Топологии микрофлюидных устройств для изучения миграции клеток в градиентах химических веществ (обзор)
Автор: Кухтевич Игорь Владимирович, Белоусов К.И., Букатин А.С., Евстрапов А.А.
Журнал: Научное приборостроение @nauchnoe-priborostroenie
Рубрика: Приборостроение для биофизики и биохимиии
Статья в выпуске: 1 т.25, 2015 года.
Бесплатный доступ
Направленная клеточная миграция играет важную роль в физиологических процессах, например таких как защита организма от инфекций и вирусов, заживление ран, метастазирования рака и др. Клеточная миграция зависит в том числе и от воздействия на клетки градиентов концентраций химических веществ. Не так давно микрофлюидные устройства начали применяться для изучения миграции клеток. Подобные устройства позволяют прецизионно конфигурировать и управлять градиентами химических веществ, открывая новые возможности при изучении сложных механизмов взаимодействия клеток как внутри популяции, так и с окружающей средой. Обзор посвящен достижениям, связанным с разработкой микрофлюидных устройств для изучения влияния градиентов химических веществ на клеточную миграцию, классификации данных устройств, а также сравнению их с "традиционными" подходами, применяемыми в клеточной биологии
Микрофлюидное устройство, клеточная миграция, градиент химического вещества
Короткий адрес: https://sciup.org/14264963
IDR: 14264963 | УДК: 57.086.8+576.08+576.5
Текст обзорной статьи Топологии микрофлюидных устройств для изучения миграции клеток в градиентах химических веществ (обзор)
Клеточная миграция является высокоупорядоченным процессом, происходящим при различных взаимодействиях, которые могут иметь место между клетками, тканью и окружающей их средой [1, 2]. Подобные взаимодействия играют важную роль в различных биологических процессах включая иммунный ответ на воспалительные процессы и заживление ран [3–6]. Биологические механизмы направленной клеточной миграции являются сложными и зависят от режима движения клеток и их типа [1, 7]. Например, бактериальные клетки плывут сквозь жидкую среду в поисках питательных веществ при помощи вращения тонкого жгутика без необходимости прикрепления к подложке, а их направление и подвижность напрямую зависят от локальных условий [8]. С другой стороны, многие типы клеток млекопитающих мигрируют сквозь твердую внеклеточную матрицу при помощи циклических процессов сложной передачи сигналов, реорганизации цитоскелета и взаимодействия клеток с данной матрицей [9].
Градиенты концентрации химических веществ относятся к факторам окружающей среды, влияющим на миграцию различных типов клеток. Данный фактор делят на две условные группы: хемотаксис — направленная миграция клеток при воздействии градиентов, созданных при растворении веществ (хемоаттрактанта); гаптотаксис — направленная клеточная миграции при создании градиентов веществ на поверхности (хемоаттрактант связан с твердой фазой). Например, белые кровяные тельца, такие как нейтрофилы и лимфоциты, следуют за градиентами хемоаттрактантов управляемых / вырабатываемых тканью для выполнения ими иммунных функций [10]. Неправильно направленные белые кровяные тельца могут быть причиной различных заболеваний и расстройств, например хронической обструктивной болезни легких, воспаления кишечника и рассеянного склероза [10, 11]. Другим примером является миграция раковых клеток в направлении градиентов выделяемых тканями хемоаттрактантов (эпидермальный фактор роста и т. д.), приводящая к образованию метастазов [12–14]. Примером также может служить миграция эпителиальных клеток при заживлении ран и миграции клеток нервного гребня при развитии эмбриона [4, 15].
Из-за высокой биологической и физиологической важности хемотаксиса понимание его механизмов является одной из главных задач клеточных исследований. На текущей момент времени следующие "традиционные" подходы получили широкое применение при проведении исследований, связанных с изучением клеточной миграции: использование проницаемых мембран, или т. н. камеры Бойдена; изучение миграции с использование агарозы; раскапывание хемоаттракатанта на капли сверхмалого объема; применение камер Зигмонда и камер Дунна, которые позволяют создавать градиенты концентрации химических ве- ществ за счет свободной диффузии [16–20]. Данные подходы, как правило, не позволяют поддерживать и контролировать создание химического градиента, а некоторые из них также не позволяют проводить в реальном времени количественную оценку клеточной миграции методами оптической микроскопии на уровне отдельных клеток.
В последнее время микрофлюидные устройства, которые изготавливаются с применением технологий микроэлектронной промышленности и состоят из микроканалов, резервуаров, смесителей и других функциональных микроэлементов, стали применяться при изучении хемотаксиса. В данных устройствах, как правило, изучается хемотаксис клеток размерами от долей микрометра (бактериальные клетки) до сотен микрон [21, 22]. Ключевыми преимуществами микрофлюидных устройств являются: возможность конфигурирования и стабилизации градиентов концентрации химических веществ; возможность миниатюризации; низкий расход реагентов; возможность постановки высокопроизводительных анализов [23]. В частности, микрофлюидные устройства позволяют проводить количественную оценку клеточной миграции при воздействии динамических градиентов хемоаттроктанатов, что является наиболее приближенным к естественным условиям [23]. Диапазон создаваемых в микрофлюидных устройствах концентраций хемоаттрактантов может варьироваться от десятых долей нанограмма до сотен нанограмм на миллилитр, или от единиц наномолей до десятков миллимолей [22, 24–26].
Таким образом, данный обзор посвящен последним достижениям, связанным с разработкой микрофлюидных устройств для прецизионного изучения влияния градиента химических веществ на клеточную миграцию (хемотаксиса), а также сравнению их с "традиционными" подходами, применяемыми в клеточной биологии.
КЛАССИФИКАЦИЯ МИКРОФЛЮИДНЫХ УСТРОЙСТВ ПО СПОСОБУ
СОЗДАНИЯ ГРАДИЕНТА
Как уже отмечалось выше, "традиционные" подходы к проведению анализа на хемотаксис позволяют создавать плохо контролируемые градиенты химических веществ на основе неконтролируемой диффузии и часто требуют большого количества реагентов и исходного образца (пробы), содержащего клеточную культуру, в то время как микрофлюидика предлагает новые устройства для количественного изучения клеточной миграции с лучшим контролем градиентов химических веществ и меньшим расходом реагентов и проб [23, 27]. Микрофлюидные устройства для формирования градиента химических веществ можно разделить на две большие группы: "проточные" (flow-based — в англоязычной литературе) и "диффузионные" (diffusion-based / flow-free — в англоязычной литературе) [21, 22].
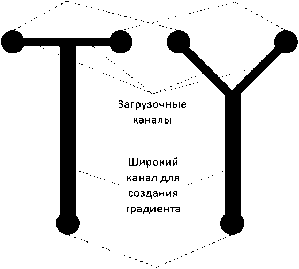
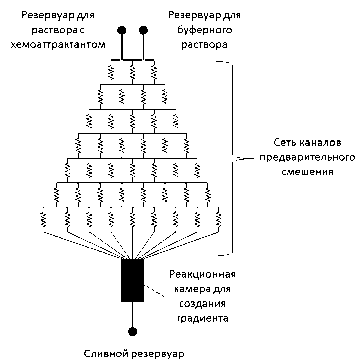
"Проточные" микрофлюидиные устройства позволяют создавать стабильные и прецизионные градиенты концентраций в широком диапазоне путем смешивания в каналах или специальных камерах ламинарных потоков растворов, содержащих разные химические вещества или одинаковые вещества, но с разной концентрацией (см. рисунок, а, б) [23, 27–30].
Резервуар для раствора с Резервуар для буферного хемоаттрактантом раствора
Сливной резервуар
а б в
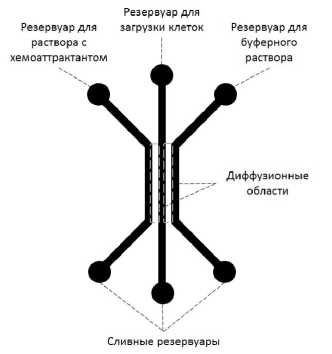
Топологии микрофлюидных устройств: а — "T" и "Y"; б — "сетевая"; в — "диффузионная"
Данные устройства позволяют осуществлять манипуляции потоками, имитируя динамические изменения градиентов хемоаттрактантов в естественных условиях. Примером таких условий могут служить периодические градиенты хемоаттрактанта, наблюдаемые при направленной миграции лейкоцитов в тканях. "Проточные" устройства широко используются для изучения хемотаксиса бактерий и клеток различных типов, таких как нейтрофилы, раковые клетки, лимфоциты и дендритные клетки [28, 31–35]. Миграция клеток также изучается и в более сложных, физиологически релевантных градиентах распределения веществ, например таких как градиенты с различной зависимостью (линейные и нелинейные градиенты, которые соответствуют стадиям процесса диффузии молекул хемоаттрактанта), сосуществующие и конкурирующие градиенты, быстро меняющиеся во времени градиенты [28, 30–31, 34]. Проведение подобных исследований представляется невозможным с применением "традиционных" подходов.
В "диффузионных" микрофлюидных устройствах используется явление диффузии молекул веществ через мембраны или гидрогели для создания диффузионных градиентов (см. рисунок, в). Например, время диффузии, требуемое флуоресцентному красителю для диффундирования поперек канала шириной 0.8 мм, составляет примерно 20 мин при коэффициенте диффузии D = 10–6 см2/с [36]. В сравнении с "проточными" устройствами "диффузионные" устройства имеют ряд очевидных преимуществ: минимизация эффектов, связанных с движением потоков, например напряжения сдвига, которое влияет на клеточную миграцию; сохранение выделяемых в ходе диффузии молекул; возможно создание 3D (объемных) градиентов; улучшенная портативность устройств за счет уменьшения требований к внешним элементам управления; возможность изучения межклеточных взаимодействий в строго определенных статичных градиентах веществ [37–40]. В частности, такое достоинство "диффузионных" устройств как портативность позволяет проводить измерения с использованием оборудования уже имеющегося в исследовательских лабораториях. С другой стороны, способность управлять градиентами в "диффузионных" устройствах хуже в сравнении с "проточными". Микрофлюидные устройства, построенные на использовании диффузионных градиентов, успешно применялись для исследования миграции и хемотаксиса клеток различных типов, например таких как иммунные и раковые клетки в 2D- и 3D-средах, имитирующих межклеточную матрицу в тканях [37–41]. Кроме того, было предложено микрофлюидное устройство, состоящее из "бифурцирующих" градиентных кана- лов — множества различных путей к источнику хемоаттрактантов, которое использовалось для тестирования нейтрофилов на направленное принятие хемочувстительных "решений" [42]. Результаты, полученные при помощи микрофлюидного устройства, показали, что клетки предпочтительней мигрируют в сторону увеличения концентрации хемоаттрактанта через наиболее короткие каналы, т. е. нейтрофилы способны делать "умные" и энергетически выгодные "решения" по выбору наиболее эффективных путей направленной миграции в "запутанных" градиентах среды.
СРАВНЕНИЕ "ТРАДИЦИОННЫХ" ПОДХОДОВ И РЕШЕНИЙ НА ОСНОВЕ МИКРОФЛЮИДНЫХ ТЕХНОЛОГИЙ
Сопоставление достоинств и ограничений "традиционных" подходов и решений на основе мик-рофлюидных технологий для изучения клеточной миграции под действием градиентов химических веществ приводится в таблице (см. Приложение).
Из анализа источников, приведенных в таблице, становится очевидным, что к характерным недостаткам устройств, построенных на "традиционных" подходах и предназначенных для изучения миграции клеток в градиентах химических веществ, следует отнести затрудненный контроль создаваемого градиента, большой расход реагентов необходимых для формирования градиента, необходимость в относительно большом объеме пробы с клетками. Напротив, микрофлюидные устройства лишены данных недостатков и предоставляют исследователям такие возможности, как визуализация миграции отдельных клеток, создание динамически меняющихся градиентов и возможность их наложения, а также создание градиентов требуемой 1D, 2D и 3D формы. Однако подобные возможности определяют и ряд ограничений, свойственных микрофлюидным устройствам. Так, "проточным" микрофлюидным устройствам свойственно наличие напряжения сдвига, которое возникает как следствие тока жидкости, и потребность во внешнем контроле (например, использование насосов). С другой стороны, "диффузионные" микрофлюидные устройства менее требовательны к осуществлению внешнего контроля, но это накладывает ограничение, связанное с худшей в сравнении с "проточными" устройствами возможностью контроля создаваемого градиента.
ТОПОЛОГИИ МИКРОФЛЮИДНЫХ УСТРОЙСТВ
Основной идей топологий "T" и "Y" (см. рисунок, а) является наличие двух загрузочных каналов, через которые подается раствор с хемоат- трактантом и без. Каналы расположены симметрично друг напротив друга и сопрягаются с широким каналом или реакционной камерой. В месте встречи двух потоков происходит смешение раствора с хемоаттрактантом и без (в том числе диффузионным путем), регулируя которое путем изменения скоростей потоков можно контролировать создаваемый градиент. Загрузочные каналы могут располагаться относительно широкого канала или реакционной камеры, предназначенной для высаживания клеток как под прямым углом — "T" топология, так и под другими углами, например — "Y" топология. Стоит отметить, что градиент концентрации вдоль длины широкого канала или реакционной камеры не постоянен и распределение клеток к концу канала является следствием суммарного воздействия всех имеющихся величин градиента, что может затруднять анализ процессов, лежащих в основе механизма хемотаксиса. Увеличение скоростей потоков частично решает указанную проблему. Данные топологии часто сравнивают с Т-сенсором за сходство с Т-образным микрофлюидным устройством, предназначенным для измерения химических концентраций и определения вязкостей [61, 62].
Одним из первых микрофлюидных устройств с "T" топологией являлось устройство для изучения подвижности клеток в градиенте химического вещества, предложенное в работе [47]. Его действие основано на объединении трех потоков внутри широкого канала. Из двух противоположных каналов подавался раствор с хемоаттрактантом и буферный раствор. За счет диффузии в процессе течения в широком канале между двумя веществами формировался градиент концентрации хемоаттрактанта. Из канала, расположенного перпендикулярно относительно оси симметрии широкого канала, подавалась суспензия бактерий в область, где градиент хемоаттрактанта достигал наибольших значений. По приоритетному смещению бактерий к концу канала, авторы судили о величине хемотаксиса.
Модификация описанного выше T-устройства была представлена в работе [48], в которой использовались только два загрузочных канала. Через один из каналов происходила подача раствора с хемоаттрактантом, а через другой буферного раствора. Бактерии растворялись в обоих растворах таким образом, что изначально их распределение в широком канале было примерно одинаковое относительно оси его симметрии. Распределение бактерий к концу канала измерялось по рассеянию ими света.
Другое микрофлюидное устройство с "Т" топологией можно встретить в работе [49]. Однако принципиальным отличием данного устройства является наличие в широком канале цилиндров диаметром 200 мкм, которые призваны имитировать почву с пористостью 40 %.
Пример микрофлюидного устройства с "Y" топологией приводится в работе [28]. Главным отличаем данного устройства от "T" конструкции является наклон обоих загрузочных каналов на 45°, а не на 90° относительно оси симметрии широкого канала. При помощи предложенного устройства авторы изучали хемотаксис T-клеток человека в ответ на один из конкурирующих градиентов хемокинов CCL19 и CXCL12.
Принцип работы "сетевой" топологии (рисунок, б) заключается в том, что раствор, содержащий хемоаттрактант, и буферный раствор подаются не напрямую в широкий канал или реакционную камеру, в которой формируется градиент, воздействующий на клетки, т. е. не так, как в случае "T" и "Y" топологий, а через многоуровневую сеть каналов предварительного смешения. Подобное решение имеет ряд ключевых преимуществ по сравнению с "T" и "Y" топологиями, т. к. позволяет создавать более стабильные градиенты химических веществ, а также создавать градиенты произвольных форм.
В работе [63] представлено микрофлюидное устройство с "сетевой" топологией, в которое добавлен загрузочный канал для того, чтобы ввод бактерий осуществлялся сразу в область линейного градиента. Данный градиент создается смесителем, представляющим собой сеть каналов, в которых происходит многостадийный процесс смешивания, позволяющий создавать профили концентрации произвольной формы, контролируя диффузионное смешение двух начальных потоков. Таким образом, устраняется возможное воздействие на бактерии ступенчатого профиля концентрации. Более того, начальная линейная форма градиента дольше сохраняется вдоль длины широкого канала, т. к. диффузионные процессы протекают медленнее вследствие полученного на выходе из смесителя градиента.
Авторами работы [64] было предложено "сетевое" микрофлюидное устройство для создания градиентов сложной формы. Данное устройство, управляемое при помощи приложения разности давления на специальные клапаны, контролируемые компьютером, содержит 16 загрузочных каналов и большую реакционную камеру для позиционирования клеточной культуры и наложения на нее градиентов. Возможно осуществлять ввод жидкости через один из 16 входов или через все, тем самым получая от одной до 64 комбинаций градиента, при этом устройство обладает субсекундным откликом по времени. Таким образом, возможно создание в реакционной камере гетерогенного ламинарного потока, градиента веществ или полностью гомогенной смеси. Для отчистки входных каналов имеется специальная система обходных каналов. Благодаря наличию боковых каналов возможно создание асимметричных или нелинейных потоков в реакционной камере, а замедление потока может достигаться путем подключения в цепь или исключения из нее сегментов с большим гидродинамическим сопротивлением. Использование всего 4 клапанов открывающих доступ потоку к каналам разной длинны, т. е. с разным гидродинамическим сопротивлением, позволяет получать до 16 различных уровней скорости потока в реакционной камере. Описываемое устройство объединило в себе большой набор функциональных возможностей, что позволило авторам создавать сложные шаблоны градиентов химических веществ и выполнять последовательную перфузию в реакционной камере.
Недавно в работе [53] было представлено гибридное полидимитилсилоксан-поликарбонатное "сетевое" микрофлюидное устройстве для изучения влияния комбинации химических и кислородных градиентов на клеточную культуру. Данное устройство состоит из двух полидимитилсилокса-новых слоев, в которых выполнены системы каналов, и тонкой разделительной мембраны из того же материала. В верхний слой также встраивается поликарбонатная пленка поверх системы кислородных каналов для генерации кислородного градиента в реакционной камере нижнего слоя, в которой размещаются клетки, во время создания химического градиента. Нижний слой включает упорядоченную систему змеевидных каналов для создания градиентов химических веществ и камеру для клеточной культуры. При помощи разработанного устройства были выполнены 48часовое исследование влияния хемокина на клетки и анализ клеточной миграции с использованием аденокарциномных эпителиальных клеток легких человека (А549) при воспроизведении различных комбинаций химических и кислородных градиентов. Результаты изучения влияния хемо-кина показали увеличение апоптоза клеток А549 из-за вызванной гипоксией цитотоксичности тира-пазамина. Анализ миграции клеток А549 продемонстрировал зависимое от концентрации кислорода поведение данных клеток, т. е. то, что градиент кислорода предположительно играет важную роль в управлении клеточной миграцией.
Существуют реализации "сетевой" топологии в которых реакционная камера для формирования градиента заменяется на массив микрокамер, или ячеек, в каждой из которых создается свой градиент или концентрация химического вещества. В работе [54] рассматривается микрофлюидное устройство, содержащее массив ячеек для культивирования клеток и наблюдения за клетками в те- чение длительного времени. Устройство было спроектировано для интегрирования стадий, которые имеют место при стандартном эксперименте, связанном с культивированием клеток. Основные функции включают повторяющиеся циклы роста, поддержания культуры в условиях, приближенных к естественным, введения реагентов и оптический анализ в реальном времени. Каждый отдельный элемент массива (ячейка) — это круглая камера с множеством узких перфузионных каналов, окружающих ее по периметру, и четырьмя каналами для доступа жидкости. Для подтверждения работоспособности предлагаемого устройства авторы работы выполняли на нем культивирование клеток HeLa с непрерывной перфузией среды при 37 °С. Время удвоения популяции наблюдалось на 1.4 ± 0.1 день. При этом пиковая плотность клеток составляла ~2.5×105 клеток/см2. Устройство также было опробовано при мониторинге локализации флюоресценции кальцеина AM от 1 мин до 10 дней после введения реагента.
Подход лежащий в основе работы микро-флюидных устройств "диффузионной" топологии (см. рисунок, в) заключается в использовании диффузии молекул химического вещества, размещенного в загрузочном канале или резервуаре, через мембрану или гидрогель в направлении широкого канала или реакционной камеры с клетками. Мембрана или гель позволяют молекулам вещества проходить сквозь них, однако при этом почти полностью блокируется адвекционное течение. Поддерживая постоянную концентрацию химического вещества в загрузочном канале или резервуаре, можно добиться создания в широком канале или реакционной камере, в которую помещены клетки, стабильного во времени и пространстве высоколинейного градиента концентрации. Стоит отметить, что устройства данной конструкции предназначены для создания стабильных градиентов в отсутствие потока жидкости в области размещения клеток.
Микрофлюидное устройство "диффузионной" топологии представлено в работе [59]. В устройстве в качестве диффузионной среды используется нитроцеллюлозная мембрана с вырезанными с помощью лазера тремя параллельными каналами одинаковой ширины. Мембрана зажималась между предметным стеклом и коллектором из плексигласа для обеспечения необходимого уплотнения. В один из боковых каналов подавался раствор хемоаттрактанта, а в другой буферный раствор. В центральный канал загружались клетки. За счет диффузии в центральном канале формировался линейный профиль концентрации хемоаттрактанта, изучение влияния которого на клетки и производилось.
Авторы работы [65] предложили заменить нитроцеллюлозную мембрану на агарозу, в которой также были сформированы три параллельных канала. Подобное решение дает ряд преимуществ: получение агарозы требуемой формы может быть быстро и просто выполнено с использованием относительно простых и дешевых технологий (например, методом отливки по шаблону); диффузия в агарозе протекает на скоростях, близких к скоростям диффузии в воде; тонкий слой агарозы достаточно прозрачен, что упрощает применение оптических методов регистрации миграции клеток.
Анализ получаемых градиентов химических веществ в трех различных конструкциях "диффузионных" микрофлюидных устройств был проведен в работе [66]. Рассматривались устройства с тремя параллельными каналами, состоящие из двух слоев, выполненных в разных материалах: полидиметилсилоксан и агароза. В первой конструкции все три канала были изготовлены в поли-диметилсилоксане. Во второй все три канала были сформированы в агарозе. В третьей конструкции боковые каналы были выполнены в агарозе, в то время как центральный был изготовлен в поли-диметилсилоксане. Авторы статьи высказали предположение, что именно последний подход к изготовлению микрофлюидных устройств является наиболее перспективным вариантом, т. к. дает возможность получать более сложные формы центрального канала из полидиметилсилок-сана, в то время как диффузия молекул химических веществ через агарозу позволяет получать стабильные градиенты.
Недавно в работе [67] было предложено "диффузионное" микрофлюидное устройство для анализа клеточной подвижности, которое позволяет проводить конкурентный отбор в одной популяции клеток под воздействием одновременно градиентов нескольких цитотоксических сигналов. Данное устройство состоит из круглой реакционной камеры, к которой по периметру подведено 6 каналов. В реакционную камеру высаживали клетки. У каждого канала в центре имеется специальный технический канал, через который происходило предварительное заполнение всего канала агарозой, играющей роль среды для создания диффузного градиента. Через выходные резервуары, расположенные в начале каждого канала, происходила загрузка хемоаттрактантов (до 6 различных типов). Таким образом, на разработанном устройстве создавались независимые и устойчивые градиенты различных цитотоксических сигналов в каждом канале. Например, мезенхимальные стволовые клетки костного мозга / стромальные клетки подвергались воздействию 15 цитокинов, которые, как правило, присутствует при артрите. Сначала цитокины, индуцирующие отбор стро- мальных клеток в многокомпонентных группах, были выделены для отбора в финальном раунде. Затем определение наиболее сильных химических сигналов выполнялось на базе оценки количества мигрировавших клеток, расстояния пройденного клетками, изменения показателей подвижности с течением времени. Было отмечено, что воздействие цитокина в ходе конкурентного отбора часто отличается от его индивидуального действия, что обосновывает необходимость изучения влияния нескольких цитокинов одновременно из-за синергетических или антагонистических эффектов.
Итак, существуют три базовые топологии мик-рофлюидных устройств для создания градиентов химических веществ: "T" / "Y"; "сетевая", "диффузионная". Конечно, встречаются микрофлюидные устройства построенные и на других, иногда более сложных топологиях. Однако данные топологии, как правило, являются либо развитием, либо сочетанием решений, свойственных для трех базовых топологий. Следовательно, данные микрофюид-ные устройства вбирают в себя достоинства и ограничения базовых топологий в зависимости от их сочетания. Примерами подобных устройств могут служить устройства, рассматриваемые в работах [42, 68–77].
ЗАКЛЮЧЕНИЕ
Микрофлюидные устройства, предназначенные для изучения миграции клеток под действием градиентов химических веществ, находят все более широкое применение при решении биологических задач (изучение процесса заживления ран, метастазирования рака, направленного принятия хемо-чувстительных "решений" и др.), требующих создания градиента определенной формы, его динамического изменения и обеспечения возможности наблюдения подвижности отдельных клеток в режиме реального времени методами оптической микроскопии, что является недостижимым при применении "традиционных" подходов. Помимо этого, микрофлюидные устройства требуют меньшего расхода реагентов и пробы для проведения исследований, что обуславливает их экономическую эффективность, особенно если необходимо использование дорогостоящих хемоаттрактантов.
Все микрофлюидные устройства для создания градиентов химических веществ можно условно разделить по принципу формирования градиента на две группы: "проточные" и "диффузионные". В устройствах первой группы градиент создается за счет смешения контролируемых потоков растворов с разной концентраций хемоаттрактанов. В устройствах второй группы градиент создается за счет диффузии молекул химических веществ, переносимых растворами через мембраны или гидрогели. Обе группы обладают своими достоинствами и недостатками. Например, "проточные" устройства, как правило, требуют применения внешних устройств для прецизионного контроля давления / скорости потока, что позволяет оперативно манипулировать формой градиента. С другой стороны, "диффузионные" устройства, как правило, не требуют применения указанных выше внешних устройств и при этом не обладают возможностью оперативного изменения формы градиента.
Существуют три базовые топологии микро-флюидных устройств для создания градиентов химических веществ: "T" / "Y"; "сетевая", "диффузионная". Каждая из этих топологий имеет свои достоинства и ограничения, которые отражены в таблице. Однако следует выделить два принци- пиальных отличия между базовыми топологиями: во всех топологиях, за исключением последней, наблюдается напряжение сдвига на границе контакта смешиваемых потоков (в случае "сетевой" топологии напряжение минимально); только "диффузионная" топология дает возможность создавать 2D и 3D градиенты химических веществ, остальные позволяют контролируемо получать только 1D градиенты. Кроме того, встречаются микрофлюидные устройства более сложных топологий, которые, как правило, являются либо развитием, либо сочетанием решений, свойственных для трех базовых топологий.
Работа выполнена при финансовой поддержке РФФИ в рамках научного проекта № 14-08-31641 мол а.
ПРИЛОЖЕНИЕ
Сравнение "традиционных" подходов к изучению хемотаксиса клеток с микрофлюидными технологиями
|
Подход |
Реализация |
Наиболее распространенные объекты исследований |
Достоинства и возможности |
Ограничения |
Ссылки на источники |
|
и и о S я S Я св ^ |
Камера Бойдена / анализ с использованием мембраны |
Лейкоциты; эпителиальные клетки; фибробласты |
Высокая производительность; возможен постанализ |
Затруднен контроль градиента; большой расход реагентов; необходимо большое количество клеток (пробы); анализ только по конечной точке |
[16, 43] |
|
Камера Зиг-монда / Дунна |
Лейкоциты; фибробласты; макрофаги |
Создание линейного градиента; возможность визуализации клеточной миграции |
Затруднен контроль градиента; большой расход реагентов |
[17, 19, 44] |
|
|
Использование агарозы |
Лейкоциты; моноциты |
Возможности: визуализация клеточной миграции; наложение градиентов |
Затруднен контроль градиента; не работает с клетками определенных типов |
[20, 45] |
|
|
Микропипе-тирование |
Макрофаги; нейроны |
Возможности: визуализация клеточной миграции; воздействие на определенные клетки; оказание динамических воздействий; наложение градиентов |
Затруднен контроль градиента |
[18, 46] |
Продолжение таблицы
|
Подход |
Реализация |
Наиболее распространенные объекты исследований |
Достоинства и возможности |
Ограничения |
Ссылки на источники |
|
s s о ч о и X о н о и rt S 2 ч •е о и S |
"T" и "Y" топологии (рисунок, а) |
Лейкоциты; бактерии; дрожжи; фибробласты; стволовые клетки |
Создание стабильного градиента; создание динамических градиентов и их наложение; небольшой расход реагентов; высокая производительность; не требуется большого количества клеток (пробы); возможна визуализация миграции отдельных клеток |
Создание градиента в 1D; ограничение по возможным формам градиента; индуцируемое потоками напряжение сдвига; требуется внешний контроль (например, использование насосов) |
[28, 31, 47–52] |
|
"Сетевая" топология (рисунок, б) |
Раковые клетки; дендритные клетки; эпителиальные клетки; нейроны; дрожжи; стволовые клетки |
Стабильность создаваемого градиента выше, чем для "T" и "Y" топологий; создание динамических градиентов и их наложение; небольшой расход реагентов; высокая производительность; не требуется большого количества клеток (пробы); возможна визуализация миграции отдельных клеток; возможность создания градиента произвольной формы |
Создание градиента в 1D; индуцируемое потоками напряжение сдвига; требуется внешний контроль (например, использование насосов); более сложная в сравнении с "T" и "Y" топология |
[26, 33, 34, 53– 57] |
|
|
"Диффузионная" тополо гия (рисунок, в) |
Раковые клетки; эндотелиальные клетки; бактерии; лейкоциты; нейроны; дрожжи; стволовые клетки |
Нет напряжения сдвига; возможна визуализация миграции отдельных клеток; возможно создание градиентов в 2D и 3D; меньший расход реагентов в сравнении с "Y", "T" и "сетевой" топологиями; высокая производительность; не требуется большого количества клеток (пробы); меньше требование к внешнему контролю в сравнении с "Y", "T" и "сетевой" топологиями |
Меньше возможностей контроля градиента в сравнении с "Y", "T" и "сетевой" топологиями |
[37–42, 58–60] |
Список литературы Топологии микрофлюидных устройств для изучения миграции клеток в градиентах химических веществ (обзор)
- Vicente-Manzanares M., Webb D.J., Horwitz A.R. Cell migration at a glance//Journal of Cell Science. 2005. Vol. 118. P. 4917-4919.
- Berzat A., Hall A. Cellular responses to extracellular guidance cues//EMBO Journal. 2010. Vol. 29. P. 2734-2745.
- Jin T., Xu X., Hereld D. Chemotaxis, chemokine receptors and human disease//Cytokine. 2008. Vol. 44. P. 1-8.
- Zhao M. Electrical fields in wound healing-an overriding signal that directs cell migration//Seminars in Cell and Developmental Biology. 2009. Vol. 20. P. 674-682.
- Campbell J., Butcher E. Chemokines in tissue-specific and microenvironment-specific lymphocyte homing//Current Opinion Immunology. 2000. Vol. 12. P. 336-341.
- Baggiolini M. Chemokines and leukocyte traffic//Nature. 1998. Vol. 392. P. 565-568.
- Fletcher D.A., Theriot J.A. An introduction to cell motility for the physical scientist//Physical Biology. 2004. Vol. 1. P. 1-10.
- Darnton N.C., Turner L., Rojevsky S., Berg H.C. On torque and tumbling in swimming Escherichia coli//Journal of Bacteriology. 2007. Vol. 189. P. 1756-1764.
- Ridley A.J., Schwartz M.A., Burridge K. et al. Cell migration: integrating signals from front to back//Science. 2003. Vol. 302. P. 1704-1709.
- Luster A., Alon R., Von A. U.H. Immune cell migration in inflammation: present and future therapeutic targets//Nature Immunology. 2005. Vol. 6. P. 1182-1190.
- Yonekawa K., Harlan J.M. Targeting leukocyte integrins in human diseases//Journal of Leukocyte Biology. 2005. Vol. 77. P. 129-140.
- Muller A., Homey B., Soto H., Ge N. et al. Involvement of chemokine receptors in breast cancer metastasis//Nature. 2001. Vol. 410. P. 50-56.
- Raman D., Baugher P.J., Thu Y.M., Richmond A. Role of chemokines in tumor growth//Cancer Letters. 2007. Vol. 256. P. 137-165.
- Beeh K.M., Kornmann O., Buhl R. et al. Neutrophil chemotactic activity of sputum from patients with COPD: role of interleukin 8 and leukotriene B4//CHEST Journal. 2003. Vol. 123. P. 1240-1247.
- McCaig C., Colin D., Rajnicek A.M., Song B., Zhao M. Controlling cell behavior electrically: current views and future potential//Physiological Reviews. 2005. Vol. 85. P. 943-978.
- Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes//Journal of Experimental Medicine. 1962. Vol. 115. P. 453-466.
- Zigmond S. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors//Journal of Cell Biology. 1977. Vol. 75. P. 606-616.
- Lohof A., Quillan M., Dan Y., Poo M. Asymmetric modulation of cytosolic cAMP activity induces growth cone turning//Journal of Neuroscience. 1992. Vol. 12. P. 1253-1261.
- Zicha D., Dunn G, Jones G. Analyzing chemotaxis using the Dunn directviewing chamber//Methods in Molecular Biology. 1997. Vol. 75. P. 49-457.
- Nelson R.D., Quie P.G., Simmons R.L. Chemotaxis under agarose: a new and simple method for measuring chemotaxis and spontaneous migration of human polymorphonuclear leukocytes and monocytes//Journal of Immunology. 1975. Vol. 115. P. 1650-1656.
- Li J., Lin F. Microfluidic devices for studying chemotaxis and electrotaxis//Trends in Cell Biology. 2011. Vol. 21, no. 8. P. 489-497.
- Chung B.G., Choo J. Microfluidic gradient platforms for controlling cellular behavior//Electrophoresis. 2010. Vol. 31. P. 3014-3027.
- Kim S., Kim H.J., Jeon N.L. Biological applications of microfluidic gradient devices//Integrative Biology. 2010. Vol. 2. P. 584-603.
- Saadi W., Wang S.-J., Lin F., Jeon N.L. Chemotaxis of metastatic breast cancer cells in parallel gradient microfluidic chambers//NSTI-Nanotech. 2005. Vol. 1. P. 15-18.
- Haessler U., Kalinin Y., Swartz M.A., Wu M. An agarose-based microfluidic platform with a gradient buffer for 3D chemotaxis studies//Biomedical Microdevices. 2009. Vol. 11. P. 827-835.
- Fernandes J.T.S., Tenreiro S., Gameiro A. et al. Modulation of alpha-synuclein toxicity in yeast using a novel microfluidic-based gradient generator//Lab on a Chip. 2014. Vol. 14. P. 3949-3957.
- Lin F. A microfluidics-based method for analyzing leukocyte migration to chemoattractant gradients//Methods in Enzymology/Tracy M.H. and Damon J.H. eds. Chapter 15. Academic Press, 2009. P. 333-347.
- Lin F., Butcher E. T cell chemotaxis in a simple microfluidic device//Lab on a Chip. 2006. Vol. 6. P. 1462-1469.
- Lin F., Saadi W., Rhee S.W., Wang S.J., Mittal S., Jeon N.L. Generation of dynamic temporal and spatial concentration gradients using microfluidic devices//Lab on a Chip. 2004. Vol. 4. P. 164-167.
- Irimia D., Liu Su-Y., Tharp W.G. et al. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients//Lab on a Chip. 2006. Vol. 6. P. 191-198.
- Lin F., Nguyen C.M., Wang S.J. et al. Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices//Annals of Biomedical Engineering. 2005. Vol. 33. P. 475-482.
- Englert D.L., Manson M.D., Jayaraman A. Investigation of bacterial chemotaxis in flowbased microfluidic devices//Nature Protocols. 2010. Vol. 5. P. 864-872
- Saadi W., Wang S., Lin F., Jeon N.L. A parallel-gradient microfluidic chamber for quantitative analysis of breast cancer cell chemotaxis//Biomedical Microdevices. 2006. Vol. 8. P. 109-118.
- Ricart B.G., John B., Lee D., Hunter C.A., Hammer D.A. Dendritic cells distinguish individual chemokine signals through CCR7 and CXCR4//Journal of Immunology. 2011. Vol. 186. P. 53-61.
- Lin F., Nguyen C.M., Wang S.J. et al. Effective neutrophil chemotaxis is strongly influenced by mean IL-8 concentration//Biochemical and Biophysical Research Communications. 2004. Vol. 319. P. 576-581.
- Diao J.P., Young L., Kim S. et al. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis//Lab on a Chip. 2006. Vol. 6. P. 381-388.
- Chung S., Sudo R., Mack P.J. et al. Cell migration into scaffolds under co-culture conditions in a microfluidic platform//Lab on a Chip. 2009. Vol. 9. P. 269-275.
- Chen Z., Chen W., Yuan B. et al. In vitro model on glass surfaces for complex interactions between different types of cells//Langmuir. 2010. Vol. 26. P. 17 790-17 794.
- Abhyankar V.V., Lokuta M.A., Huttenlocherbc A., Beebe D.J. Characterization of a membrane-based gradient generator for use in cell-signaling studies//Lab on a Chip. 2006. Vol. 6. P. 389-393.
- Chung B., Lin F., Jeon N.L. A microfluidic multi-injector for gradient generation//Lab on a Chip. 2006. Vol. 6. P. 764-768.
- Huang C.P., Lu J., Seon H. et al. Engineering microscale cellular niches for three-dimensional multicellular co-cultures//Lab on a Chip. 2009. Vol. 9. P. 1740-1748.
- Ambravaneswaran V., Wong I.Y., Aranyosi A.J. et al. Directional decisions during neutrophil chemotaxis inside bifurcating channels//Integrative Biology. 2010. Vol. 2. P. 639-647.
- Rhoads D.S., Nadkarni S.M., Song L. et al. Cell migration: developmental methods and protocols/Ed. J.-L. Guan. V. 294. Totowa (NJ): Humana Press Inc., 2004.
- Wells C.M., Ridley A.J. Analysis of cell migration using the Dunn chemotaxis chamber and time-lapse microscopy//Methods in Molecular Biology. 2005. Vol. 294. P. 31-41.
- Mandarino G.L., Suarez A.F., Hirata A.A., Ward P.A. Chemotaxis under agarose utilizing human serum depleted of C-5 derived peptides//Journal of Immunological Methods. 1981. Vol. 45. P. 283-299.
- Wu H.-J., Liu Y.-J., Li H.-Q. et al. Analysis of microglial migration by a micropipette assay//Nature Protocols. 2014. Vol. 9. P. 491-500.
- Mao H.B., Cremer P.S., Manson M.D. A sensitive, versatile microfluidic assay for bacterial chemotaxis//Proceedings of the National Academy of Sciences of the United States of America. 2003. Vol. 100. P. 5449-5454.
- Lanning L.M., Ford R.M., Long T. Bacterial chemotaxis transverse to axial flow in a microfluidic channel//Biotechnology and Bioengineering. 2008. Vol. 100. P. 653-663.
- Long T., Ford R.M. Enhanced transverse migration of bacteria by chemotaxis in a porous T-sensor//Environmental Science & Technology. 2009. Vol. 43. P. 1546-1552.
- Moore T.I., Chou C.S., Nie Q. et al. Robust spatial sensing of mating pheromone gradients by yeast cells//PLoS One. 2008. Vol. 3. P. e3865.
- Chung B. G., Manbachi A., Saadi W. et al. A gradient-generating microfluidic device for cell biology//Journal of Visualized Experiments. 2007. V. 7. P. 271
- Park J.Y., Kim S.K., Woo D.H. et al. Differentiation of neural progenitor cells in a microfluidic chip-generated cytokine gradient//Stem Cells. 2009. V. 27. P. 2646-2654.
- Chang W., Cheng Y.-J., Tu M. et al. A polydimethylsiloxane-polycarbonate hybrid microfluidic device capable of generating perpendicular chemical and oxygen gradients for cell culture studies//Lab on a Chip. 2014. Vol. 14. P. 3762-3772.
- Hung P.J., Lee P.J., Sabounchi P. et al. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays//Biotechnology and Bioengineering. 2005. Vol. 89. P. 1-8.
- Wang S.J., Saadi W., Lin F. et al. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis//Experimental Cell Research. 2004. V. 300. P. 180-189.
- Chung B.G., Flanagan L.A., Rhee S.W. et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device//Lab on a Chip. 2005. Vol. 5. P. 401-406.
- Dertinger S.K.W., Jiang X.Y., Li Z.Y. et al. Gradients of substrate-bound laminin orient axonal specification of neurons//Proceedings of the National Academy of Sciences of the United States of America. 2002. Vol. 99. P. 12 542-12 547.
- Paliwal S., Iglesias P.A., Campbell K. et al. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast//Nature. 2007. Vol. 446. P. 46-51.
- Diao J.P., Young L., Kim S. et al. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis//Lab on a Chip. 2006. Vol. 6. P. 381-388.
- Abhyankar V.V., Beebe D.J. Spatiotemporal micropatterning of cells on arbitrary substrates//Analytical Chemistry. 2007. Vol. 79. P. 4066-4073.
- Keenan T.M., Folch A. Biomolecular gradients in cell culture systems//Lab on a Chip. 2008. Vol. 8. P. 34-57.
- Kamholz A.E., Weigl B.H., Finlayson B.A., Yager P. Quantitative analysis of molecular interaction in a microfluidic channel: the T-sensor//Analytical Chemistry. 1999. Vol. 71, no. 23. P. 5340-5347.
- Englert D.L., Manson M.D., Jayaraman A.//Applied and Environmental Microbiology. 2009. Vol. 75. P. 4557-4564.
- Cooksey G.A., Sip С.G., Folch A. A multi-purpose microfluidic perfusion system with combinatorial choice of inputs, mixtures, gradient patterns, and flow rates//Lab on a Chip. 2009. Vol. 9, no. 3. P. 417-426.
- Cheng S.Y., Heilman S., Wasserman M. et al. A hydrogel-based microfluidic device for the studies of directed cell migration//Lab on a Chip. 2007. Vol. 7. P. 763-769.
- Ahmed T., Shimizu T.S., Stocker R. Bacterial chemotaxis in linear and nonlinear steady microfluidic gradients//Nano Letters. 2010. Vol. 10. P. 3379-3385.
- Mendelson A., Cheung Y.K., Paluch K. et al. Competitive stem cell recruitment by multiple cytotactic cues//Lab on a Chip. 2013. Vol. 13. P. 1156-1164.
- Law A.M.J., Aitken M.D. Continuous-flow capillary assay for measuring bacterial chemotaxis//Applied and Environmental Microbiology. 2005. Vol. 71. P. 3137-3143.
- Atencia J., Morrow J., Locascio L.E. The microfluidic palette: A diffusive gradient generator with spatio-temporal control//Lab on a Chip. 2009. Vol. 9. P. 2707-2714.
- Kim T., Pinelis M., Maharbiz M.M. Generating steep, shear-free gradients of small molecules for cell culture//Biomedical Microdevices. 2009. Vol. 11. P. 65-73.
- Scherber C., Aranyosi A.J., Kulemann B. et al. Epithelial cell guidance by self-generated EGF gradients//Integrative Biology. 2012. Vol. 4. P. 259-269.
- Choi E., Chang H.-K., Lim C.Y. et al. Concentration gradient generation of multiple chemicals using spatially controlled self-assembly of particles in microchannels//Lab on a Chip. 2012. Vol. 12. P. 3968-3975.
- Cho H., Hamza B., Wonga E.A., Irimia D. On-demand, competing gradient arrays for neutrophil chemotaxis//Lab on a Chip. 2014. Vol. 14. P. 972-978.
- Jin B.-J., Ko E.-A., Namkung W., Verkman A.S. Microfluidics platform for single-shot dose -response analysis of chloride channel-modulating compounds//Lab on a Chip. 2013. Vol. 13. P. 3862-3867.
- Baker B.M., Trappmann B., Stapleton S.C. et al. Microfluidics embedded within extracellular matrix to define vascular architectures and pattern diffusive gradients//Lab on a Chip. 2013. Vol. 13. P. 3246-3252.
- Xu B.-Y., Hu S.-W., Qian G.-S. et al. A novel microfluidic platform with stable concentration gradient for on chip cell culture and screening assays//Lab on a Chip. 2013. Vol. 13. P. 3714-3720.
- Sip C.G., Bhattacharjeea N., Folcha A. Microfluidic transwell inserts for generation of tissue culture-friendly gradients in well plates//Lab on a Chip. 2014. Vol. 14. P. 302-314.


