Wheat genome sequence opens new opportunities to understand the genetic basis of frost tolerance (FT) and marker-assisted breeding in wheat (Triticum aestivum L.)
Автор: Kumar Pardeep, Patni Babita, Singh Machiavelli
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 1 т.18, 2022 года.
Бесплатный доступ
The population is increasing day by day and agricultural land is occupied by urban cities because of the shortage of residential areas and by increasing the industries the pollution is at peak level that causes climate change and uses of chemical fertilizers the soil fertility is decreased. Due to these environmental conditions the overall yield of cereals crops is reduced. The wheat is a major food source for all over the world. But the production of wheat nowadays is more far from the requirement. After the availability of whole genome sequence of bread wheat opens many new opportunities for scientists. This whole genome helps in deep analysis and to formulate new breeding technology and to develop an advanced variety of bread wheat that fulfills the need of requirement. The environment conditions (abiotic and biotic stresses) have a severe impact on wheat growth and development that reduces the overall yield. In winter wheat the cold stress causes delay in the fruiting process and break in growth. After the availability of the whole genome of wheat the deep analysis showed the presence of some frost tolerance genes. The over expression of these genes in Arabidopsis showed the increase in frost tolerance. This deep analysis helps breeders to come up with more stress tolerance variety. It also helps in finding new molecular markers that help in marker assisted breeding and in speed breeding. The advancement of Genomic and Proteomic drive the crop biotechnology to resolve the shortage of food for a large population.
Wheat, genome sequence, frost tolerance
Короткий адрес: https://sciup.org/143178335
IDR: 143178335
Текст научной статьи Wheat genome sequence opens new opportunities to understand the genetic basis of frost tolerance (FT) and marker-assisted breeding in wheat (Triticum aestivum L.)
The genomics are becoming day by day more advanced due to whole genome sequencing technology being developed. Therefore, availability of genomic sequence of any organisms provides more deep insights for understanding the phenotype and genotype to reveal the mystery of various phenomenon’s. The population size day by day is increasing at a very high rate and it causes more severe problems like availability of food, health and many other concerns but availability of food is a major concern in front of us, to ensure food for everyone requires deep analysis of genome sequences of crops.
The Triticum aestivum is a major crop all over the world to provide food to a huge population. The availability of the genome sequence of Triticum aestivum resolved the many obstacles to understanding the problem on genetic basis. The re-sequencing of genes may help in deep understanding of various metabolic pathways of wheat. The Triticum aestivum ( bread wheat) is a winter crop and in the winter season the temperature falls below the level of requirement for growth of plants. The low temperature causes various types of abiotic stress that reduce the yield of crops and that is also a major concern to reduce the abiotic stresses and enhance the net production of crops.
The study of the genomic sequence of Arabidopsis provides a good number of genes that are activated during cold stress. The homology of these genes is also conferred in Triticum aestivum so it is a good opportunity for scientists to tackle the obstacle of cold stress in wheat. The wheat has the largest genome sequence, the deep analysis of this sequence may open various opportunities. The mystery of cold stress resistance lies deep inside of genes to resolve this mystery requiring deep analysis of genomic sequence.
The tolerance of cold stress depends on various metabolic pathways and activation of many chemical molecules. It is studied that the cold signal in plants is conveyed to activate CBF-dependent (C-repeat binding factor-dependent) and CBF independent transcriptional pathway, of which CBF-dependent pathway activates CBF regulator. These mechanisms required expression of numbers of genes simultaneously. To develop the resistance to cold stress crops have been adapted by SNPs and various other pathways but it is not enough to deal with the requirement, so it is required over expression of responsible genes. The technologies are already more advanced and require only deep knowledge and analysis of genomic information to tackle obstacles like cold stress tolerance.
Mystery of largest genomes.
To understand the complexity of wheat genome the genomics is not enough; it required various other perspectives like botany, archeology and a knowledge of the geographical distribution of the various grass species that have some contribution to its evolution and cytogenetics also required to understand the patterns of chromosomes.
Bread wheat is one of the major staple food crops in the world. In the world, China is the largest producer of wheat ( Figure. 1) . As per say the population is increasing the production should be increased every year. To achieve this goal the wheat genome plays a major significant role in improving nutrient quality and quantity.
The wheat has various ecotypes grown in the different seasons and adapted to climate change conditions. The wheat genome is very long (~17 gigabases, Gb) than major crops, which consist of more than 80% repetitive sequence Babben et al. , 2015). This makes the assembly of the genome very complex. It took many years to sequence the whole genome of bread wheat.
The International Wheat Genome Sequencing Consortium (IWGSC) established in 2005 to resolve the mystery of complex genomes. After establishing this consortium various project designs and many sequencing methodologies selected to open the largest genome. The first chromosome to be sorted was chromosome 3B Paux et al. , 2008). Within the two years, IWGSC published the draft sequence of Chinese Spring wheat based on chromosomes. Now all chromosome sequences of wheat are publicly available for further analysis. By the comparative analysis of the genomic data genome-wide sequence rearranged
Zimin et al, 2017). It is near to complete wheat genome assembly. The chromosome level, sequencing of wild emmer, the tetraploid ancestor of common wheat, was reported in July 2017 Avni et al. , 2017).
In the long past history two species of wild grass ( T. urartu and T. speltoide ) hybridized with each other to create emmer wheat( Triticum dicoccum ) and was tetraploide (2n = 4x = 28) chromosomes. After humans domesticated this plant and planted it in their fields, a third grass species ( Triticum turgidum ) inadvertently joined the mix. Nucleotide diversity in the AABB and DD genomes is substantially decreased than ancestral species, indicating a major diversity bottleneck on the transition to cultivated lines Brenchley et al. , 2012).This convoluted history has left modern bread wheat with three pairs of every chromosome, one pair from each of the three ancestral grasses. In technicality terms defined as hexaploid genome. The genomes of ancient wheat, such as emmer wheat, contain more of the DNA base pairs required to create proteins than that of humans. Domesticated hybrids, like bread wheat, are even larger. Furthermore, the DNA of ancient wheat contained a huge amount of duplication. This means that bread wheat not only contains an enormous size of genetic information, but that much of it is repeated. That makes decoding its genome complex. Aegilops tauschii Wholegenome sequence analysis revealed expansion in agronomically relevant gene families that were associated with disease resistance, abiotic stress tolerance. Aegilops tauschii genome served as the source for many grain quality genes in hexaploid wheat Guo-Liang Jiang, 2013).
How the hexaploid genome formed.
The Bread wheat (Triticum aestivum) is an allopolyploid developed from two separate hybridisation consequences. The first hybridisation, that takes place approximately 10,000 years ago, is thought to have been between the two grass species Triticum urartu (the genome donor A), and Triticum speltoides (the genome donor B). This new species would have been tetraploid (four complete genome complements) Hexaploid wheat arose as a result of a second hybridisation between the new tetraploid and a third diploid species, Aegilops tauschii (the genome donor D), whole process described by flow chart in Figure. 2. Again, chromosome doubling must have occurred to produce a fertile individual. This new species has 42 chromosomes; that is, six complete genomes each of 7 chromosomes.
The biotic and abiotic stress.
The both biotic and abiotic stress have various impacts on plant growth and development. The many plants serve as nutrient sources or food sources for humans and animals. The various crops are grown to get food. These crops considered as a major source of food include wheat, rice, cereals, and pulses. The wheat is the most important crop grown worldwide for food. The wheat has a long history of evolution and that helps it to compete with abiotic and biotic stresses. Bread wheat is a winter wheat so in winter temperature falls low sometimes and in some areas of earth temperature is very low that has a large impact on wheat and reduces the overall yield. But the wheat evolved and adapted to low temperature stress and the genomic evolution helped them to compete with cold stress. wheat responds and adapts to cold and low temperature stress to survive under stress conflicts at the molecular and cellular levels as well as at the metabolic levels.
Low temperature has various impacts on plant growth and metabolism, which reduces the significant crop yield. Plants differ in their tolerance to chilling (0-15 ºC) and freezing (< 0ºC) temperatures. The plants from various geographic conditions have adapted to compete with the specific stresses. Those plants that survive in cold conditions have some chilling tolerance and they can increase their cold stress tolerance by rearrangement of genetic makeup or by recombination to overexpress specific genes. Thus, in current analysis of genomic data of various crops showing changes in expression of genes, composition of membrane. To reduce the impact of frost stress, wheat requires acclimatization to low temperatures, which prevents premature transition to the reproductive phase, and this must happen before the threat of freezing stress during winter Chinnusamy et al. , 2007).
The genomic study of model plant Arabidopsis thaliana conferred the expression of some genes to compete with cold stress. TaMYB56 (on chromosomes 3B and 3D) in wheat was identified as a cold stress- related gene Zhang et al., 2012). The expression rate of TaMYB56-B and TaMYB56-D were highly induced by cold stress, but lightly induced by salinity stress in wheat. Deep analysis of the Arabidopsis transgenic plants that overexpressed TaMYB56-B revealed that TaMYB56-B is conferred involvement in responses to freezing and salt stresses. The expression of some cold stress-responsive genes, such as DREB1A/CBF3 and COR15a, were found to be upregulated in the TaMYB56-B-overexpressing Arabidopsis plants compared to wild-type Zhang et al., 2012). TaMYB3R1 is another MYB gene which has been potentially involved in wheat response to drought, salt and cold stress Cai et al., 2011) TaMYB2A transgenics had enhanced tolerance to abiotic stress, which were confirmed by the up-regulated expressions of abiotic stress-responsive genes. TaMYB2A is a multifunctional regulatory factor. Thus TaMYB2A has the potential for utilization in transgenic breeding to improve abiotic stress tolerances in crops Mao et al, 2011). The low-temperature also induced TaNAC4 expression, suggesting a role of Ta-NAC4 as a transcriptional activator during biotic and abiotic stresses responses in wheat Xia et al., 2010).
The cold stress causes delay flowering during reproduction. The reproductive phase meiosis is more susceptible than the vegetative, while male reproductive organs have severe harm than to female organs. The entire processes of reproduction and maturation of seeds are susceptible to cold stress and also cause dysfunction on several metabolic processes such as photosynthesis, water and mineral intake, respiration, and fresh and dry biomass (Farooqi et al. , 2018). Under the cold stress plants compete to modulate expression of various genes controlling cell membrane lipid composition, mitogen-activated protein kinase cascade, total soluble proteins, polyamines, glycine, betaine, proline, reactive oxygen species (ROS) scavengers, cryoprotectants, and a large number of cold responsive factors John et al. , 2016).
Freezing stress significantly alters gene expression profiles of more than 400 wheat genes, and some of the up-regulated genes encode kinases, phosphatases, calcium trafficking-related proteins and gly- cosyltransferases in the crown tissues of cold-acclimated plants Skinner et al., 2009).
The plant protein phosphatase 2Cs (PP2Cs) play significant roles in phytohormone signaling pathways, developmental processes, and both biotic and abiotic stress responses. All TaPP2C genes showed varied expression patterns after cold treatment and many genes in group A were significantly up-regulated after drought and cold treatments but not in response to heat treatment, both conserved and divergent expression patterns of PP2C genes exist not only in wheat, but also between wheat and Arabidopsis. By the method of genome-wide analysis of the PP2C gene family in hexaploid wheat, the 257 TaPP2C gene homoeologous were identified and were phylogenetically classified into 13 groups (Yu et al,. 2019). By the method of screening wheat cDNA yeast library, and recognized 4,695 freezing-related genes, 2,641 saltiness stress-related genes, and 2,771 osmotic pressure related genes. The quality capacity in stress resistance was affirmed by investigation of a covering quality TaPR-1-1 distinguished in each of the three pressure medicines through overexpression in Arabidopsis and yeast Wang et al. , 2019). TaAREB3 gene articulation was instigated with abscisic acid (ABA) and low temperature stress, and its protein was restricted in the nucleus when transiently expressed in tobacco epidermal cells and stably expressed in transgenic Arabidopsis. Overexpression of TaAREB3 in Arabidopsis improved ABA affectability, yet in addition fortified dry spell and freezing resistance. TaAREB3 likewise activated RD29A, RD29B, COR15A, and COR47 by binding to their promoter regions in transgenic Arabidopsis. These outcomes showed that TaAREB3 assumes a significant job in dry spell and freezing resistances in Arabidopsis Wang et al. ,2016).
The conventional breeding has not been fully successful in developing cold tolerant crop cultivars. However, biotechnological, and molecular basis approaches, including whole genome sequencing and modification of transgenic development, provide an insight to understand and access the complex cold tolerance process functional at the central dogma John et al., 2016). A few pressure responsive qualities have been distinguished and effectively brought into other yields to make transgenic crops with upgraded pressure resilience. In any case, it is imperative to call attention to here that during the improvement of a transgenic crop assortment, care is taken to present qualities that bring about improved resistance to numerous burdens, explicitly at the entire plant level. This requires the advancement of sets of markers planned to improve pressure resilience Ahanger et al., 2017).
Frost tolerance in wheat
The CBF regulatory group has a major role in configuring the low-temperature transcriptome and in conditioning freezing tolerance Thomashow et al. , 2010). The genetics studies on frost tolerance in wheat ( Triticum aestivum L.) reveal new highly conserved amino acid substitutions in CBF-A3, CBF-A15, VRN3 and PPD1 genes Babben et al,. 2018). The frost is a formation of an ice layer on the leaves of plants and it is a complex trait regulated by a number of genes and many gene families. The availability of the genomic sequence of bread wheat opens new opportunities for analyzing diversity of responsible genes for frost tolerance.
The induction of the Cor (cold-responsive)/Lea (late-embryogenesis-abundant) gene family is important process for development of freezing tolerance and QTLs for ABA sensitivity do not correspond to Fr-1 and Fr-2, and the two Fr loci are postulated to act independently of ABA signal transduction pathways. There-fore, multiple signal pathways are involved during cold acclimation to develop freezing tolerance Yokota et al. , 2015). The expression QTL region for Cor/Lea and CBF genes on 5AL, which play significant roles in developing freezing tolerance in common wheat Motomura et al. , 2013).
The mechanisms of signaling and transcriptional regulation of cold stress
The frost tolerance is a complicated biological pathway and consists of two metabolic reaction chains regulated by many genes. The rigidification of membranes is a type of signal induced by cold stress and in this process the Ca2+ influx is activated and recognized by calcium binding proteins (CBPs). The activation of this process leads to the activation of inducers of CBF promoters and also results in expression of responsible genes. The transcription factor CBF bind to the C-repeat/dehydration-responsive element and induce the expression of cold-responsive/late embryogenesis abundant genes Winfield et al., 2010). The frost tolerance involved flowering pathways that consist of vernalization and photoperiod response genes that contributed to low temperature acclimatization Dhillon et al., 2010). Translation factors (TFs) are viewed as the most significant controllers that control genes and gene bunches. Plants have developed modern stress reaction techniques, and genes that encode translation factors (TFs) that are ace controllers of stress-responsive genes are a significant possibility for crop improvement (Baillo et al., 2019).
Status of frost tolerance in wheat.
The frost resistance 1 (FR1) and frost resistance 2 (FR2), were two important loci identified on the long arm of chromosome 5A of wheat Kocsy et al. , 2010). The independence of the FR1 gene not analyzed whether it is based or not on a pleiotropic effect of VRN1. There are many homologous VRN1 genes and their impact on vernalisation are based on their dominance/recessive nature. The dominance of VRN-A1 makes the spring wheat and recessive traits (vrn-A1alleles) make winter wheat. This difference is characterized by only the difference of C/T single nucleotide polymorphism (SNP). The winter wheat has a special nucleotide y (C/T0 are conferred higher frost tolerance than genotype having VRN-B1 or VRN-D1 allele Chen et al. , 2010, Eagles et al. , 2011). The various analysis of wheat genome conferred FR-A2 most significant locus of chromosome 5A involved in wheat frost tolerance [10]. This locus consists of approximately eleven CBF genes located nearly 30cM proximal to VRN1 Båga et al. , 2006).
Two independent analyses illustrate that the FR-A2 locus has CBF-A3 gene, having major significance in wheat frost tolerance (Yu et al., 2019). The other one analysed the FR-Am2 locus of diploid T. monococcum and identified three CBFs (CBF12, CBF14 and CBF15) expressed in wheat frost tolerance Miller et al., 2005, Vágújfalvi et al., 2005). (Soltesz et al., 2013) confirmed this for Triticum aestivum. Additionally, Sutton et al., 2009) conferred three genes, Tacr7 (Triticum aestivum cold-regulated 7), Cab (calcium-binding EF-hand family protein-like) and Dem (Defective embryo and meristems) being differentially expressed during cold stress in wheat. The transcriptome level frost tolerance signaling is much more complex and Hundreds to thousands of wheat genes were identified to be significantly up- or downregulated under low temperature Knox et al., 2008). Genome-wide associations contemplate recognized regions in chromosomes 4A and 6A related with higher scopes and freezing resilience, separately. Landraces with freezing resistance might be valuable in growing new germplasm as novel wellsprings of more prominent cold toughness Sthapit et al., 2018).
TOP 10 WHEAT PRODUCING NATION FROM 2000TO 2018 (FAOSTAT 2020)
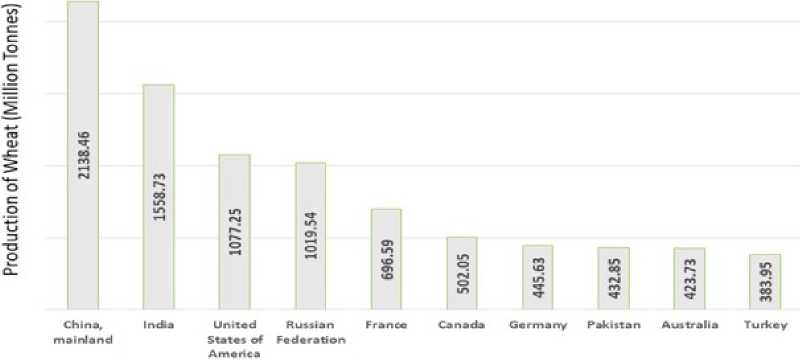
Figure 1. The production of wheat: Top 10 producing nations (FAOSTAT, 2020).
Markers assisted breeding
The techniques of developing or improving the crops takes a very long time that are not compatible with the requirements of foods. Biotechnology reduced the time and with the advanced technology the crops were modified within the 4-5 years. The genomic selection of desired phenotype and genotype are isolated and inserted in crops for improvement of nutrition value and also yield. The environment conditions (abiotic and biotic stresses) have a severe impact on crops that reduce the overall production. The naturally crops adapted to tackle these stresses but they still have large impacts.
To tackle the abiotic stress required advancement of technology. Molecular biology is the major field that deals with this kind of situation. The marker assisted breeding, a technique that helped the scientist to develop the crops that fulfill the requirement of food.
MAS is a strategy where phenotypic determination is made based on the genotype of a marker. MAS is a molecular breeding procedure that assists with staying away from the troubles worried about conventional plant breeding. CRISPR innovation has reformed the plant breeding and genetics and scientists are concentrating on altering the genomes of all financially significant plants. The coming years are probably going to see proceeds with developments in molecular marker innovation to make it increasingly exact, profitable and cost viable so as to research the basic science of different attributes of intrigue Nadeem et al., 2017).The molecular markers are strings or segments of DNA located near the desired genes. The presence of desired genes in plants limits the scope of genetic engineering because desired genes are present within the genome of plants.
Marker-assisted breeding should be empowered through the recognizable proof of powerful QTLs, the plan of dependable marker frameworks to choose for these QTLs, and the conveyance of these QTLs into tip top genomic foundations to empower their utilization without related hereditary drag Cobb et al., 2018).
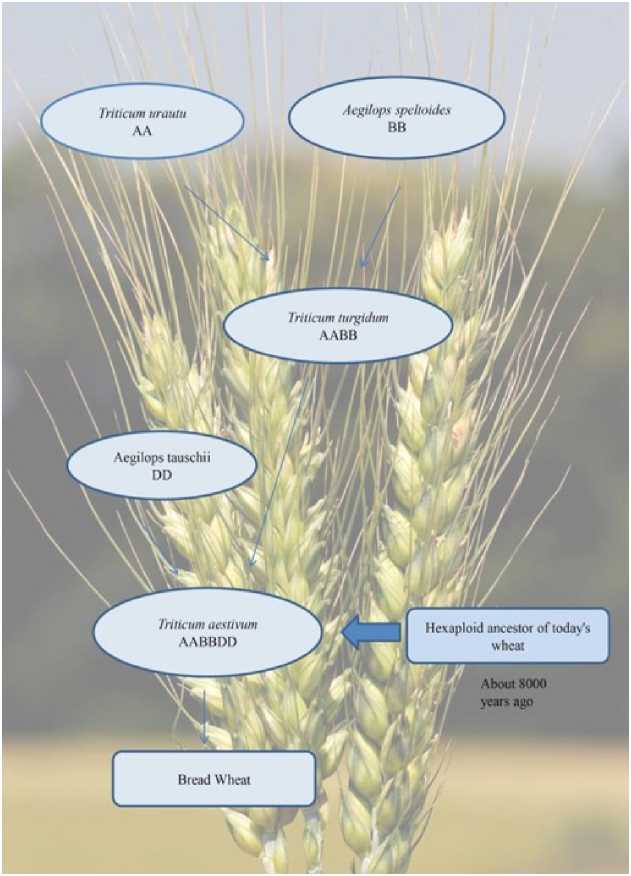
Figure 2. The historical development of bread wheat
The improvement of traditional breeding techniques requires molecular breeding or Marker-assisted selection. By the help of molecular markers, the desired trait improved in early generations of the breeding process. The benefits of molecular markers are they are not affected by environment conditions and can be detected in every stage of plants. The genetic linkage between loci is a major focusing area of marker-assisted selection. The closely related loci are always inherited together. Molecular markers are focused at sight on chromosomes that are closer to desired genes. This process is based on recognizing different polymorphism between various individuals. The genetic linkage highlights that the linked allele is present or not.
Speed breeding
To improve the profitability and steadiness of crops, there is a need to fast track research and increment the pace of assortment advancement. The generation time of most plant species speaks to a bottleneck in applied analysis projects and breeding, making the requirement for innovations that quicken plant advancement and generation turnover. The 'Speed breeding' (SB) abbreviates the reproducing cycle and quickens crop research through fast generation advancement. SB can be performed from various perspectives, one of which includes increasing the duration of plants' every day exposure to light, joined with early seed collect, to cycle rapidly from seed to seed, along with the decreasing generation times for some long day (LD) or day-neutral crops Ghosh et al., 2018 .
The CRISPR/Cas framework currently empowers promising new chances to make genetic diversity for breeding in an exceptional manner. Because of its multiplexing capacity, different targets can be changed at the same time in an effective manner, empowering prompt pyramiding of numerous helpful characteristics into a tip top foundation inside one generation. By focusing on regulatory components, a selectable scope of transcriptional alleles can be created, empowering exact calibrating of alluring genes. The calibrating of gene expression and de-novo domestication currently give plant raisers with energizing new chances to produce genetic diversity for rearing in a phenomenal manner Wolter et al. , 2019).
CONCLUSION
The bread wheat has the largest and complex genome of three ancestral species. The past year's hybridisation of these species makes the wheat genome (AABDD) very complex and very long in length. In recent past years the advancement of sequencing technology opens this complex mystery and created many opportunities for next generation breeders (marker assisted breeding and speed breeding). Bread wheat is a major food source for all over the world and China is the largest producer of wheat, after this the large population not having enough food so it requires advancement of analysis of molecular biology of wheat for reaching the production requirement.
The wheat has many varieties all over the world according to the environment conditions. The environment conditions have many impacts on plant growth and development that reduce the overall yield.
The abiotic and biotic stress defined the environment conditions of crops. In India the winter wheat is cultivated all over the nation but in the winter season for a short time temperature falls very low causing the cold stresses or frost tolerance.
More accurate and precise phenotyping procedures are important to enable high-goals linkage mapping and genome-wide affiliation reads and for preparing genomic determination models in plant improvement. Powerful phenotyping frameworks are expected to portray the full set-up of hereditary components that add to quantitative phenotypic variety across cells, organs and tissues, formative stages, years, situations, species and examination programs.
The conventional breeding technology in current advancement of technology does not fit well because it is not able to reach its goal in a short time. To tackle this problem requires a molecular approach that helps to increase the overall yield. Marker-assisted breeding is one of technology that fits well in this framework. The molecular markers help in finding important QTLs in a plant's genome. The identification of various genes expressed during the stressful conditions are a major concern of today’s scientists. This thought helps in developing better varieties of wheat that fulfill the need of large production.
ACKNOWLEDGMENT
The authors acknowledge the information shared in the webinar series “Next generation Genomics and Integrated Breeding for Crop Improvement (VII-NGGIBCI) workshop on Genomics for food, health and nutrition organized by ICRISAT's Center of Excellence in Genomics and Systems Biology (CEGSB) on 14 May, 2020.
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.
Список литературы Wheat genome sequence opens new opportunities to understand the genetic basis of frost tolerance (FT) and marker-assisted breeding in wheat (Triticum aestivum L.)
- Ahanger, M. A., Akram, N. A., Ashraf, M., Alyemeni, M. N., Wijaya, L., & Ahmad, P. (2017). Plant responses to environmental stresses—from gene to biotechnology. AoB PLANTS, 9(4). https://doi.org/10.1093/aobpla/plx025
- Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., Hale, I., Mascher, M., Spannagl, M., Wiebe, K., Jordan, K. W., Golan, G., Deek, J., Ben-Zvi, B., Ben-Zvi, G., Himmelbach, A.,
- MacLachlan, R. P., Sharpe, A. G., Fritz, A. Distelfeld, A. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science, 357(6346), 93-97. https://doi.org/10.1126/science.aan0032
- Babben, S., Perovic, D., Koch, M., & Ordon, F. (2015). An efficient approach for the development of locus specific primers in bread wheat (Triticum aestivum L.) and its application to re-sequencing of genes involved in frost tolerance. PLOS ONE, 10(11), e0142746. https://doi.org/10.1371/journal.pone.0142746
- Babben, S., Schliephake, E., Janitza, P., Berner, T., Keilwagen, J., Koch, M., Arana-Ceballos, F. A., Templer, S. E., Chesnokov, Y., Pshenichnikova, T., Schondelmaier, J., Börner, A., Pillen, K., Ordon, F., & Perovic, D. (2018). Association genetics studies on frost tolerance in wheat (Triticum aestivum L.) reveal new highly conserved amino acid substitutions in CBF-A3, cbf-a15, vrn3 and PPD1 genes. BMC Genomics, 19(1). https://doi.org/10.1186/s12864-018-4795-6
- Baga, M., Chodaparambil, S. V., Limin, A. E., Pecar, M., Fowler, D. B., & Chibbar, R. N. (2006). Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Functional & Integrative Genomics, 7(1), 53-68. https://doi.org/10.1007/s10142-006-0030-7
- Baillo, Kimotho, Zhang, & Xu. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes, 10(10), 771. https://doi.org/10.3390/genes10100771
- Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L., D'Amore, R., Allen, A. M., McKenzie, N., Kramer, M., Kerhornou, A., Bolser, D., Kay, S., Waite, D., Trick, M., Bancroft, I., Gu, Y., Huo, N., Luo, M.,
- Sehgal, S., Gill, B..... Hall, N. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature, 491(7426), 705-710. https://doi.org/10.1038/nature11650
- Cai, H., Tian, S., Liu, C., & Dong, H. (2011). Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene, 485(2), 146-152. https://doi.org/10.1016/j.gene.2011.06.026
- Chen, Y., Carver, B. F., Wang, S., Cao, S., & Yan, L. (2010). Genetic regulation of developmental phases in winter wheat. Molecular Breeding, 26(4), 573582. https://doi.org/10.1007/s11032-010-9392-6
- Chinnusamy, V., Zhu, J., & Zhu, J. (2007). Cold stress regulation of gene expression in plants. Trends in Plant Science, 12(10), 444-451. https://doi.org/10.1016/j.tplants.2007.07.002
- Cobb, J. N., Biswas, P. S., & Platten, J. D. (2018). Back to the future: Revisiting MAS as a tool for modern plant breeding. Theoretical and Applied Genetics, 132(3), 647-667. https://doi.org/10.1007/s00122-018-3266-4
- Dhillon, T., Pearce, S. P., Stockinger, E. J., Distelfeld, A., Li, C., Knox, A. K., Vashegyi, I., Vagujfalvi, A., Galiba, G., & Dubcovsky, J. (2010). undefined. Plant Physiology, 153(4), 1846-1858. https://doi.org/10.1104/pp.110.159079
- Eagles, H. A., Cane, K., & Trevaskis, B. (2011). Veery wheats carry an allele of Vrn-A1 that has implications for freezing tolerance in winter wheats. Plant Breeding, 130(4), 413-418. https://doi.org/10.1111/j.1439-0523.2011.01856.x
- Farooqi, M. Q., Zahra, Z., & Lee, J. K. (2018). Molecular genetic approaches for the identification of candidate cold stress tolerance genes. Cold Tolerance in Plants, 37-51. https://doi.org/10.1007/978-3-030-01415-5_2
- Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suarez, M., Simmonds, J., Wells, R., Rayner, T., Green, P., Hafeez, A., Hayta, S., Melton, R. E., Steed, A., Sarkar, A., Carter, J., Perkins, L., Lord, J., Tester, M..... Hickey, L. T. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nature Protocols, 13(12), 2944-2963. https://doi.org/10.1038/s41596-018-0072-z
- Guo-Liang Jiang (2013). Molecular Markers and Marker-Assisted Breeding in Plants, Plant Breeding from Laboratories to Fields, Sven Bode Andersen, IntechOpen, DOI: 10.5772/52583. https://www.intechopen.com/books/plant-breeding-from-laboratories-to-fields/molecular-markers-and-marker-assisted-breeding-in-plants
- John, R., Anjum, N. A., Sopory, S. K., Akram, N. A., & Ashraf, M. (2016). Some key physiological and molecular processes of cold acclimation. Biologia plantarum, 60(4), 603-618. https://doi.org/10.1007/s10535-016-0648-9
- Knox, A. K., Li, C., Vagujfalvi, A., Galiba, G., Stockinger, E. J., & Dubcovsky, J. (2008). Identification of candidate CBF genes for the frost tolerance locus Fr-A M 2 in triticum monococcum. Plant Molecular Biology, 67(3), 257-270. https://doi.org/10.1007/s11103-008-9316-6
- Kocsy, G., Athmer, B., Perovic, D., Himmelbach, A., Szucs, A., Vashegyi, I., Schweizer, P., Galiba, G., & Stein, N. (2010). Regulation of gene expression by chromosome 5a during cold hardening in wheat. Molecular Genetics and Genomics, 283(4), 351363. https://doi.org/10.1007/s00438-010-0520-0
- Mao, X., Jia, D., Li, A., Zhang, H., Tian, S., Zhang, X., Jia, J., & Jing, R. (2011). Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in arabidopsis. Functional & Integrative Genomics, 11(3), 445-465. https://doi.org/10.1007/s10142-011-0218-3
- Miller, A. K., Galiba, G., & Dubcovsky, J. (2005). A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-A M 2 in triticum monococcum. Molecular Genetics and Genomics, 275(2), 193-203. https://doi.org/10.1007/s00438-005-0076-6
- Motomura, Y., Kobayashi, F., Iehisa, J. C., & Takumi, S. (2013). A major quantitative trait locus for cold-responsive gene expression is linked to frost-resistance gene Fr-A2 in common wheat. Breeding Science, 63(1), 58-67. https://doi.org/10.1270/jsbbs.63.58
- Nadeem, M. A., Nawaz, M. A., Shahid, M. Q., Dogan, Y., Comertpay, G., Yildiz, M., Hatipoglu, R., Ahmad, F., Alsaleh, A., Labhane, N., Ozkan, H., Chung, G., & Baloch, F. S. (2017). DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment, 32(2), 261-285. https://doi.org/10.1080/13102818.2017.1400401
- Paux, E., Sourdille, P., Salse, J., Saintenac, C., Choulet, F., Leroy, P., Korol, A., Michalak, M., Kianian, S., Spielmeyer, W., Lagudah, E., Somers, D., Kilian, A., Alaux, M., Vautrin, S., Berges, H., Eversole, K., Appels, R., Safar, J..... Feuillet, C. (2008). A physical map of the 1-Gigabase bread wheat chromosome 3B. Science, 322(5898), 101-104. https://doi.org/10.1126/science.1161847
- Skinner, D. Z. (2009). Post-acclimation transcriptome adjustment is a major factor in freezing tolerance of winter wheat. Functional & Integrative Genomics, 9(4), 513-523. https://doi.org/10.1007/s10142-009-0126-y
- Soltesz, A., Smedley, M., Vashegyi, I., Galiba, G., Harwood, W., & Vagujfalvi, A. (2013). Transgenic Barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. Journal of Experimental Botany, 64(7), 1849-1862. https://doi.org/10.1093/jxb/ert050
- Sthapit Kandel, J., Huang, M., Zhang, Z., Skinner, D., & See, D. (2018). Genetic diversity of Clinal freezing tolerance variation in winter wheat landraces. Agronomy, 8(6), 95. https://doi.org/10.3390/agronomy8060095
- Sutton, F., Chen, D., Ge, X., & Kenefick, D. (2009). Cbf genes of the Fr-A2 allele are differentially regulated between long-term cold acclimated crown tissue of freeze-resistant and - susceptible, winter wheat mutant lines. BMC Plant Biology, 9(1), 34. https://doi.org/10.1186/1471-2229-9-34
- Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway: Figure 1. Plant Physiology, 154(2), 571-577. https://doi.org/10.1104/pp.110.161794
- Vagujfalvi, A., Aprile, A., Miller, A., Dubcovsky, J., Delugu, G., Galiba, G., & Cattivelli, L. (2005). The expression of several Cbf genes at the Fr-A2 locus is linked to frost resistance in wheat. Molecular Genetics and Genomics, 274(5), 506-514. https://doi.org/10.1007/s00438-005-0047-y
- Wang, J., Li, Q., Mao, X., Li, A., & Jing, R. (2016). Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. International Journal of Biological Sciences, 12(2), 257-269. https://doi.org/10.7150/ijbs.13538
- Wang, J., Mao, X., Wang, R., Li, A., Zhao, G., Zhao, J., & Jing, R. (2019). Identification of wheat stress-responding genes and tapr-1-1 function by screening a cDNA yeast library prepared following abiotic stress. Scientific Reports, 9(1). https://doi.org/10.1038/s41598-018-37859-y
- Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A., & Edwards, K. J. (2010). Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnology Journal, 8(7), 749-771. https://doi.org/10.1111/j.1467-7652.2010.00536.x.
- Wolter, F., Schindele, P., & Puchta, H. (2019). Plant breeding at the speed of light: The power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biology, 19(1). https://doi.org/10.1186/s12870-019-1775-1
- Xia, N., Zhang, G., Liu, X., Deng, L., Cai, G., Zhang, Y., Wang, X., Zhao, J., Huang, L., & Kang, Z. (2010). Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Molecular Biology Reports, 37(8), 3703-3712. https://doi.org/10.1007/s11033-010-0023-4
- Yokota, H., Iehisa, J. C., Shimosaka, E., & Takumi, S. (2015). Line differences in cor/Lea and fructan biosynthesis-related gene transcript accumulation are related to distinct freezing tolerance levels in synthetic wheat hexaploids. Journal of Plant Physiology, 176, 78-88. https://doi.org/10.1016/jjplph.2014.12.007
- Yu, X., Han, J., Wang, E., Xiao, J., Hu, R., Yang, G., & He, G. (2019). Genome-wide identification and Homoeologous expression analysis of PP2C genes in wheat (Triticum aestivum L.). Frontiers in Genetics, 10. https://doi.org/10.3389/fgene.2019.00561
- Zhang, L., Zhao, G., Xia, C., Jia, J., Liu, X., & Kong, X. (2012). Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic arabidopsis. Gene, 505(1), 100-107. https://doi.org/10.1016/j.gene.2012.05.033
- Zimin, A. V., Puiu, D., Hall, R., Kingan, S., Clavijo, B. J., & Salzberg, S. L. (2017). The first near-complete assembly of the hexaploid bread wheat genome, triticum aestivum. GigaScience, 6(11). https://doi.org/10.1093/gigascience/gix097


