Unraveling NAC family transcription factors and their expression analysis under high temperature and drought stress in peanut
Автор: Suchithra B., Shafia Hoor F., Puspha T.C., Nagesh Babu R.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 2 т.21, 2025 года.
Бесплатный доступ
Transcription factors play pivotal roles in the conversion of stress signal perception to stress-responsive gene expression. NAC (NAM, ATAF1/2 and CUC2) domain proteins are plant-specific transcriptional factors known to play diverse roles in various plant developmental processes and received considerable attention as regulators in stress signaling. Considering the relatively large number of NAC transcription factors from different plants and their diverse roles under complex environmental stimuli remains a challenge. In this study, phylogenies, genome localizations, gene structure and expression profiles of NAC genes under high temperature and drought treatments, with a focus on Peanut ( Arachis hypogaea ) genotypes ( A. duranensis & A. ipaensis ) was performed. Thirty eight NAC genes from each genotype were detected, including eight membrane-bound members which includes AdNAC26, AdNAC36, AiNAC16, AiNAC17, AiNAC37, AdNAC14, AiNAC12 & AiNAC29. Majority of the identified NAC proteins had atleast four NAC domain containing conserved motifs and were found to be localized to nucleus. AdNAC21 and AiNAC3 were found to be positive regulators in drought and high temperature responses. Our results provides foundation for selection of promising stress- responsive NAC candidates for detailed plant functional analysis, leading to development of transgenic Peanut varieties with improved productivity under drought and high temperature.
Nac transcription factor, phylogenetic analysis, qrt-pcr, peanut
Короткий адрес: https://sciup.org/143184708
IDR: 143184708
Текст научной статьи Unraveling NAC family transcription factors and their expression analysis under high temperature and drought stress in peanut
Abiotic stresses such as high temperature and drought greatly limit the growth and crop productivity worldwide. everal NAC (NAM, ATAF1/2, and CUC2) proteins have been documented as important regulators in stress responses. The NAC TFs function as important components in complex signaling progresses during plant stress responses. Until recently, the possible involvement of TF NAC proteins in abiotic stress responses was deduced indirectly from transcription profiling; recent functional analyses, however, have provided some direct evidence. The tight regulation and fine-tuning of NAC genes during plant stress responses contribute to the establishment of complex signaling webs, and the important roles of NAC genes in stress responses make them potential candidates for imparting stress tolerance. Plant-specific NAM, ATAF1/2, and CUC2 (NAC) proteins constitute one of the largest transcription factor (TF) families and are characterized by a well-conserved N terminal NAC domain (Olsen et al. , 2005, Puranik et al. , 2012). The NAC domain, which comprises nearly 160 amino acid residues, can be divided into five sub domains (A to E) based on its motif distribution (Aida et al. , 1997, Ooka et al, 2003). The highly conserved sub-domains C and D may be responsible for binding to DNA, sub domain A may be involved in homo- and hetero- dimerization, and the divergent sub-domains B and E may be implicated in the functional diversity of NAC proteins (Ooka et al. , 2003; Jensen et al. , 2010; Chen et al. , 2011). Genes encoding NAC proteins were regulated (i) transcriptionally by upstream TFs such as ABREs (ABA-responsive elements) and DREs (Dehydration-responsive elements), (ii) post transcriptionally by micro-RNAs or alternative splicing, and (iii) post-translationally by ubiquitinization, dimerization, phosphorylation or proteolysis (Puranik et al. , 2012). As an additional feature, some NAC proteins comprise a helical transmembrane motif for anchoring to plasma membrane or endoplasmic reticulum NAC proteins have been implicated in a wide range of plant developmental processes, including lateral root formation (He et al. , 2005), shoot branching (Mao et al. , 2007), flowering ( ablowski, Meyerowitz, 1998) and leaf senescence
(Guo, Gan, 2006). In particular, numerous NAC domain proteins have also been implicated in various defense responses such as drought (Jeong et al. , 2010), salinity (Zheng et al. , 2009), cold (Aslam et al. , 2012), fungal and bacterial pathogens (Wang et al. , 2009). Extensive investigation aided by the availability of several complete plant genomic sequences has identified more than 100 NAC genes in Arabidopsis, rice, soybean, foxtail millet, Chinese cabbage, 74 in grape ( Vitis vinifera ) and 88 in pigeonpea (Wu et al. , 2015) etc. Abiotic stress triggers a wide range of plant responses, from the alteration of gene expression and cellular metabolism to changes in plant growth, development, and crop yield. Thus, understanding the complex mechanism of drought and high temperature tolerance is important for agriculture production (Nuruzzaman et al. , 2013). Although the genes encoding the transcription factors just accounts for a little portion in the whole genome, transcription factors are important in the regulated networks (Hobert, 2008).
Peanut (Arachis hypogaea L.) is an oilseed crop cultivated worldwide and one of the major grain legumes in tropical and subtropical regions. However, its productivity is strongly affected by drought one of the most serious constraints to crop production and, associated with the predicted consequences of global climate change, increases the need for drought-adapted varieties. To date, 80 species in genus Arachis were identified and have been classified into nine taxonomic sections (Bertiol et al., 2011). Wild species are diploid, but cultivated peanut is allotetraploid (AABB). The wild ancestral species of cultivated peanut are generally considered to be duranensis and ipaënsis, which contributed the A and B sub-genomes, based on morphology, cytology, fertility of the interspecific hybrid and molecular studies (Kochert et al., 1996; Ramos et al., 2006). NAC genes in plants have been investigated in great detail for their role in various stress and developmental aspects; thus, they could be important candidates for the improvement programs. Detailed analysis of NAC domain genes using the assembled Peanut genome including prediction of gene functions, evolutionary significance and investigation of their expression profiles is necessary to understand function of NAC gene family. Our results provide a comprehensive genome-wide knowledge of NAC proteins in peanut and a preliminary knowledge of specific NAC proteins potentially involved in drought and high temperature response in peanut. Through these analyses, we have increased knowledge concerning the evolution and function of Peanut NAC genes.
MATERIALS AND METHODS
Plant materials and treatment eeds of Peanut (Luhua14) were surface-sterilized and grown under controlled conditions at 28 °C day/25 °C night with a 12-h light/12-h dark photo period. After 10 days of germination, drought was imposed by withholding water for 5 days and for high temperature stress seedlings were exposed to high-temperature [42 °C for 2h (induction) followed by 48 °C for 6h]. After the stress treatment, control and stress exposed shoot were harvested immediately and stored at -80 °C for further analysis.
Identification, characterization and subcellular localization of NAC genes
The NAC domain containing protein sequences of Peanut were retrieved from the Plant Transcription Factor Database ver. 2.0. and Arachis genome (Peanut Base) for the hidden Markov model (HMM) profile of the NAC domain downloaded from the Pfam database using HAMMER (ver. 3.0). All redundant sequences were removed and the collected data were further curated by examining the presence of the conserved NAC domain with the help of Pfam , MART and
InterPro can can/) web server. The length, molecular weight and pI of each deduced polypeptide were calculated using ExpasyProtParam tool
. Further, CELLO and WOLF P ORT tools were used to predict the subcellular localizations. Amino acid sequences of NAC TFs belonging to Peanut were imported to BioEdit v7.2.5 (Hall, 1999) and multiple sequence alignment was performed with NAC protein sequences from Arabidopsis and Glycine max using
ClustalW with default parameters. The NAC sequences along with Arabidopsis sequences were imported into MEGA v6.06 (Tamura et al , 2013) to construct a phylogenetic tree by Neighbor-Joining method and the bootstrap test was performed with 1000 iterations. Finally, TMHMM erver ver. 2.0 was used to predict the membrane bound NAC members.
Genome wide distribution pattern, Gene structure and identification of conserved motifs
The chromosomal location of AdNAC and AiNAC genes were obtained from peanut base website and the map was generated using MapInspect
. Gene tructure Display erver from Center for Bioinformatics, Peking University, was used to display the intron exon junctions . The genomic and mRNA sequences of these NACs were downloaded and used as query for generating its gene structure. A number of introns and exons were estimated based on this alignment and confirmed by the coordinates given in the sequences. The MEME uite tool v4.9.1 (Bailey et al., 2009) was utilized for analysis of the conserved motifs.
TRING 10 , computational tool was used to predict the functional protein association network gene ontology annotation (GO) of the NAC proteins in Arabidopsis with the default parameters. These interactions were derived from genomic context, high-throughput experiments and co-expressions studies.
Expression analysis of NAC genes
Total RNA was isolated from control and stress tissue using TRizol (Invitrogen) according to the manufacturer’s instructions and then treated with RNAase- free DNAase I (Promega). All RNA samples were quantified by Nanodrop 2000 (Thermo cientific). cDNA was synthesized by reverse transcription with 500ng of total RNA using Prime cript RT Reagent Kit (Takara) according to the manufacturer’s instructions. Gene specific primers were designed using Primer3 software (Additional Resource 1). qRT- PCR reactions were performed using YBR Green PCR Master mix
(Takara) on Lightcycler96 Real time PCR (Roche). Each PCR reaction (20 μl) included 2 μlcDNA, 1x YBR Green Master mix, 0.5 μl sequence-specific forward primer (10 μM), 0.5 μl universal reverse primer (10 μM), and 7 μl sterile water. The NAC expression was normalized against actin as reference gene. The reactions conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 55 °C for 30s and 72 °C for 15s. All reactions were run with three technical and two biological replicates and the data was analyzed using 2-ΔΔCT method.
RESULTS
Protein features, Multiple sequence alignment and Phylogenetic analysis
To identify all the NAC transcription factors, we retrieved all the predicted NAC genes from Plant TFDB and Peanut Base . Basic information like molecular weight and pI are depicted in Additional Resource 2. The average polypeptide length was 347.1 residues with the length ranging from 158 aa (AdNAC17) to 698 aa (AiNAC37). The pI values range from 4.43 to 9.46. The subcellular localization results revealed that majority of the proteins were localized to nucleus and 9/76 were predicted to be localized in cytoplasm. The multiple alignment of AdNACs, AiNACs and NACs from Arabidopsis and Glycine max indicated that all of the Peanut NACs shared a highly conserved N-terminal DNA binding NAC domain, which consists of five consensus sub-domains (A-E), and a variable C-terminal transcriptional regulation domain. Additionally, a conserved bipartite nuclear localization signal was also found in the D subdomain of the majority of Peanut NACs, suggesting that these NACs may be localized to the nucleus (Additional Resource 3) (Greve et al., 2003). To examine the structure and phylogenetic relationships of Peanut NAC TFs identified in our study, a combined phylogenetic tree was constructed with the aligned NAC domains from Peanut, Arabidopsis and soybean (Fig. 1). Examination of the phylogenetic tree analysis emphasis that the Peanut NAC TFs can be classified into nine major groups: Group I (14) Group II (8), Group III (17), Group IV (5), Group V (14), Group VI (4), Group VII (2), Group VIII (4) and Group IX (8). Phylognetic trees constructed from the AdNACs, AiNACs and AdNAC-
Identification, chromosomal distribution and gene structure of NAC members
The keyword, HMM profile and BLA T search predicted that the Peanut genome encodes about 38 NAC proteins. A total of 38 NAC genes were identified from both A. duranensis and A. ipaënsis . They were named as AdNAC1 to AdNAC38 , and AiNAC1 to AiNAC38 , respectively. The genome of Peanut comprises of 20 chromosomes (10 from duranensis and 10 from ipensis ) varying in their length, shortest being chromosome 8 (48.94 Mb) and longest is the chromosome 3 (133.14 Mb) in A. duranensis while in A. ipaënsis , shortest being chromosome 2 with 108.64 Mb and longest being chromosome 5 with 149.44 Mb in size (Fig. 3). In silico mapping of NACs indicated an uneven distribution of the genes on all the chromosomes. The exact position (in bp) of each AdNACs and AiNACs on Peanut chromosomes is given in Additional Resource 2. The gene structures were investigated through genomic annotation to determine the structural diversity. All NAC genes harbored at least two exons except AiNAC16 being the shortest not having intron. In addition, a separate phylogenetic tree was generated from the complete protein sequences of all the NAC genes (Fig. 2).
Identification of conserved motifs and Gene annotation
The MEME (Multiple Expectation Maximization for Motif Elicitation) server was used for exploring motif distribution in 38 AdNAC and 38 AiNAC proteins. Nine different conserved motifs were identified, of which most of them had at least four NAC domain-encoding motifs, and 54 shared a highly conserved typical NAC domain containing five consensus sub domains (motifs 2, 4, 1, 5 and 3) in the same order (Fig. 4). The motif sequence logos are depicted in the Additional Resource 4C. Prediction of functional protein association network of NAC proteins in Arabidopsis using TRING program revealed the interaction of NAC083 (AdNAC15) with VND1, VND7, NAC1, NAC41, ANAC026 and NAC007; XND1 (AdNAC5) with NAC073 and NAC010; NAC090 (AiNAC14) with NAC044 and NAC036; BTF3 (AiNAC2)
with NACA2 and AT3G12390 (Additional Resource 4D). Gene ontology (GO) annotation of NAC proteins showed the involvement of these proteins in different biological processes, cellular and molecular functions (Additional Resource 4E). The membrane transcription factors proteins are stored in their dormant forms in association with the intracellular membranes. When plants are exposed to abrupt environmental changes, they are released from the membranes through proteolytic cleavage events and enter the nucleus, where they regulate expression of genes involved in perception of stress signals, stress signaling (Kim et al. , 2010). Among
76 NACs (38 AdNACs& 38 AiNACs), 08 members (AdNAC14, 26, 36, AiNAC16, 17, 29 and 37) were identified as membrane-associated NTLs using the TMHMM v2.0 (Table 1), of which 5 (AdNAC26, AdNAC36, AiNAC16, AINAC17 and AiNAC37) and 3 (AdNAC14, AiNAC12 and AiNAC29) members contain one and two TMHs, respectively. The phylogenetic tree constructed with membrane associated NTLs identified from Peanut, Arabidopsis and Rice indicated that the Peanut NTLs were scattered into different groups (Additional Resource 4B).
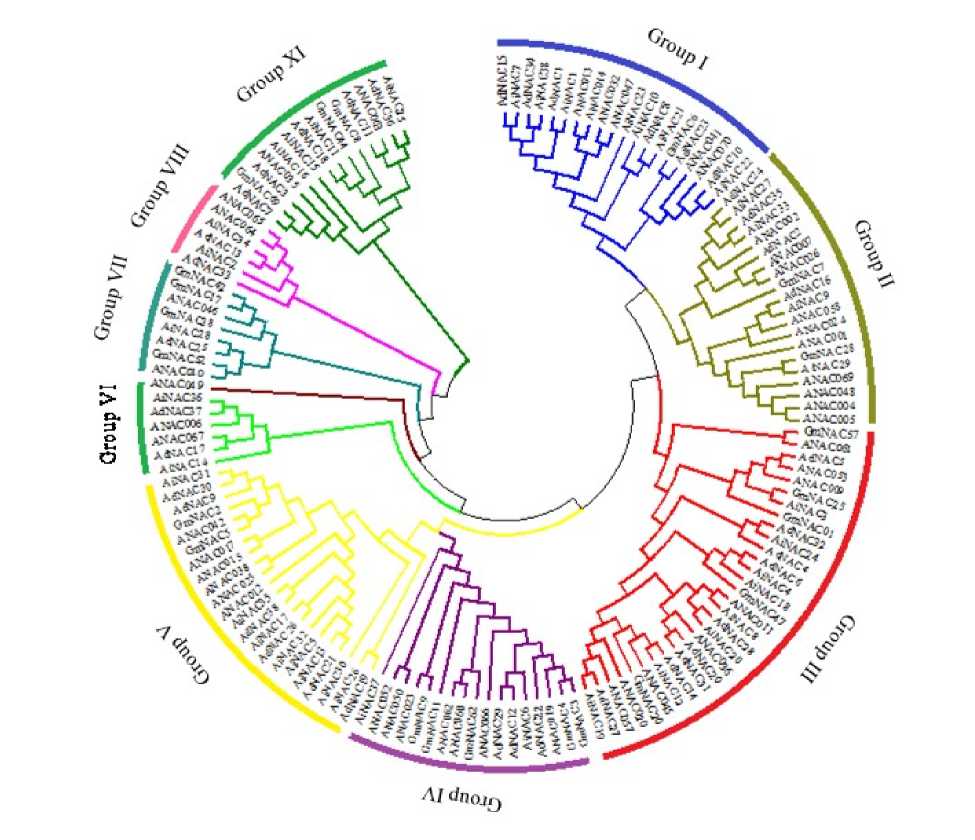
Figure 1: Unrooted phylogenetic tree constructed using the neighbor-joining (NJ) method, and the bootstrap test was carried out with 1,000 iterations representing the relationships between the Peanut, Arabidopsis and Glycine max NAC domain proteins.
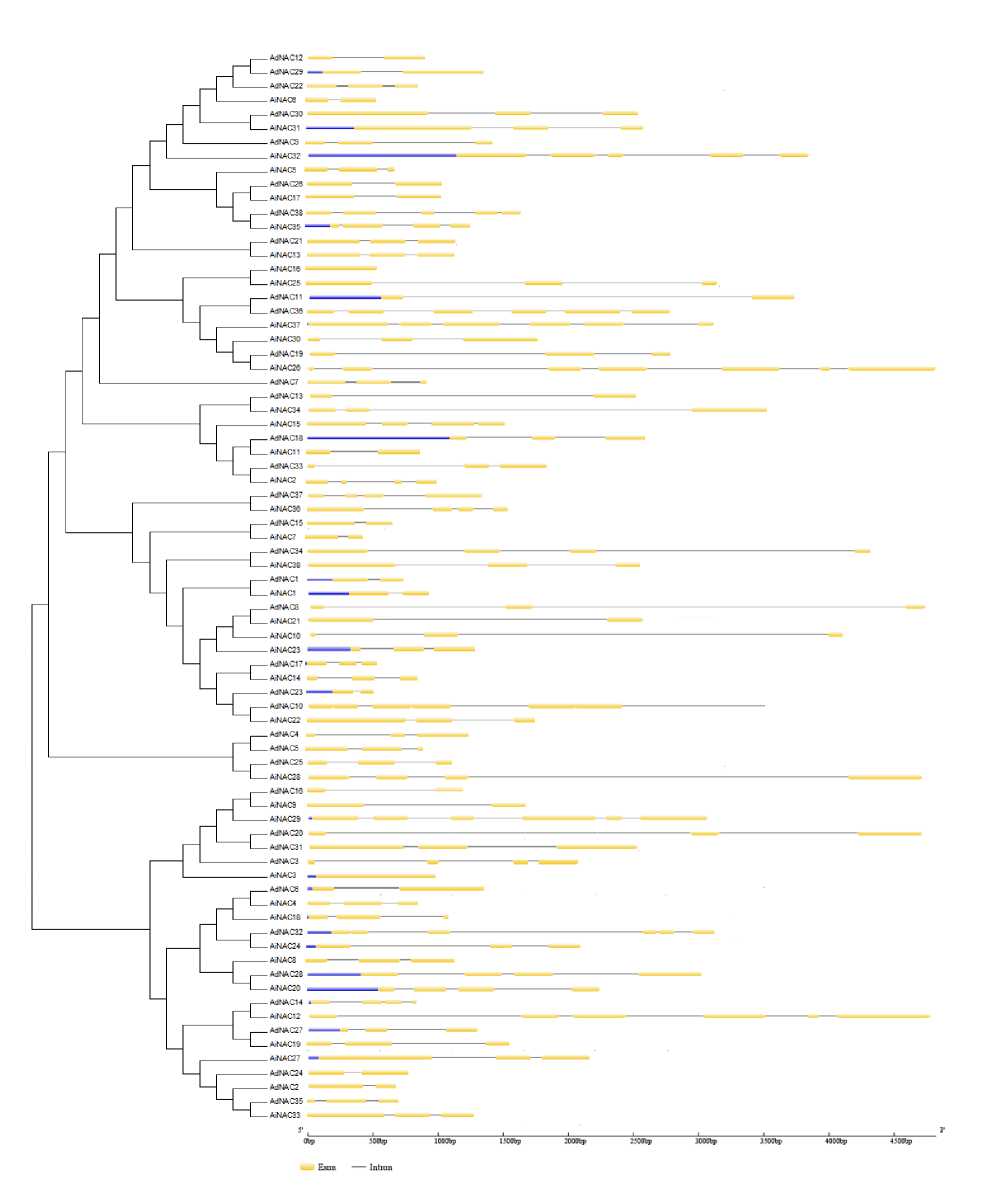
Figure 2. Phylogenetic relationship and gene structure of the NAC genes. Phylogenetic tree was constructed with MEGA6.0 using the neighbor-joining (NJ) method with 1,000 bootstrap replicatesbased on a multiple alignment of 76 amino acid sequences of NAC genes from Arachis duranensis & Arachis ipaënsis. Exon/ intron structure of NAC genes are represented by boxes and black lines, respectively.
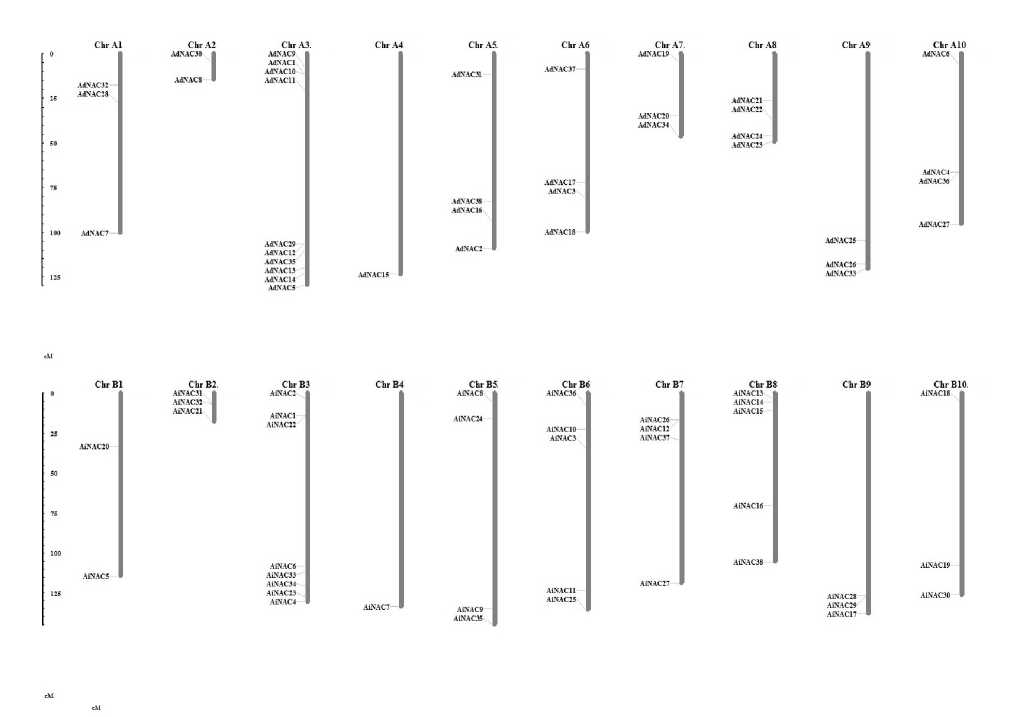
Figure 3: Distribution of 76 NAC genes on Peanut chromosomes and physical locations of each NAC gene on the ten chromosomes from each species (positions in cM).
Table 1: Putative membrane- bound Peanut NTLs
|
Gene Name |
Membrane bound member |
Length (aa) |
Transmembrane sequences |
Expected number of AAs in TMHs |
Expected number, first 60 AAs |
|
AdNAC14 |
AdNTL1, AdNTL2 |
633 |
525…..544 612…..631 |
40.88286 |
0 |
|
AdNAC26 |
AdNTL3 |
551 |
523…..545 |
22.13992 |
0.00091 |
|
AdNAC36 |
AdNTL4 |
592 |
569…..591 |
22.56383 |
0.01596 |
|
AiNAC12 |
AiNTL5, AiNTL6 |
678 |
582…..604 656…..675 |
41.84687 |
0 |
|
AiNAC16 |
AiNTL7 |
216 |
5…..27 |
20.81172 |
20.80519 |
|
AiNAC17 |
AiNTL8 |
559 |
531…..553 |
22.14053 |
0.00084 |
|
AiNAC29 |
AiNTL9, AiNTL10 |
583 |
531…..553 558…..580 |
40.01905 |
0 |
|
AiNAC37 |
AiNTL11 |
698 |
669…..691 |
22.56604 |
0.01787 |
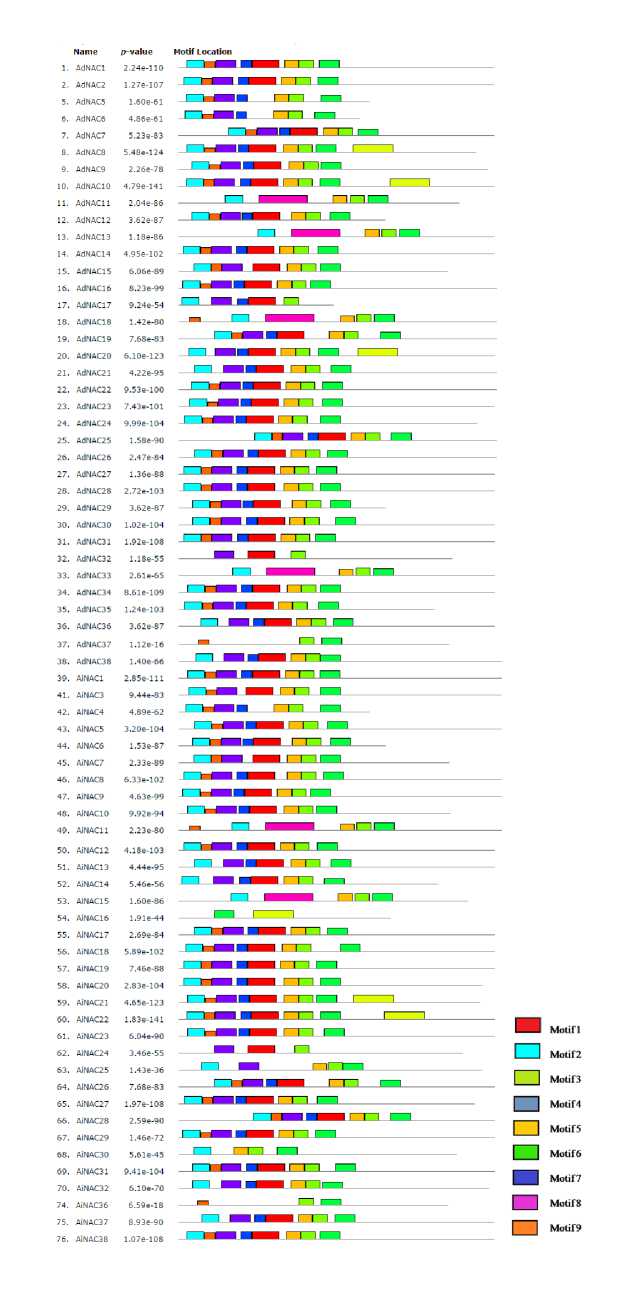
Figure 4: chematic representation of conserved motifs in the AdNAC and AiNAC proteins predicted by MEME. Each motif is represented by a number in the colored box. The black lines represent non-conserved sequences.
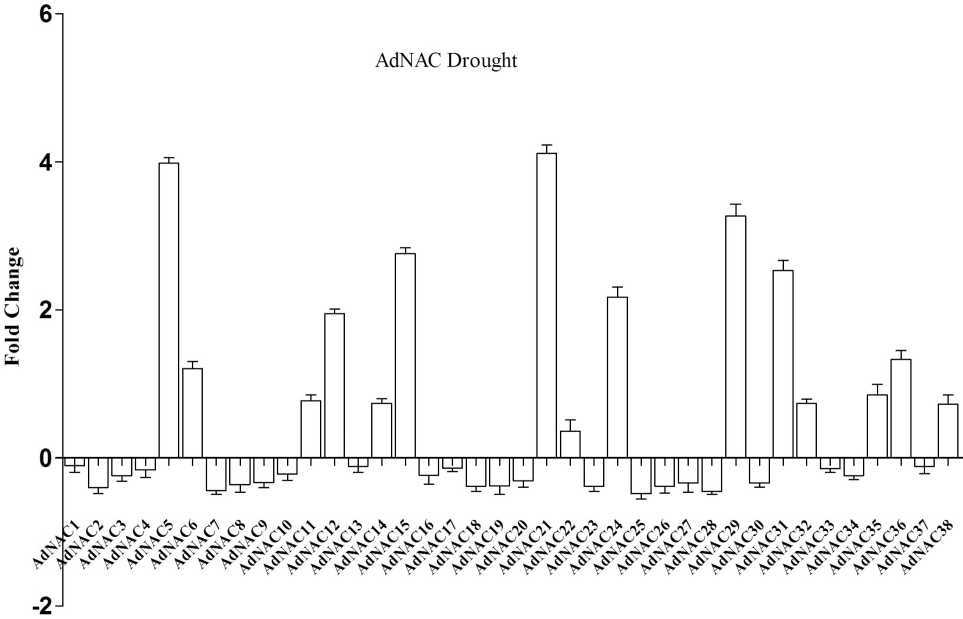
Figure 5: Expression profile of AdNAC genes obtained by RT-qPCR of treated (drought) and well watered (WW) control shoot samples
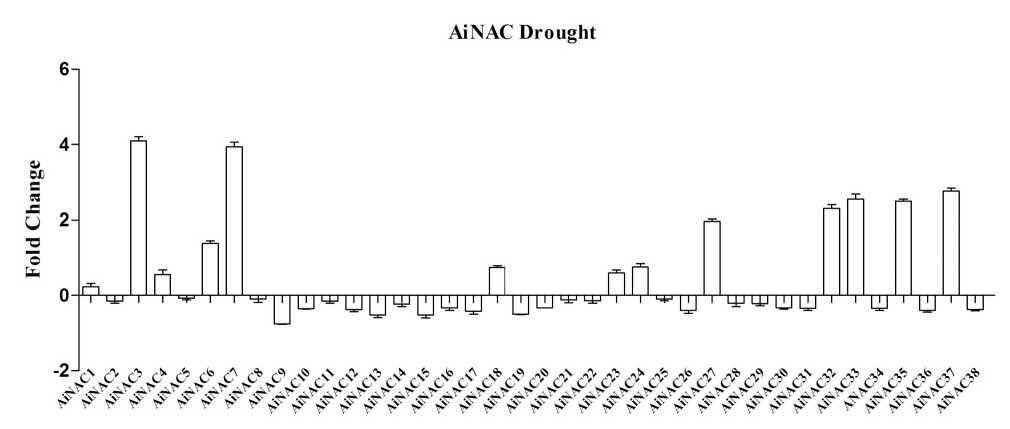
Figure 6: Expression profile of AiNAC genes obtained by RT-qPCR of treated (drought) and well watered (WW) control shoot samples
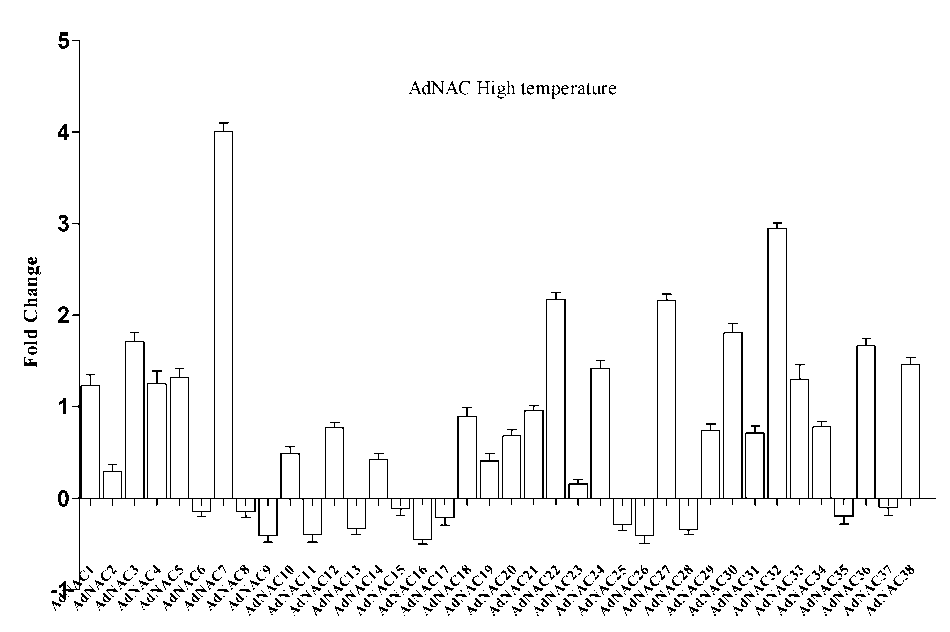
Figure 7: Expression profile of AdNAC genes obtained by RT-qPCR of treated (high temperature) and well watered (WW) control shoot samples
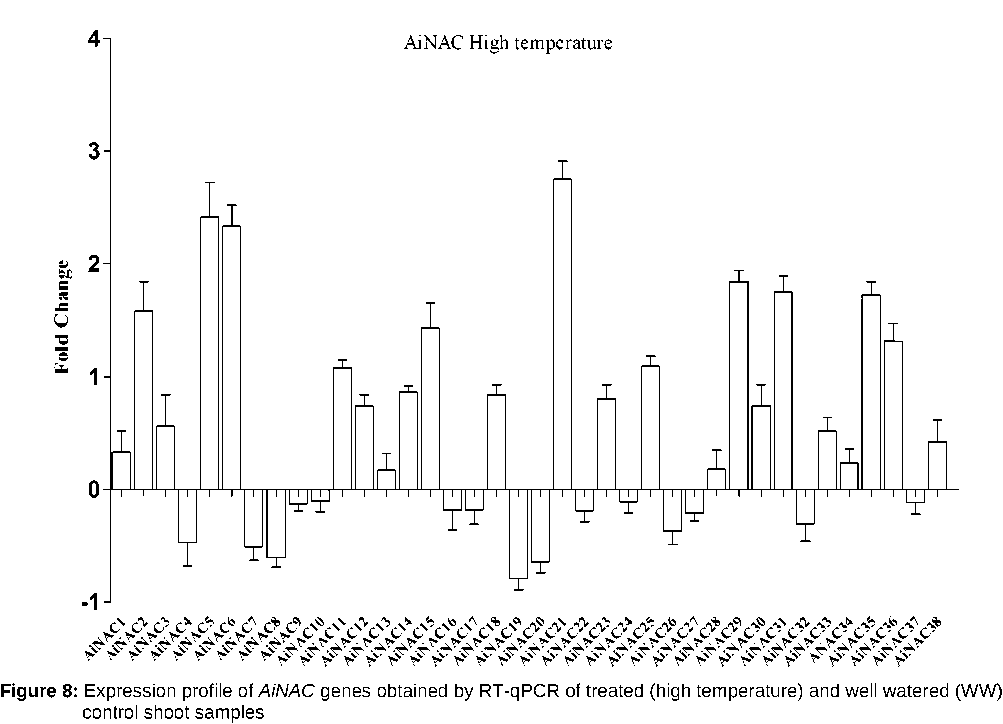
Expression profiles of AdNAC and AiNAC genes during high temperature and drought stress
NAC proteins are plant-specific TFs that have been shown to function in abiotic stress responses (Nakashima et al , 2014; Puranik et al , 2012). To investigate the responses of NAC genes to drought stress, we investigated the expression profiles of NAC genes in seedlings and expressed the results as fold changes with respect to the controls. During drought stress NACs belonging to Group V NACs such as AdNAC32, AdNAC36, AdNAC38, AiNAC24, AiNAC32, AiNAC35 and AiNAC37 were found to be up regulated by 0.68, 1.45, 0.85, 0.75, 2.4, 2.49 and 2.76 folds respectively. ANAC069 belonging to the same group encodes a membrane-bound NAC protein that integrates auxin and salt signals to regulate seed germination in Arabidopsis. In Group VI, AdNAC5, AdNAC6, AdNAC12, AdNAC15, AdNAC21, AdNAC22, AdNAC24, AdNAC29, AdNAC35 were induced by 4.06, 1.3, 2.01, 2.84, 4.01, 0.51, 2.03, 2.43 and 0.71 folds respectively (Fig. 5 & 6). During high temperature, 48 genes were up-regulated and among them AdNAC7, 22, 32, AiNAC5, 6, 21 were found to be induced by 4.1, 2.1, 3.01, 2.11, 2.52, 2.59 folds respectively. The other up-regulated genes showed fold change between 1 and 3. All the other genes were down regulated with the fold changes ranging between 0.5 to 1 (Fig7 & 8). AdNAC21 and AiNAC3 showed increased expression under both drought and high temperature stress.
DISCUSSION
The plant-specific NAC transcription factors (TFs) play important roles in regulation of diverse biological processes, including development, growth, cell division and responses to environmental stimuli. To cope with these stresses, plants have evolved a range of physiological and biochemical responses and a complex of signaling transduction pathways (You et al , 2015). AdNAC and AiNAC genes were mapped to the Peanut genome according to their position information from Peanut Base. Among all, chromosome 3 contains the highest number of NACs (26%), while only one gene was located on chromosome 4 (0.026%). There was no positive correlation between the chromosome length and the number of NAC genes. The ends of chromosome exhibited stronger synteny than the central regions of chromosomes (Nagy et al , 2012). It is well known that gene structural diversity is a possible mechanism for the evolution of multi-gene families. Investigation of gene structures reveals that the most closely related members in the same subfamilies shared similar exon/ intron structures in terms of intron number and exon length suggesting, these members may be evolved early and represent the ancestral form. All well-known NAC domain proteins bind specifically to the CATGTG motif of the promoter region (Tran et al , 2004) or act as a functional motif or activation domain (Oh et al , 2005). The relationship among the 76 Peanut NAC genes was investigated through constructing phylogenetic trees using Neighbour Joining method and the tree topology revealed several pairs of NAC proteins with a high degree of homology in the terminal nodes of each subfamily. According to Fig. 1, 76 NAC genes formed 9 clades and they were designated groups I to IX, the largest group was Group III which has 17 members. everal putative trans-membrane helices have been identified in other plant species such as Chickpea (Ha et al , 2014), Glycine max (Le et al , 2011), Arabidopsis (Kim et al , 2010), Rice (Kim et al , 2007), Maize ( hiriga et al , 2014), Potato ( ingh et al , 2013), Foxtail millet (Puranik et al , 2013), Chinese cabbage (Liu et al , 2014) and Tomato (Kou et al , 2014). Out of 11 putative GmNTLs of soybean and 08 CaNTLs of Chickpea, 2 and 4 members possess two TMHs respectively whereas all the NTLs predicted in other plant species possess only one TMH, suggesting that the existence of doubled TMHs might be specific to leguminous plants. A phylogenetic tree constructed from the NTLs from Peanut, Arabidopsis (NTLs/ANACs) and rice (OsNTLs/ONACs) indicated that the Peanut NTLs were scattered into 4 major groups (Additional Resource 4B). The membrane bound TF can immediately regulate the downstream genes upon stress perception and activation. In Arabidopsis a membrane bound NAC protein, NTL6, has been to shown to get activated upon cold stress as the membrane fluidity changes and induces the expression of pathogenesis related proteins. In addition, the plant hormone ABA also activates the NTL6 ( eo, Park, 2010), thus indicating the involvement in biotic and abiotic stress responses. Membrane bound NAC proteins have been implicated as major players in biotic and abiotic stress response affects major physiological processes like flowering (Kim et al , 2007), seed germination (Kim et al, 2008), leaf senescence (Lee et al , 2012) and also cell division (Kim et al., 2006). Considering the varied functions of membrane bound NAC genes in crops, the identification of four membrane bound NAC genes would be useful in understanding their specific function in Peanut. Gene ontology annotation reveals that, a majority of these proteins were predicted to be involved in response to stress as well as cellular, metabolic and biosynthetic processes. The molecular functions of these proteins corresponded to transcription regulator activity. Further, cellular component analysis revealed the localization of these gene products in nucleus. Multiple sequence alignment and identification of conserved motifs using MEME tool indicates that most of the NAC proteins possessed A to E subdomains in the N termini that conferred the DNA-binding activities. The motif composition of these NAC sequences may provide clues for further functional analysis of these TFs. However, the biological significance of most of the putative motifs remains to be elucidated.
everal reports demonstrated that NAC genes were involved in regulating plant development at different growth stages causing us to further associate the biological functions of NAC genes (Wang et al , 2013). In Arabidopsis, ANAC002/ ATAF1 was induced by longterm treatment with ABA and/ or during age-dependent senescence. ANAC019, ANAC055, and ANAC072 in Arabidopsis showed up-regulation at transcription levels after drought, high salinity and abscisic acid (ABA) treatments, and those over-expression results in increased tolerance to drought (Tran et al , 2004). In Glycine max, GmNAC2, GmNAC3 and GmNAC4 were strongly induced by osmotic stress. GmNAC3 and GmNAC4 were also induced by ABA, JA and salinity but differed in their response to cold (Guilherme et al., 2009). In addition, GmNAC2-overexpressing tobacco lines were developed and found to be hypersensitive to drought, high salinity, and cold stress indicating GmNAC2 functions as a negative regulator during abiotic stress, and participates in RO signaling pathways through modulation of the expression of genes related to RO -scavenging (Jin et al , 2013). TaNAC2L over-expression activated the expression of heat-related genes in the transgenic Arabidopsis, suggesting that TaNAC2L may improve heat tolerance by regulating the expression of stress-responsive genes (Guo et al , 2015). In Rice, SNAC3 was ubiquitously expressed and its transcript level was induced by drought, high temperature, salinity stress, and abscisic acid (ABA) treatment. Over-expression of SNAC3 in rice resulted in enhanced tolerance to high temperature, drought, and oxidative stress caused by methyl viologen (MV), whereas suppression of SNAC3 by RNAi resulted in increased sensitivity to these stresses (Fang et al., 2015). AdNAC21 and AiNAC3 were found to be up-regulated under both drought and high temperature stress, suggesting these genes may be positive regulators of abiotic stress responses in Peanut. Overall, qRT-PCR analysis demonstrated that all the genes displayed variations in their expression behavior in response to drought and high temperature stress, suggesting these genes may be involved in regulation of stress responsive genes and further characterization will help in understanding their role in imparting abiotic stress tolerance in Peanut.
CONCLUSIONS
The NAC TFs has been proposed as important arbitrators of various plant processes and have been subjected to intensive characterization, especially in well-known model plants. Our study to identify and characterize NAC TFs in Peanut genome using genome-wide survey, expression analysis coupled with molecular tools provides foundation of our understanding of their regulatory roles. The comprehensive genome-wide analysis led to identification of 76 NAC TF genes. A uniform nomenclature was provided to the identified genes and proteins, followed by their comparative phylogenetic analysis with Arabidopsis and Glycine max NAC TFs. Functional conservation within a sub-family and serves as an initial platform in facilitating a better understanding of the structure-function relationship between individual members. The variability in gene expression patterns implies that NACs may regulate a complex web of pathways to perform different physiological functions for acclimatizing towards multiple challenges. This understanding will prove useful in improving drought and high temperature tolerance since these differentially expressed genes are probably involved in abiotic tolerance in Peanut. Thus, the analysis provides preliminary indications of putative function of several Peanut NAC genes, which would help in channelizing directional efforts for their functional characterization.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.
ACKNOWLEDGEMENT
This work was supported by ERB (Reference no: B/EMEQ- 141/2013), New Delhi, India


