A novel germline mutation of the PALB gene in a young Yakut breast cancer woman
Автор: Gervas P.A., Molokov A.Yu., Zarubin A.A., Ivanova A.A., Tikhonov D.G., Kipriyanova N.S., Egorov A.N., Zhuikova L.D., Shefer N.A., Topolnitskiy E.B., Belyavskaya V.A., Pisareva L.F., Choynzonov E.L., Cherdyntseva N.V.
Журнал: Сибирский онкологический журнал @siboncoj
Рубрика: Лабораторные и экспериментальные исследования
Статья в выпуске: 4 т.21, 2022 года.
Бесплатный доступ
Background. Breast cancer (Bc) is the most common female malignancy worldwide. partner and localizer of BRCA2 gene ( PALB2 ) is directly involved in dNa damage response. germline mutation in PALB2 has been identified in breast cancer and familial pancreatic cancer cases, accounting for approximately 1-2% and 3-4%, respectively. the goal of this report was to describe new PALB2 mutation in a young Yakut breast cancer patient with family history of cancer. material and methods. genomic dNa were isolated from blood samples and used to prepare libraries using a capture-based target enrichment kit, Hereditary cancer solution™ (sopHia geNetics, switzerland), covering 27 genes ( ATM, APC, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, FAM175A, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PIK3CA, PMS2, PMS2CL, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53 and XRCC2 ). paired-end sequencing (2 × 150 bp) was conducted using Nextseq 500 system (illumina, usa). Results. Here we describe a case of a never-before-reported mutation in the PALB gene that led to the early onset breast cancer. We report the case of a 39-year-old breast cancer Yakut woman with a family history of pancreatic cancer. Bioinformatics analysis of the Ngs data revealed the presence of the new PALB2 gene germinal frameshift deletion (Nm_024675:exon1:c.47dela:p.K16fs). in accordance with dbpubmed clinVar, new mutation is located in codon of the PALB2 gene, where the likely pathogenic donor splice site mutation (Nm_024675.3:c.48+1delg) associated with hereditary cancer-predisposing syndrome has been earlier described. Conclusion. We found a new never-before-reported mutation in palB2 gene, which probably associated with early onset breast cancer in Yakut indigenous women with a family history of pancreatic cancer.
Germline mutation, breast cancer, asian ancestry ethnic groups in Russia, yakut
Короткий адрес: https://sciup.org/140295754
IDR: 140295754 | УДК: 618.19-006.6:575.113.1 | DOI: 10.21294/1814-4861-2022-21-4-72-79
Текст научной статьи A novel germline mutation of the PALB gene in a young Yakut breast cancer woman
Breast cancer (BC) is the most common female malignancy worldwide. BRCA1 and BRCA2 are the most widely known breast cancer susceptibility genes. However, mutations in BRCA1/2 genes account for no more than half of all hereditary breast cancers [1]. DNA damage response genes, such as ATM, BRIP1, CDH1, CHEK2, NBN, NF1, PALB2, RAD51C, RAD51D and others are also involved in a significant number of hereditary breast and ovarian cancer syndromes. Cancer genetic advances allow identification of new mutations associated with breast cancer (BC). Next generation sequencing allows detection of DNA damage of five levels of pathogenicity: not pathogenic, likely not pathogenic, uncertain, likely pathogenic, and pathogenic. The interpretation of new variants is a major challenge, and its misinterpretation can lead to serious clinical mistakes for patients and their families [2, 3]. In addition, given the fact that BRCA gene mutations are ethnospecific, the search and interpretation of new mutations remains open for little-studied populations. For example, there are no reports concerning hereditary BC associated mutations in BC patients from Yakut ethnic group of Asian ancestry living in of the Far North. The Yakuts (Sakha) are a unique indigenous Turkic ethnic group who lives in the Republic of Sakha, which is the largest and coldest region of the Russian Federation (permafrost zone) (Fig. 1). The Yakuts make up about half of the Republic of Sakha population, the population of Yakutsk is about 330,000 (2020) on the 122 sq. km [4].
Results of a comprehensive assessment of breast cancer incidence among indigenous peoples and newcomers in Sakha Republic (Yakutia) were presented by L.F. Pisareva et al. (2007) and M.P. Kirillina et al. (2012) [5, 6]. The breast cancer incidence was found to decrease among indigenous peoples, whereas it tended to increase among newcomers. The breast cancer incidence rate was higher in nonindigenous residents of the Sakha Republic (Yakutia) than in representatives of the indigenous population (60 vs 40%). The disease was more common in women aged 50–59 years (32.6 %) and 60–69 years (21.8 %), regardless of ethnicity. The data obtained point to the most informative criteria for breast tumor malignancy: low expression of estrogen and progesterone receptors, high expression of Ki67 and p53, and low expression of bcl-2. In the published literature there are no data on the frequency of occurrence of BRCA1/2 gene mutations among patients with breast cancer of the Yakut ethnic group.
The goal of this report was to describe new PALB2 mutations in a young Yakut breast cancer patient with family history of cancer.
Material and Methods
The eligibility criteria were as follows: early age of onset and/or family history (2 or more close relatives with BC) and/or the presence of synchronous or metachronous multiple primary tumors [7]. Exclusion criteria included the presence of well-known BC-associated BRCA gene mutations.
Blood was collected in blood tubes containing K2EDTA. Genomic DNA was extracted from the peripheral blood lymphocytes using the phenol/chlo-roform method. DNA library were prepared using the Hereditary Cancer SolutionTM kit (Sophia GENETICS, Switzerland) to cover 27 genes, such as ATM, APC, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, FAM175A, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PIK3CA, PMS2, PMS2CL, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53,
Fig. 1. Republic of Sakha is a federal subject of Russia
Рис. 1. Республика Саха Якутия – субъект Российской Федерации and XRCC2. Paired-end sequencing (2 × 150 bp) was conducted using NextSeq 500 system (Illumina, USA) [8].
Bioinformatics analysis
Sequencing data were analyzed according to the GATK (Genome Analysis Toolkit) best practice recommendation for Whole Exome Sequencing using GRCh37 as a reference for Burrows-Wheeler alignment. The obtained variants were annotated with AN-NOVAR software and ranged according to population frequency (genomic exome, gnomAD genome, and ExAC), ClinVar, CADD, and literature data [9–11]. Detected sequence variants were annotated using PolyPhen2, Mutation Taster, and SIFT [12–14].
ProteinPaint tool was used to complement existing cancer genome portals and provide a comprehensive and intuitive view of cancer genomic data with advanced visualization features cloud/proteinpaint).
Results and Discussion
We report the case of a 39-year-old Yakut woman with invasive ductal carcinoma in the left breast, with a family history of cancer (mother with pancreatic cancer). Immunohistochemistry testing detected estrogen receptor (0), progesterone receptor (0), and positive HER2 (+). After 10 years of follow-up, she is alive with no evidence of disease progression.
Based on the NGS data, mutational analysis revealed the presence of the new germinal frameshift deletion (NM_024675:exon1:c.47delA:p.K16fs) in exon 1 of PALB gene (Fig. 2).
In accordance with dbPubMed ClinVar, a new mutation is located in codon of the PALB2 gene, where the likely pathogenic donor splice site mutation (NM_024675.3:c.48+1delG) associated with hereditary cancer-predisposing syndrome has been earlier described. The c.48+1delG intronic variant results from a deletion of one nucleotide within intron 1 of the PALB2 gene. This rare variant was not reported in population based cohorts in the databases as Database of Single Nucleotide Polymorphisms (dbSNP), NHLBI Exome Sequencing Project (ESP),
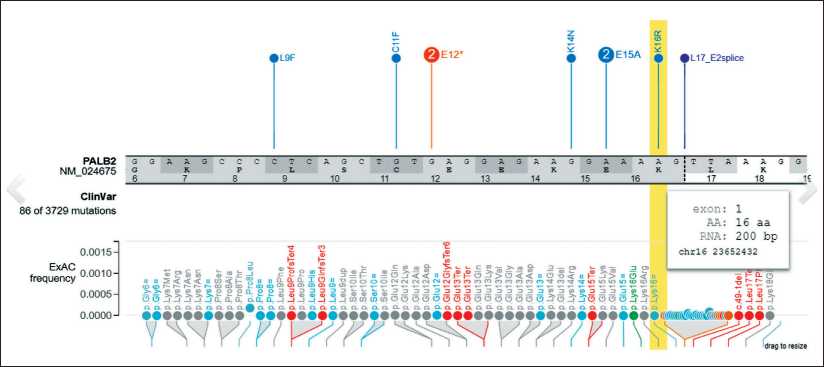
Fig. 2. Visualization of the position of germinal frameshift deletion (NM_024675:exon1:c.47delA:p.K16fs) in the PALB gene by
ProteinPaint tool (hg19)
Рис. 2. Визуализация местоположения новой мутации (NM_024675:exon1:c.47delA:p.K16fs) в гене PALB с помощью программы
ProteinPaint (hg19)
and 1000 Genomes Project and the NHLBI GO Exome Sequencing Project (ESP). This likely pathogenic donor splice site mutation has been detected with an allele frequency of approximately 0.001 % (more than 130000 alleles tested). In accordance to the BDGP and ESEfinder splice site prediction tools, it was reported that this alteration was predicted to destroy the native splice donor site. A splice site mutation is a genetic mutation that inserts and deletes or changes a number of nucleotides in the specific site at which splicing takes place during the processing of precursor messenger RNA into mature messenger RNA.
Thus, a new mutation of the PALB gene (NM_024675:exon1:c.47delA:p.K16fs) is located in a donor splice site. In accordance to ACMG Recommendations, alterations that disrupt the canonical splice donor site are typically deleterious in nature [15]. However, direct evidence is unavailable. In our study, this variant was classified by us as likely pathogenic, taking into account the early-onset BC woman with a family history of pancreatic cancer. Moreover, our data are consistent with those published in Gastroenterology Journal in 2009 and Fam Cancer Journal in 2011 and others [16, 17]. M.D. Tischkowitz et al. (2009) found that the prevalence of PALB2 mutation does not exceed 1 % in patients with pancreatic cancer not selected by family history. S. Jones et al. (2009) and E.P. Slater et al. (2010) found an overall prevalence of 3.1 % of PALB2 mutation in patients with familial pancreatic cancer [18, 19]. Thus, PALB2 gene mutations appear to be prevalent in patients with a family history of pancreatic cancer. E.W. Hofstatter et al. (2011) reported that PALB2 mutations occur with a prevalence of 2.1 % in a population of BRCA1/2 -negative breast cancer patients specifically selected for a personal and/or family history of pancreatic cancer [16, 17].
Partner And Localizer of BRCA2 gene (PALB2) is directly involved in DNA damage response. Germline mutations in PALB2 have been identified in breast cancer and familial pancreatic cancer cases, accounting for approximately 1–2 % and 3–4 %, respectively [20]. It is well known that homologous recombination (HRR) genes, such as ATM, BRIP1, CDH1, CHEK2 and others are involved in a significant number of hereditary breast and ovarian cancer syndromes [21]. Hereditary Breast and Ovarian Cancer Syndrome associated with BRCA1/2 mutations is the most common hereditary cancer syndrome. Patients who carry a mutation in BRCA1/2 genes are well respond to platinum chemotherapy (cisplatin) or PARP inhibitors (PARPi). The identification of molecular alterations in homologous recombination (HRR) genes is an area of active research to expand the use of PARPi. Several studies are ongoing to evaluate the efficacy of PARPi (ClinicalTrials.gov identifiers: NCT02401347 and NCT03330847) in women with germline or somatic mutations in HRR pathway genes including PALB2. T. Grellety et al. (2020) reported on the effective PARPi treatment of a PALB2-deficient breast cancer in a 42-year-old woman [22]. In our study, we report the case of a 39-year-old Yakut woman with a family history of pancreatic cancer and new detected likely pathogenic germinal mutation of PALB gene (NM_024675:exon1:c.47delA:p.K16fs). In the case of disease progression, the patient may be considered for the appointment of PARP inhibitors.
Conclusion
This report is the first to describe the new germinal frameshift deletion (NM_024675:exon1:c.47delA:p. K16fs) in the PALB gene in a Yakut BC patient with young-onset and familial BC. This mutation likely can be associated with Hereditary Breast and Ovarian Cancer Syndrome. Further studies are required to confirm pathogenicity of new frameshift mutation of the PALB gene in Yakut breast cancer patients.
Список литературы A novel germline mutation of the PALB gene in a young Yakut breast cancer woman
- Gifoni A.C.L.V.C., Gifoni M.A.C., Wotroba C.M., Palmero E.I, CostaE.L.V., Dos Santos W., AchatzM.I. Hereditary Breast Cancer in the Brazilian State of Ceara (The CHANCE Cohort): Higher-Than-Expected Prevalence of Recurrent Germline Pathogenic Variants. Front Oncol. 2022; 12. doi: 10.3389/fonc.2022.932957.
- Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., GreenblattM.S., HogervorstF.B., Hoogerbrugge N., Spurdle A.B., Tavti-gian S.V.; IARC Unclassified Genetic Variants Working Group. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008; 29(11): 1282-91. doi: 10.1002/humu.20880.
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L.; ACMGLaboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17(5): 405-24. doi: 10.1038/gim.2015.30.
- Kononova S., Vinokurova D., Barashkov N.A., Semenova A., Sofronova S., Oksana S., Tatiana D., Struchkov V., Burtseva T., Romanova A., Fedorova S. The attitude of young people in the city of Yakutsk to DNA-testing. Int J Circumpolar Health. 2021; 80(1). doi: 10.1080/22423982.2021.1973697.
- Kirillina M.P., Loskutova K.S., Lushnikova E.L., Nepomnyash-chikh L.M. Expression of molecular biological markers in breast cancer under conditions of the Sakha Republic (Yakutia). Bull Exp Biol Med. 2014; 157(5): 623-7. doi: 10.1007/s10517-014-2630-x.
- Писарева Л.Ф., Одинцова И.Н., Иванов П.М., Николаева Т.И. Особенности заболеваемости раком молочной железы коренного и пришлого населения Республики Саха (Якутия). Сибирский онкологический журнал. 2007; (3): 69-72. [Pisareva L.F., Odintsova I.N., Ivanov P.M., Nikolaeva T.I. Breast cancer incidence among indigenous peoples and newcomers in Sakha republic (Yakutia). Siberian Journal of Oncology. 2007; (3): 69-72. (in Russian)].
- Eccles D.M. Hereditary cancer: guidelines in clinical practice. Breast and ovarian cancer genetics. Ann Oncol. 2004; 15(4): 133-8. doi: 10.1093/annonc/mdh917.
- Slatko B.E., Gardner A.F., Ausubel F.M. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol. 2018; 122(1): 59. doi: 10.1002/cpmb.59.
- Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella, K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline, Curr Protoc Bioinformatics. 2013; 43(1110): 11.10.1-11.10.33. doi: 10.1002/0471250953.bi1110s43.
- DePristoM.A., BanksE., PoplinR., GarimellaK. V, Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKennaA., Fennell T.J., KernytskyA.M., SivachenkoA.Y., CibulskisK., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011; 43(5): 491-8. doi: 10.1038/ng.806.
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., GarimellaK., AltshulerD., Gabriel S., Daly M., DePristoM.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20(9): 1297-303. doi: 10.1101/gr.107524.110.
- AdzhubeiI.A., SchmidtS., PeshkinL., Ramensky V.E., GerasimovaA., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7(4): 248-9. doi: 10.1038/nmeth0410-248.
- Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. Mutation-Taster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014; 11(4): 361-2. doi: 10.1038/nmeth.2890.
- Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009; 4(7): 1073-81. doi: 10.1038/nprot.2009.86.
- Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E.; Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008; 10(4): 294-300. doi: 10.1097/ GIM.0b013e31816b5cae.
- Hofstatter E.W., Domchek S.M., Miron A., Garber J., Wang M., Componeschi K., Boghossian L., Miron P.L., Nathanson K.L., Tung N. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer. 2011; 10(2): 225-31. doi: 10.1007/s10689-011-9426-1.
- Tischkowitz M.D., SabbaghianN., HamelN., BorgidaA., Rosner C., Taherian N., Srivastava A., Holter S., Rothenmund H., Ghadirian P., Foulkes W.D., Gallinger S. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009; 137(3): 1183-6. doi: 10.1053/j.gastro.2009.06.055.
- Jones S., Hruban R.H., Kamiyama M., Borges M., Zhang X., Parsons D.W., Lin J.C., Palmisano E., Brune K., Jaffee E.M., Iacobuzio-Donahue C.A., Maitra A., Parmigiani G., Kern S.E., Velculescu V.E., KinzlerK. W., VogelsteinB., Eshleman J.R., GogginsM., KleinA.P. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009; 324(5924): 217. doi: 10.1126/science.1171202.
- Slater E.P., Langer P., NiemczykE., Strauch K., Butler J., Habbe N., Neoptolemos J.P., GreenhalfW., Bartsch D.K. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010; 78(5): 490-4. doi: 10.1111/j.1399-0004.2010.01425.x.
- Hanenberg H., Andreassen P.R. PALB2 (partner and localizer of BRCA2). Atlas Genet Cytogenet Oncol Haematol. 2018; 22(12): 484-90. doi: 10.4267/2042/69016.
- Practice Bulletin No 182: Hereditary Breast and Ovarian Cancer Syndrome. Obstet Gynecol. 2017; 130(3): 110-26. doi: 10.1097/ AOG.0000000000002296.
- Grellety T., Peyraud F., Sevenet N., Tredan O., Dohollou N., Barouk-Simonet E., Kind M., Longy M., Blay J.Y., Italiano A. Dramatic response to PARP inhibition in a PALB2-mutated breast cancer: moving beyond BRCA. Ann Oncol. 2020; 31(6): 822-3. doi: 10.1016/j. annonc.2020.03.283.


