Спиральные параметры регулярных-спиралей в белках (часть 2)
Автор: Батхишиг Д., Муиддорж Б., Энхбаяр П.
Журнал: Вестник Бурятского государственного университета. Химия. Физика @vestnik-bsu-chemistry-physics
Рубрика: Химия
Статья в выпуске: 4, 2016 года.
Бесплатный доступ
Α-Cпираль, 310-спираль, π-спираль и -спираль наблюдались в белковых структурах. Они составляют 32% от остатков, 4%, 0,3% и 0,2%, соответственно. Однако эти проценты зависят от разрешения решаемых структур и способу присвоения вторичных структур. Возможно 2016, из отобранного набора в данных банк белков (PDB), содержащих 2901 белковые цепи с менее чем 25% идентичности последовательности и 1.6Å разрешающей способности (R-значения 0.25), использовать в этом анализе. Вторичные задания структуры выполняются DSSP, STRIDE и SECSTR для π-спиралей. Спиральные параметры шага, остатки на оборот, радиусы, хиральности и р = RMSD/(N-1)1/2 для p-спиралей определяются программой HELFIT. р-Значения, оценивающие спиральную регулярность и все π -спиралей с р 0.10Å, были идентифицированы как регулярные. Спиральные параметры белка p-спиралей сравнивались с данными канонических p-спиралей и других типов белковых спиралей.
Α-спиралей, π-спираль, спиральные параметры, регулярные спирали, белковые структуры
Короткий адрес: https://sciup.org/148317763
IDR: 148317763 | УДК: 579.519.6 | DOI: 10.18101/2306-2363-2016-4-17-25
Текст научной статьи Спиральные параметры регулярных-спиралей в белках (часть 2)
Helix is one of two main types of secondary structures in proteins. Helices are usually designated as in based on the number of residues per turn (i) and the number of atoms in the ring joined by the backbone hydrogen bond (n) [1]. Pauling and Corey first hypothesized the α-helix (3.613) and the γ-helix (5.l17) structures [2]. Donohue later considered the possibility of other types of helices (2.2, 310, 4.314 and 4.416) [3]. Low and Baybutt also suggested the possibility of the 4.416-helix or π-helix [3]. The main stabilizing factor for helical structures in polypeptides is re- peated hydrogen bonds between main chain carbonyl oxygen (C=O) and amide hydrogen (NH) groups with the α-helix characterized by an (i ← i+4) pattern, the 310 and the π-helix by repealing (i ← i+3) and (i ← i+5) hydrogen bonds, respectively [4].
There are several programs perform assignments of secondary structures based on three-dimensional (3D) atomic coordinates of proteins [4-6]. Among these, DSSP [4] and STRIDE [5] are the most widely used [7]. DSSP identifies helices based on the repeating ( i←i+n ) hydrogen bonds with corresponding to n of 3, 4 and 5 for 3 10 , α- and π-helices, respectively [4, 8]. STRIDE uses both hydrogen bonds and main chain dihedral angles to define secondary structures [5]. DSSP program identified only 9 unique π-helices from the database of more than 6000 of proteins [9]. Fodje and Karadaghi defined 116 π-helices using their home made program, SECSTR, from the database of 932 high resolution 3D structures of proteins [7].
These different results can be explained by the following two reasons: 1) Number of solved 3D structures was insufficient by this time 2) Programs to assign of secondary structures use different methods.
We studied helical parameters of protein helices with HELFIT program and compared with the parameters of canonical π -helices.
Materials and Methods
Composition of database
The 16 May 2016 culled PDB data set, containing 2969 protein chains with less than 20% sequence identity and resolution ≤ 1.6 Å ( R -value ≤ 0.25), was used in this analysis.
DSSP program
DSSP performs secondary structure assignments by the bonding energy Е ≤-0.5 kcal/mоl between C=O of residue i and N–H residue n ( i ← i+n ). The optimal hydrogen bonding energy for mainchain-mainchain N—H···O hydrogen bonds Е m < -3 kcal/mol. Hydrogen bond energy depends on both electrostatic interaction N—H···O of atoms and of hydrogen bonds angle θ [4].
STRIDE program
STRIDE program is designed for protein secondary structure assignment from 3D atomic coordinates based on the combined use of hydrogen bond energy and statistically derived backbone torsional angle information [7]. The hydrogen bond energy E hb is calculated using the empirical energy function derived from the analysis of experimental data on hydrogen bond geometries in crystal structures of amino acids in polypeptide chains [10].
SECSTR program
SECSTR is a new addition to the DSSP program that is dedicated to identifying π-helices, which were seldom assigned by older versions of DSSP and STRIDE [7]. The secondary structure assignment methods based on hydrogen bond assignments (DSSP, STRIDE, and SECSTR) produced nearly identical assignments, with more than to 90% [6].
HELFIT program
HELFIT enables to calculate simultaneously all five of the helix parameters with high accuracy. The minimum number of data points required for the analysis is only four. HELFIT also calculates a parameter, p = RMSD / ( N -1)1/2, which estimates the regularity of helical structures independent of the number of data points, where RMSD is the root mean square distance from the best-fit helix to data points and N is the number of data points [11].
Results and Discussion
We identified 27, 22 and 340 π-helices from 2901 high resolution protein structures by DSSP, STRIDE and SECSTR programs, respectively. All π-helices are divided into two groups, regular and irregular, with p -value: p ≤ 0.10 Å regular and p > 0.10 Å irregular. 7 of 27, 5 of 22, and 76 of 340 helices are grouped as regular by the HELFIT program. In order to compare protein π-helices with the canonical π-helices the only parameters of regular π-helices are used for the further analysis (Table 1).
Table 1
Helical parameters of 86 regular π-helices in proteins identified by DSSP, STRIDE and SECSTR program
|
PDB _ID |
Chain_Po sition |
P (Å) |
n |
Δ z (Å) b |
r (Å) |
V c (Å3) a |
p (Å) |
Identified Program |
|
1DJ0 |
A_81-87 |
5.01 |
4.18 |
1.20 |
2.58 |
25.06 |
0.10 |
SECSTR |
|
1DK8 |
A_242-249 |
5.12 |
4.36 |
1.17 |
2.69 |
26.70 |
0.10 |
SECSTR |
|
1ELK |
A_95-101 |
5.24 |
4.42 |
1.19 |
2.70 |
27.15 |
0.10 |
SECSTR |
|
1JET |
A_301-308 |
4.82 |
4.44 |
1.09 |
2.80 |
26.74 |
0.09 |
SECSTR |
|
1KJQ |
A_119-125 |
5.10 |
4.30 |
1.19 |
2.67 |
26.56 |
0.09 |
SECSTR |
|
1KK O |
A_199-205 |
4.99 |
4.53 |
1.10 |
2.81 |
27.33 |
0.09 |
DSSP, STRIDE, SECSTR |
|
1NU Y |
A_1276-1282 |
5.30 |
4.64 |
1.14 |
2.87 |
29.56 |
0.10 |
SECSTR |
|
1RK6 |
A_386-393 |
5.30 |
4.47 |
1.19 |
2.80 |
29.20 |
0.06 |
DSSP |
|
1RK6 |
A_387-393 |
5.14 |
4.41 |
1.17 |
2.74 |
27.49 |
0.04 |
STRIDE |
|
1RK6 |
A_384-393 |
5.22 |
4.37 |
1.19 |
2.71 |
27.56 |
0.06 |
SECSTR |
|
1W5 R |
A_58-64 |
5.17 |
4.37 |
1.18 |
2.73 |
27.70 |
0.08 |
SECSTR |
|
1XG0 |
A_105-111 |
5.32 |
4.55 |
1.17 |
2.84 |
29.63 |
0.10 |
SECSTR |
|
1XG K |
A_266-272 |
5.02 |
4.31 |
1.16 |
2.70 |
26.67 |
0.07 |
SECSTR |
|
2BF D |
A_109-115± |
5.20 |
4.50 |
1.16 |
2.80 |
28.46 |
0.10 |
SECSTR |
|
2CI1 |
A_51-57 |
5.12 |
4.33 |
1.18 |
2.68 |
26.68 |
0.09 |
SECSTR |
|
2DPL |
A_68-74 |
5.17 |
4.42 |
1.17 |
2.77 |
28.20 |
0.03 |
SECSTR |
|
2GZS |
A_163-169 |
5.31 |
4.53 |
1.17 |
2.83 |
29.49 |
0.09 |
SECSTR |
|
PDB _ID |
Chain Po Δ z V Identified Pro P (Å) n r (Å) c p (Å) sition (Å) b (Å3) a gram |
|
2H1V |
2A7_4264- 5.15 4.21 1.22 2.62 26.38 0.09 SECSTR |
|
2JIS 2O0A |
A_28-35 5.15 4.42 1.17 2.75 27.68 0.07 SECSTR A 474- _ SFCSTR 4A3_0424- 5.13 4.48 1.15 2.79 28.00 0.08 SECSTR |
|
2P51 |
2A1_3207- 5.15 4.32 1.19 2.71 27.51 0.08 SECSTR |
|
2P6 W 2PB D 2POF 2PY Q 2PY X 2PY X 2PY X 2RB K 2VL A 2WQ F 2XR Y 2Y53 3A0Y |
1A6_0154- 5.18 4.38 1.18 2.73 27.69 0.08 SECSTR A_88-94 5.24 4.42 1.19 2.77 28.58 0.09 SECSTR A_37-43 5.29 4.52 1.17 2.81 29.03 0.10 SECSTR B_61-67 5.17 4.39 1.18 2.76 28.18 0.02 DSSP A 737- DSSP 2A3_9232- 5.09 4.42 1.15 2.75 27.36 0.06 DSSP 2A3_8232- 5.06 4.42 1.14 2.78 27.80 0.05 STRIDE 2A3_9232- 5.08 4.42 1.15 2.75 27.31 0.06 SECSTR 1A2_9122- 5.25 4.49 1.17 2.77 28.19 0.09 SECSTR A_68-77 5.27 4.00 1.32 2.79 32.22 0.09 SECSTR A_59-65 5.36 4.55 1.18 2.81 29.22 0.10 SECSTR 3A0_6300- 5.20 4.34 1.20 2.70 27.44 0.08 SECSTR A_48-54 5.11 4.23 1.21 2.62 26.05 0.10 SECSTR 7A2_9723- 5.11 4.32 1.18 2.70 27.09 0.06 SECSTR |
|
3BH Q 3H9C |
1A3_4128- 5.07 4.40 1.15 2.77 27.78 0.08 SECSTR 3A9_1382- 5.28 4.44 1.19 2.75 28.25 0.09 SECSTR |
|
3IT3 3OAJ 3OCJ |
A_56-63 5.16 4.45 1.16 2.80 28.56 0.04 SECSTR A_24-30 4.97 4.29 1.16 2.72 26.93 0.07 SECSTR 2A5_9253- 5.13 4.62 1.11 2.88 28.93 0.10 SECSTR |
|
3OY V 3PB6 3PJP |
2A3_3227- 5.49 4.41 1.24 2.74 29.36 0.08 SECSTR X_93-99 5.25 4.54 1.16 2.83 29.10 0.06 SECSTR А 1334- _ SFCSTR 1A3_410334- 5.18 4.44 1.17 2.77 28.12 0.10 SECSTR |
|
3Q28 |
2A8_6280- 5.30 4.50 1.18 2.81 29.22 0.08 SECSTR |
|
3RRI 3S5 M 3T4L |
A_22-28 5.17 4.43 1.17 2.77 28.13 0.06 SECSTR 6A9_8692- 5.24 4.45 1.18 2.76 28.18 0.09 SECSTR 1A7_4168- 5.29 4.45 1.19 2.77 28.66 0.05 SECSTR |
|
3VE N 3WA 2 |
A 437- _ SFCSTR 4A4_3437- 5.14 4.41 1.17 2.75 27.69 0.10 SECSTR X 797- _ DSSP 3X0_3297- 5.04 4.25 1.19 2.68 26.76 0.10 DSSP |
|
PDB _ID |
Chain_Po sition |
P (Å) |
n |
Δ z (Å) b |
r (Å) |
V c (Å3) a |
p (Å) |
Identified Program |
|
3ZB O |
A_94-100 |
5.22 |
4.50 |
1.16 |
2.82 |
28.98 |
0.08 |
SECSTR |
|
4AY O |
A_122-128 |
5.18 |
4.28 |
1.21 |
2.65 |
26.70 |
0.10 |
SECSTR |
|
4B1Y |
B_88-94 |
5.22 |
4.42 |
1.18 |
2.76 |
28.26 |
0.09 |
SECSTR |
|
4BR C |
A_359-365 |
5.06 |
4.35 |
1.16 |
2.76 |
27.84 |
0.05 |
SECSTR |
|
4CB U |
A_89-95 |
5.22 |
5.07 |
1.03 |
2.72 |
23.93 |
0.06 |
SECSTR |
|
4CD5 |
A_248-254 |
5.11 |
4.38 |
1.17 |
2.75 |
27.72 |
0.07 |
SECSTR |
|
4CD5 |
A_350-356 |
5.25 |
4.55 |
1.15 |
2.81 |
28.62 |
0.09 |
SECSTR |
|
4DJA |
A_305-311 |
4.99 |
4.32 |
1.16 |
2.73 |
27.05 |
0.09 |
SECSTR |
|
4DJA |
A_405-412 |
5.10 |
4.39 |
1.16 |
2.75 |
27.60 |
0.10 |
SECSTR |
|
4ES M |
A_137-143 |
5.36 |
4.14 |
1.29 |
2.59 |
27.28 |
0.07 |
SECSTR |
|
4EZI |
A_128-135 |
5.02 |
4.35 |
1.15 |
2.72 |
26.82 |
0.09 |
SECSTR |
|
4GV F |
A_231-239 |
5.19 |
4.46 |
1.16 |
2.76 |
27.85 |
0.06 |
DSSP |
|
4GV F |
A_232-238 |
5.14 |
4.46 |
1.15 |
2.80 |
28.39 |
0.07 |
STRIDE |
|
4GV F |
A_229-239 |
5.16 |
4.44 |
1.16 |
2.77 |
28.01 |
0.05 |
SECSTR |
|
4I3G |
A_257-264 |
5.13 |
4.36 |
1.18 |
2.73 |
27.55 |
0.08 |
DSSP |
|
4I3G |
A_257-263 |
5.12 |
4.40 |
1.16 |
2.74 |
27.45 |
0.09 |
STRIDE |
|
4I3G |
A_253-264 |
5.21 |
4.38 |
1.19 |
2.71 |
27.44 |
0.09 |
SECSTR |
|
4JA8 |
A_66-72 |
5.29 |
4.53 |
1.17 |
2.85 |
29.80 |
0.10 |
SECSTR |
|
4LRT |
A_267-273 |
5.28 |
4.46 |
1.18 |
2.78 |
28.74 |
0.09 |
SECSTR |
|
4ME 2 |
A_192-198 |
4.86 |
4.47 |
1.09 |
2.81 |
26.97 |
0.09 |
SECSTR |
|
4QB3 |
A_66-72 |
5.08 |
4.53 |
1.12 |
2.84 |
28.42 |
0.09 |
SECSTR |
|
4R75 |
A_311-318 |
5.11 |
4.38 |
1.17 |
2.74 |
27.52 |
0.09 |
SECSTR |
|
4U9H |
L_127-133 |
5.07 |
4.54 |
1.12 |
2.85 |
28.50 |
0.07 |
SECSTR |
|
4W7 L |
A_373-379 |
5.23 |
4.53 |
1.15 |
2.81 |
28.64 |
0.08 |
SECSTR |
|
4WRI |
A_65-71 |
5.18 |
4.38 |
1.18 |
2.73 |
27.69 |
0.06 |
SECSTR |
|
4XE M |
A_120-126 |
5.15 |
4.33 |
1.19 |
2.69 |
27.04 |
0.08 |
SECSTR |
|
4XFJ |
A_68-74 |
5.30 |
4.45 |
1.19 |
2.74 |
28.09 |
0.10 |
SECSTR |
|
4XQ7 |
A_217-223 |
5.17 |
4.49 |
1.15 |
2.79 |
28.16 |
0.09 |
SECSTR |
|
4Z5S |
A_108-115 |
5.22 |
4.40 |
1.19 |
2.73 |
27.78 |
0.09 |
SECSTR |
|
4ZG W |
A_115-121 |
5.29 |
4.53 |
1.17 |
2.80 |
28.76 |
0.10 |
SECSTR |
|
PDB _ID |
Chain_Po sition |
P (Å) |
n |
Δ z (Å) b |
r (Å) |
V c (Å3) a |
p (Å) |
Identified Program |
|
5A0Y |
A_314-324 |
5.09 |
4.46 |
1.14 |
2.77 |
27.51 |
0.10 |
SECSTR |
|
5AZ B |
A_203-210 |
5.15 |
4.41 |
1.17 |
2.74 |
27.54 |
0.09 |
SECSTR |
|
5BSR |
A_240-247 |
5.12 |
4.33 |
1.18 |
2.68 |
26.68 |
0.10 |
SECSTR |
|
5DA W |
A_89-95 |
5.32 |
4.37 |
1.22 |
2.72 |
28.30 |
0.09 |
SECSTR |
|
5DP2 |
A_143-149 |
5.18 |
4.44 |
1.17 |
2.78 |
28.33 |
0.06 |
SECSTR |
|
5E8X |
A_442-448 |
5.09 |
4.35 |
1.17 |
2.74 |
27.60 |
0.08 |
SECSTR |
|
5EJ8 |
A_485-491 |
5.22 |
4.55 |
1.15 |
2.81 |
28.46 |
0.10 |
SECSTR |
|
5HZ7 |
A_280-286 |
5.25 |
4.49 |
1.17 |
2.80 |
28.80 |
0.08 |
SECSTR |
|
Average |
5.17±0 |
4.42±0 |
1.17±0 |
2.75±0 |
27.89± |
0.08±0 |
||
|
.11 |
.13 |
.04 |
.06 |
1.09 |
.02 |
|||
|
Canonical π-helix |
5.16 |
4.40 |
1.15 |
2.68 |
25.9 |
|||
|
a Voronoi volume ( V c = π· |
r 2·Δz); |
b Helix rise per residue Δ z |
=P / n ; |
|||||
Total of 88 regular π-helices are 7, 5 and 76 identified by DSSP, STRIDE and SECSTR program respectively. The π-helix is identified at position 199-205 of A chain in 1KKO protein by the three programs [12-18].
Helix radius and Voronoi volume of real π-helices are larger than that of canonical π-helix. The other helix parameters are close to the parameters of canonical π-helix. Average length is 7.47 residues and length is in range of 7-12 residues (Table 2).
Table 2
Average of helical parameters for regular π-helices in proteins and standard deviations
|
Average |
‹ P › (Å) |
‹ n › |
‹Δ z ›(Å) |
‹ r › (Å) |
‹ V c ›(Å3) |
‹ p › (Å) |
|
π-helices |
5.13±0. |
4.41±0. |
1.16±0. |
2.76±0. |
27.75±0. |
0.07±0. |
|
(DSSP) |
10 |
09 |
03 |
04 |
78 |
03 |
|
π-helices |
5.09±0. |
4.44±0. |
1.15±0. |
2.77±0. |
27.69±0. |
0.07±0. |
|
(STRIDE) |
06 |
05 |
02 |
03 |
38 |
02 |
|
π-helices |
5.17±0. |
4.42±0. |
1.17±0. |
2.75±0. |
27.90±1. |
0.08±0. |
|
(SECSTR) |
11 |
13 |
04 |
06 |
13 |
02 |
Standard deviations of helical parameters for π-helices identified by SECSTR program are larger than DSSP and STRIDE programs. Also, average values of the helix radius r and number of residue per turn n are approximate to each for the three programs.
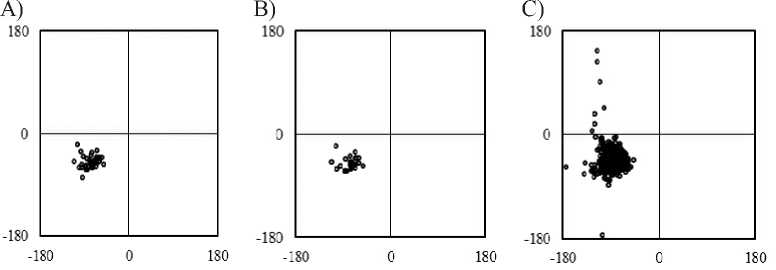
Fig. The Ramachandran-map of regular π-helices in proteins. The φ, ψ angles are indicated in panels which regular π-helices identified by A) DSSP, B) STRIDE and C) SECSTR, respectively. The abscissa is φ; the ordinate axis is ψ. The φ, ψ of residues at N c and C c are not shown.
Average dihedral angles of regular π-helices were determined at each for DSSP (-77°±14°, -50°±11°), STRIDE (-77°±15°, -51°±12°) and SECSTR (-81°±18°, -44°±21°) programs. The average values of backbone dihedral angles (φ, ψ) of all regular π-helices observed were found to be (φ, ψ) = (-81°, -45°) with standard deviations (σ φ , σ ψ ) = (17°, 20°). The average of dihedral angle is larger than canonical π-helix (-57°, -70°). The φ, ψ angles of regular π-helices are located on an allowed regions for other residues except for glycine, were removed from the calculation (Fig.).
Conclusion
-
• 2901 3D structures of high resolution protein structures were downloaded
from Protein Data Bank (PDB) and there are 389 π-helices. In average, every protein contains 0.13 π-helices.
-
• All π-helices are divided into two groups, regular and irregular. 89 π-helices are regular among the total of 389 π-helices, 4.37%. Helix parameters of all regular π-helices are used for further analysis.
-
• Radii of all π-helices and Voronoi volume are larger than that of canonical π-helices and all the helical parameters are comparable with those of canonical helices.
Список литературы Спиральные параметры регулярных-спиралей в белках (часть 2)
- Donohue J. Hydrogen Bonded Helical Configurations of the Polypeptide Chain // Proc. Natl. Acad. Sci. USA. - 1953. - V. 39, № 6. - P. 470-478.
- Pauling L., Corey R. B., Branson H. R. The structure of proteins; two hydrogen- bonded helical configurations of the polypeptide chain // Proc. Natl. Acad. Sci. USA. - 1951. - V. 37, № 4. - P. 205-211.
- Low B. W., Baybutt R. B. The π-helix a hydrogen bonded configuration of the polypeptide chain // J. of the American Chemical Society. - 1952. - V. 74(22). - P. 5806-5807.
- ВЕСТНИК БУРЯТСКОГО ГОСУДАРСТВЕННОГО УНИВЕРСИТЕТА ХИМИЯ. ФИЗИКА Вып. 4. 2016
- Kabsch W., Sander C. How good are predictions of protein secondary structure? // FEBS Lett. - 1983. - 155(2). - P. 179-82.


