A Novel Hierarchical Document Clustering Framework on Large TREC Biomedical Documents
Автор: Pilli. Lalitha Kumari, M. Jeeva, Ch. Satyanarayana
Журнал: International Journal of Information Technology and Computer Science @ijitcs
Статья в выпуске: 3 Vol. 14, 2022 года.
Бесплатный доступ
The growth of microblogging sites such as Biomedical, biomedical, defect, or bug databases makes it difficult for web users to share and express their context identification of sequential key phrases and their categories on text clustering applications. In the traditional document classification and clustering models, the features associated with TREC texts are more complex to analyze. Finding relevant feature-based key phrase patterns in the large collection of unstructured documents is becoming increasingly difficult, as the repository's size increases. The purpose of this study is to develop and implement a new hierarchical document clustering framework on a large TREC data repository. A document feature selection and clustered model are used to identify and extract MeSH related documents from TREC biomedical clinical benchmark datasets. Efficiencies of the proposed model are indicated in terms of computational memory, accuracy, and error rate, as demonstrated by experimental results.
Similarity, Retrieval, Clustering and Classification, Hierarchical Methods, Phrase patterns
Короткий адрес: https://sciup.org/15018437
IDR: 15018437 | DOI: 10.5815/ijitcs.2022.03.02
Текст научной статьи A Novel Hierarchical Document Clustering Framework on Large TREC Biomedical Documents
Published Online June 2022 in MECS
Three phases have been implemented to solve the traditional feature prediction problem. Keyword-based matching is used to identify TREC documents in the datasets. Similarity measures are characterized by consistency and conciseness. As a third measure, information retrieval techniques are used to judge the quality of text features. In order to explain the relationship between features and measures, linear mixed-effects regression models [1] are applied. The process of key phrase extraction traditionally attempts to summarize the peer documents by selecting relevant phrases and sentences. An inverse term frequency is a way of ranking a term or phrase based on characteristics that might be predefined in a peer report, like term frequency. The row-column matrix representation of a document collection is traditional in document keyphrase extraction services with each row representing a phrase or ID and the corresponding column representing a word or phrase. This method ignores the context information in the text and assumes that each phrase or sentence is independent of other documents. There are two classes of automatic topic keyphrase extraction methods: supervised and unsupervised. There are three ways of implementing the traditional information retrieval system: peer-to-peer initialization, topic clustering, and topic summarization [2]. It is primarily the developer's job to investigate documents in TREC systems as they are used to fix document classifications based on feature extraction. Identification of new TREC documents requires two vital tasks. As a first step [3], identify the features of the TREC training data.
There are a number of documents clustering techniques in use today that make use of feature vector spaces. These spaces are often used to train text clustering and classification models for documents. The documents within each feature vector space are all character vectors with terms that occur multiple times in each document collection set. There are words and phrases associated with every document feature vector [4]. The similarity is measured using document similarity metrics such as the Jaccard measure and the cosine measure, which are based on feature vectors or word frequencies. In the case of clustering techniques based on these vector spaces, each gram is interpreted separately. Neither word neighborhoods nor phrase-based clustering is employed.
This paper makes the following main contributions:
-
a) A clustering algorithm for gene/protein terms extraction was proposed for the TREC medical documents.
-
b) On the large biomedical TREC documents, we proposed an innovative mesh-based clustering approach.
-
2. Related Work
-
3. Methodology
The following is the outline for this paper. Text mining and biomedical documents are discussed in Section 2. Using a large TREC dataset, Section 3 illustrates a hierarchical clustering approach [5]. The results and analysis of the study are described in Section 4. In the final section of this paper, we conclude the discussion.
The modern approach to feature extraction attempts to model more sophisticated documents with a variety of methods. A number of the methods are developed from other NLP research areas and are only tailored for application to feature extraction. Feature extraction with Non-negative Matrix Factorization (NMF) appears to be one of the newest applications of NMF. In addition to focusing on subtopics, this method is also claimed to be quite effective. As the application of Latent Dirichlet Allocation (LDA) to feature extraction has been successful in many other related tasks, it has not previously been applied to this specific task [6]. Automated feature extraction has also been developed using graph-based approaches. Many of the early studies on graph-based feature extraction are based on research done in other areas of Natural Language Processing, particularly in Information Retrieval. This method was applied to this field because document feature extraction is the same as discovering the most relevant documents in a search engine query because extracting the most important sentences is similar to retrieving the most relevant documents. The first system, known as LexRank, measures similarity between sentences by analyzing multiple documents' content. Text Rank was also developed around the same time as graph-based feature extraction. In the same vein as LexRank, TextRank is based on PageRank and uses pointless graphs, however, unlike LexRank, TextRank is limited to single documents and is a general, unsupervised extractive feature extraction system as part of the initial tokenization, the text is annotated with parts-of-speech tags, but individual words are only considered as possible additions to the graph [7]. Using this proposed method, a text unit will recommend other text units that fit within the same theme. Recursively, the strength of the recommendation is calculated. Overlap is the number of tokens common to both sentences represented in their lexical form. To avoid biasing longer sentences, a normalizing factor is used. Training data is not required nor is a specific language preferred. Feature extraction is an unsupervised, extractive, and generic process. Its computation and stationary distribution are not changed, unlike PageRank, as the similarity graph between sentences has no direction [8]. The bag-of-words assumption means that LexRank cannot care about the order in which words appear in the document, but it does care about whether they are included in a document. Inverse Document Frequency (IDF) is used to calculate the similarity between two sentences [9]. LexRank ranked first in more than one task in the Document Understanding Conferences (DUC) evaluation. Utilizing Random Walks for Question-focused Sentence Retrieval significantly expands LexRank to be query-focused. The underlying algorithm remains the same, but individual sentences are obtained from complex news stories based on a specific question.
SVM is a classification scheme based on the SVM model. SVDD is one of the most popular clustering algorithms [10]. The SVDD algorithm traverses a high-dimensional feature space by mapping data objects. When mapping is performed, a nonlinear transformation function is used. Additionally, it contains methods for detecting outliers whose entire data set is arranged in a specific feature space. In TREC, document features are selected and categorized using a variety of IR methods with a limited set of documents. Among the intensive computational semantic methods are Latent Dirichlet Allocation and Latent Semantic Indexing. A straightforward method for lexical matching is the vector space model. When dealing with high-dimensional datasets, latent semantic indexing (LSI) and Latent Dirichlet Allocation (LDA) may perform inaccurately due to noisy information. The LDA method provides the ability to model documents within corpora as collections of topics, which is an effective improvement over other methods. The Probabilistic Latent Semantic Indexing (PLSI) method [11] is also aimed at predicting new documents based on previously unseen documents, but LDA uses a hidden random variable.
The TREC topics are the two basic methods like static and dynamic methods in localization of the TREC topics. By examining the execution trace, breakpoints, and program data, the dynamic approach looks for feature topics. A program that runs successfully or fails under certain inputs is examined, and its differences are determined. Because dynamic methods require those features to be run, they may not be able to handle errors spanning over small datasets. In static methods, the function is to detect a feature in textual documents using information retrieval. These are some examples of content: events, domain concepts, system attributes, features, and exceptions. They are independent of specific languages and are less expensive to compute than dynamic methods.
A large volume of data has been extracted from TREC sources, resulting in text reports that have been preprocessed and extracted. Lucene and Stanford NLP are used as preprocessing libraries in the proposed model. As part of pre-processing, the sentences and terms are separated, stop words are filtered, and stemming is done. Using stemming reduces vocabulary size and eliminates duplication, which may lead to verbs being duplicated. Noise is represented by pronouns, prepositions, and special characters. Mallet provided the list of stop words. The results of an
Information Retrieval approach will largely be influenced by how well the training files are interpreted, and how well the TREC report is analyzed. Normalizing texts, removing stops, and stemming texts are the three steps of preparing an information system. Pre-processed TREC reports and training files are created, and subsequently, analysis terms are created, followed by the calculation of similarity. During normalization, punctuation symbols are removed, terms are tokenized, and identifiers are split. Abstract Syntax Trees are used in the sources to parse identifiers. Combining, analyzing, and scoring are terms used to describe the method "combine Analyzed Score." Following that, all nonessential words from a stop-word list are eliminated, which reduces the number of noisy matches and increases accuracy. Examples Stemming is the repetition of words that are derived from a common root word. Includes the words "goes" and "going". Using the statistics collected and stored, the training files are indexed after preprocessing, these include the number of documents containing the topic being studied (DF), and the number of instances of the term within the document (TF). Inverse DF, called IDF, is used to compare the content of source files and TREC reports. TREC reports and training files can be represented vector-wise using IDF and TF.
A biomedical MeSH entity is extracted from the TREC repository of biomedical entities initially in the proposed model. Tokenization, stemming, and stop word removal are applied to all TREC biomedical documents. As shown in Figure 1, each document is pre-processed, and its MeSH features are extracted using the MeSH tagger.
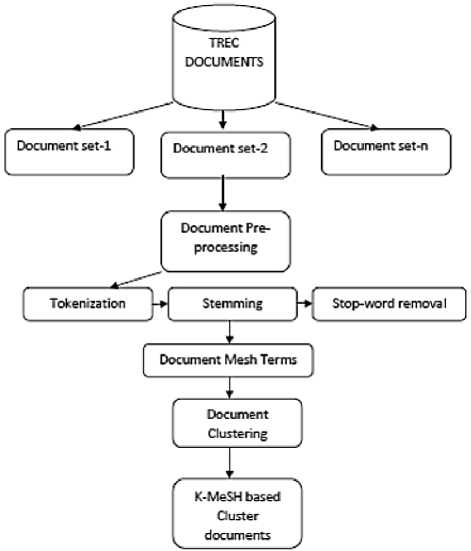
-
Fig.1. Block diagram of the Proposed Model.
Hybrid TREC Document Clustering Technique Algorithm:
Input: MeSH entities with biomedical significance Tgp, token-sets Tk, biomedical datasets Sen, * minimum threshold; Output: Clusters of MeSH documents.
T gp =Read (MeSH_Tags)
Tk= DatasetTokenize=DatasetTokenize for each tg in Tgp
Compute MeSH token probability
end
For each token t in Tk
For each s in Sentence-set
(&&()) ∈ > iftsgetprobt then
D ^ Pmid,Entropy_weight,
Sentence_id,token,Synonyms,
Data,Title,Positive Class
Else
D ^ Pmid,Entropy_weight,Sentence_id token,Synonyms,Data,Title,
Negative Class
End
End
For each feature-Pair in D
Dis tan ce ( CS i , CS j) = 0.5 x (1 - p ij )
d
Z ( csij - csi )( csii - cl
P = ii
i = 0
dd
E ( c s4 - cs i ) Z ( csii - csj)
i = 1 j = 1
-
a. Cluster pairs are initialized at level 0 with sequence no = 0 and sequence position = 0.
-
b. In order of smallest measurements to largest measurements of distance, rank the cluster pairs.
-
c. Step 1 should be repeated for each document and each cluster.
In the case where c >=0, o Based on all the clusters computed, the median node of the hierarchical tree is calculated.
o The median cluster object is used to divide the hierarchical tree into two halves; the left branch is the left branch and the right branch is the right branch.
o A method is developed for calculating the smallest disjoint cluster pairs on the right (rc) and left (lc) of a cluster.
o In order to calculate the distance between cluster pairs [(rc),(lc)], min[c(i),c(j)] should be used.
-
o A cluster with at least one common object is merged into one over all pairs of clusters if the left and right branches share a cluster object.
Else
Calculate the maximum dissimilarity between clusters
Maximum distance between points (rc), (lc) = cu(i),(j)
-
d. Add one to the count by using p = p+1. Put these two clusters into one cluster with level p. Level(p) = merge[lc,rc] specifying this cluster's level.
-
e. By deleting the old nodes associated with clusters lc and rc, create a new cluster named T for the merged node. A distance between two clusters of size (p, (nc,oc)) equals the minimum distance between them, d[(nc,(oc)] is the closest neighbor between new and old clusters of size (nc and oc). The distance between NC and OC is equal to d[(NC, OC)] represents the minimum distance between NC and OC.
Then, if the distance less than 0
All objects in the cluster should be merged together, stop.
Else go to step b.
A document contextual measure between a gene and a protein refers to the sum of the means square errors between the two similar pairs of genes and proteins in the merging process. The similarity cluster SSE is calculated by (3).
55E Z = ^ || x j - A ci |2 (3)
X j e C
Document Clustering Algorithm Using Hybrid TREC:
Input: MeSH entities with biomedical significance Tgp, token-sets Tk, biomedical datasets Sen, * minimum threshold;
Output: MeSH document clusters.
Input: The similarity measure is used to cluster hierarchical documents.
Output: Patterns in the top K.
Getclusters (T) = Cluster-set;
Cluster-set contains a cluster called c
Cluster 1=find genes and proteins in cluster (c).
Cluster c genes/proteins with their synonyms The getsync(c) for cluster c is C2.
-
C1 and C2 have similar gene/protein sequences.
Measuring similarity between genes and proteins:
∑
cos ine ( C 1, C 2)
C ∈ Cluster
C ∈ synkeyword
End for
Calculate the similarity between c1 and c2.
Use c1 and c2 pairs to retrieve biomedical abstracts from the biomedical repository.
4. Results and Analysis
The experiment [13] examines a large collection of TREC document sets collected from the TREC website. For the process of ranking documents, different TREC datasets are used, including Biomedical and Biomedical datasets A pre-processing procedure is used to remove undefined features and noisy content from the datasets. A graph-based sequential pattern mining algorithm is used to cluster and classify each document, following the pre-processing phase.
Table 1. On the TREC Medline data set, the Proposed Model was compared with what is typically used in the traditional document cluster.
Models
Biomedical
Sample set-1
Biomedical
Sample set-2
Biomedical
Sample set-3
Biomedical
Sample set-4
LDA Model
64.75
119.3
162.65
198.24
SVM+Kmeans
55.36
112.93
158.24
184.65
Proposed Model
30.64
100.25
132.64
162.45
Table 2. Cluster accuracy of Proposed Model with the Traditional document cluster models on Biomedical dataset.
Models
Biomedical
Sample set-1
Biomedical
Sample set-2
Biomedical
Sample set-3
Biomedical
Sample set-4
LDA Model
0.83
0.864
0.856
0.875
SVM+Kmeans
0.89
0.925
0.9154
0.935
Proposed Model
0.973
0.986
0.974
0.986
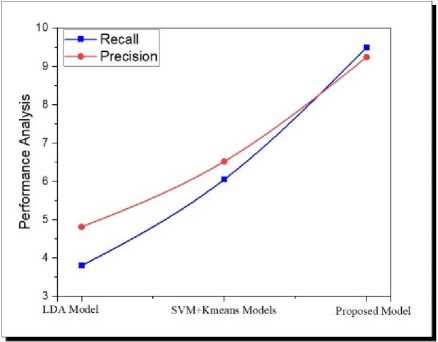
-
5. Conclusions
Fig.2. Compared to the existing models, the proposed model performance.
Fig.2 illustrates the comparative analysis of the proposed model to the existing models in terms of recall and precision are concerned. From Fig 1, it is clearly observed that the proposed model has high computational efficiency in terms of recall and precision than the existing models.
The present paper proposes a novel clustering model for the TREC Biomedical dataset. It is extremely difficult to manually process raw TREC datasets because they are so numerous. Hence, features extraction and context identification are the most relevant factors for mining interesting patterns on unstructured datasets. When processing large corpus data, it becomes more difficult to extract features and represent documents as the size of the document corpus grows. A big advantage of large clinical databases is their sparsity, as well as their ability to identify the evidence-based feature vectors. The traditional clustering and classification of TREC text documents is complex and difficult to assess its similarity to other documents. Since there are more and more unstructured documents in the TREC repository, finding feature-based key phrase patterns is becoming more challenging. Using large TREC data repositories, the authors propose and implement a novel hierarchical document clustering framework. TREC biomedical clinical benchmark datasets are used for finding and extracting MeSH related documents, using proposed document feature selection and clustering models. As demonstrated in Fig.2. Based on the experimental results, the proposed model is more accurate and has better computational memory.
Acknowledgment
Список литературы A Novel Hierarchical Document Clustering Framework on Large TREC Biomedical Documents
- W. Dai, G. Xue, Qi. Yang and Y. Yu, "Co-clustering based Classification for Out-of-domain Documents", Proceedings of the 13th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, pp.210-219, 2007.
- S. W. Chan and M. W Chong, "Unsupervised clustering for non-textual web document classification", Decision Support Systems, pp.377-396, 2004.
- D. Curtis, V. Kubushyn, E. A. Yfantis and M. Rogers, "A Hierarchical Feature Decomposition Clustering Algorithm for Unsupervised Classification of Document Image Types", Sixth International Conference on Machine Learning and Applications, pp.423-428, 2007.
- I. Diaz-Valenzuela, V. Loia, M. J. Martin-Bautista, S. Senatore and M. A. Vila, "Automatic constraints generation for semi-supervised clustering: experiences with documents classification", Soft Computing 20, no. 6, pp. 2329-2339, 2016.
- C. Hachenberg and T. Gottron, "Locality Sensitive Hashing for Scalable Structural Classification and Clustering of Web Documents", Proceedings of the 22nd ACM international conference on Information & Knowledge Management. ACM, pp.359-363, 2013.
- S. Jiang, J. Lewis, M. Voltmer and H. Wang, "Integrating Rich Document Representations for Text Classification", IEEE Systems and Information Engineering Design Conference (SIEDS '16), pp.303-308, 2016.
- W. Ke, "Least Information Document Representation for Automated Text Classification", Proceedings of the American Society for Information Science and Technology 49.1, pp.1-10, 2012.
- B. Lin and T. Chen, "Genre Classification for Musical Documents Based on Extracted Melodic Patterns and Clustering", Conference on Technologies and Applications of Artificial Intelligence, pp. 39-43, 2012.
- L. N. Nam and H. B. Quoc, "A Combined Approach for Filter Feature Selection in Document Classification", IEEE 27th International Conference on Tools with Artificial Intelligence, pp.317-324, 2015.
- S. Shruti and L. Shalini, "Sentence Clustering in Text Document Using Fuzzy Clustering Algorithm", International Conference on Control, Instrumentation, Communication and Computational Technologies (ICCICCT), pp.1473-1476, 2014.
- Michalis, "Clustering Validity Assessment: Finding the optimal partitioning of a data set", Data Mining, 2001. ICDM 2001, Proceedings IEEE International Conference.


