making the database available for the experiments .
Список литературы Segmentation of Pre-processed Medical Images: An Approach Based on Range Filter
- Sinha U, Bui A, Taira R . A Review of Medical Imaging Informatics. Annals of the New York Academy of Sciences, 2002, 980(1):168-197.
- Zhu J, Yang X, Du X, Song L. Pre-processing for MRI. International Journal of Computer Engineering, 2001, 27(2):25–26.
- Muller H, Michoux N, Bandon D, Geissbuhler A. A Review of Content-based Medical Image Retrieval Systems in Medical Application - Clinical Benefits and Future Directions. International Journal of Medical Informatics, 2004, 73(1):1-23.
- Smeulders A, Worring M, Santini S, Gupta A, Jain R. Content-Based Image Retrieval at the End of the Early Years. IEEE Transaction on Pattern Analysis and Machine Intelligence, 2000, 22(12):1380-1394.
- Peng F, Yuan K, Feng S, Chen W. Pre-processing of CT Brain Images for Content-Based Image Retrieval. In: Proceedings of International Conference on BioMedical Engineering and Informatics, 2008, 208-212.
- Červinka T, Provazník I. Pre-processing for Segmentation of Computer Tomography Images. In: Proceedings of RADIOELEKTRONIKA,2005, 167-170.
- Hussein ZR, Rahmat RW, Nurliyana L, Saripan MI, Dimon MZ. Pre-processing Importance for Extracting Contours from Noisy Echocardiographic Images. International Journal of Computer Science and Network Security (IJCSNS),2009, 9 (3): 134-137.
- Suzuki H, Toriwaki J . Automatic Segmentation of Head MRI Images by Knowledge Guided Thresholding. Journal of Computer Medical Imaging Graph, 1991, 15(4):233-240 .
- Robb R A. Biomedical Imaging, Visualization and Analysis. edited by Wiley-Liss, USA, 2000 .
- Williams DJ, Shah M. A Fast Algorithm for Active Contours and Curvature Estimation. CVGIP: Image Understanding, 1991, 55:14-26.
- Hall Lo, Bensaid AM, Clarke LP, Velthuizen RP, Silbeger MS, Bezdek J. A Comparison of Neural Network and Fuzzy Clustering Techniques in Segmenting Magnetic Resonance Images of the Brain. IEEE Transaction on Neural Networks, 1992, 26(4): 479-486.
- Cohen LD, Cohen I. Finite-element Methods for Active Contour models and Balloons for 2D and 3D Images. IEEE Transaction on Image Processing on Pattern Analysis & Machine Intelligence, 1993, 25:1131-1147.
- Adalsteinsson D, Sethian JA. A Fast Level Set Method for Propagating Interfaces. Journal of Computing Physics, 1995, 118:269-277.
- Li N, Liu M, Li Y . Image Segmentation Algorithm using Watershed Transform and Level Set Method. IEEE Transaction International Conference on Acoustics, Speech and Signal Processing,2007,1: 613-616.
- Felzenszwalb PF, Daniel P. Efficient Graph-Based Image Segmentation. International Journal of Computer Vision, 2004, 59 (2):167-181.
- Lu Y, Quan Y, Zhang Z, Wang G . MST Segmentation for Content-Based Medical Image Retrieval. In: Proceedings of International Conference on Computational Intelligent and Software Engineering, 2009, 1-4.
- Cheng J, Xue R, Lu W, Jia R. Segmentation of Medical Images with Canny Operator and GVF Snake Model. In: Proceedings of 7th World Congress on Intelligent Control and Automation, 2008, 1777-1780.
- Chuang C, Lie W. A Downstream Algorithm Based on Extended Gradient Vector Fellow Field for Object Segmentation. , IEEE Transaction on Image Processing, 2004, 13:1379-1392.
- Wu J, Poehlman S, Nosewrthy M, Kamath MV . Texture Feature based Automated Seeded Region Growing in Abdominal MRI Segmentation. In; Proceedings of International Conference on Biomedical Engineering and Informatics, 2008,263-267.
- Chang-ming Z, Guo-chang G, Hai-bo L, Jing S, Hualong Y . Segmentation of Ultrasound Image Based on Texture Feature and Graph Cut. In: Proceedings of International Conference on Computer Science and Software Engineering, 2008, 795-798.


