Detection MEC a gene and susceptibity some various antibiotic of Staphlococcus aureus
Автор: Al-karaawi A.S.A., Alhadrawi H.A.N.
Журнал: Cardiometry @cardiometry
Рубрика: Original research
Статья в выпуске: 30, 2024 года.
Бесплатный доступ
Through the study conducted in Najaf Governorate, bacterial samples were collected from different places. We notice during the study that the results were divided into two parts. The first was that the antibiotic Vancomycin had the highest rate of sensi-tivity to most of the bacterial samples, and this can be explained as follows. The bacterial isolates do not possess resistance genes or phenotypic factors that are highly effective against this antibi-otic. Therefore, the effect of the antibiotic was significant on most of the bacterial isolates. At the same time, some bacterial isolates showed resistance to the antibiotic Vancomycin. This can be ex-plained by the fact that these isolates possessed resistance genes or Phenotypic factors are resistant to this antibiotic, so resistance has been shown in some bacterial isolates. As for the second section, we note that the bacterial isolates had a very high rate of resistance to all other types of anti-biotics. This can be explained by the fact that these bacterial isolates possessed resistance genes, virulence factors, or both, which made them significantly more resistant to this antibiotic... and through review. Based on other research at the level of Iraq, as well as other countries, we notice that Staphylococcus au-reus bacteria is constantly evolving in resistance to antibiotics. We also note that the places where samples are collected have a role in resistance and sensitivity to antibiotics
Pcr, isolates, meca
Короткий адрес: https://sciup.org/148328215
IDR: 148328215 | DOI: 10.18137/cardiometry.2024.30.109113
Текст научной статьи Detection MEC a gene and susceptibity some various antibiotic of Staphlococcus aureus
Ali Salam Ali Al-Karaawi, Hazim A. N. Alhadrawi. Detection mec A gene and Susceptibity some various antibiotic of Staphlococ- cus aureus. Cardiometry; Issue No. 30; February 2024; p. 109113; DOI: 10.18137/cardiometry.2024.30.109113; Available from:
Staphylpococcus aureus is a major global public health problem causing serious, often life threatening infections in the community and hospital setting that are becoming more difficult to manage with current antibiotics therapy regimens ( Brea et al .,2010).
The pathogen Staphylococcus aureus is a gram-positive, gold-colored bacterium of which 20.8% is resistant to methicillin and all available β-lactam antibiotics [1-3]. About 20% of the population is always colonized with S. aureus, 60% are intermittent carriers, and 20% never carry the organism [4,5].
Staphylococcus aureus expresses many potential virulence factors: surface proteins that promote colonization of host tissues, Invasins that promote bacterial spread in tissues (leukocidin, kinases, hyaluronidase), Surface factors that inhibit phagocytic engulfment (capsule, Protein A), biochemical properties that enhance their survival in phagocytes (carotenoids, catalase production),
METHED
Specimens Collection
From August 2023 to February 2024, a total of 300 clinical specimens were collected during this study. Were taken from patients suffering from different infections and visited Al-Furat Al-Awsat, Al-Najaf Hospital and Al-Hakim General Hospital and the Central Laboratory. were aged from 1 to 60 years. a total of 300 clinical specimens were collected during this study From Patients who suffered from urinary tract infections( 80 sample), vaginal infections (50 sample), prostatitis (40 sample) and pyogenic infections namely postoperative wound infections(60 sample) and burn wound infections formed ( 70 sample). Swab was taken from infection area and then transported by transport media to the lab. Data about type of infection, sex and age of patients were recorded on the swab. Specimens were collected from patients who did not receive antibiotic for one week before sample. Midstream urine Samples were collected in sterile wide mouthed con- tainers and then streaked by loop on plates of blood agar and mannitol salt agar.
Bactrrial isolate:(Routine Test)
Primary Identification of S. aureus Isolate:
Staphylococcus aureus bacteria appear in mannitol Salt agar test If surrounding the growth the media has turned from pink to yellow, the bacteria has fermented the mannitol and produced acid. The acid lowers the pH of the media and the phenol red turns yellow. If yellow, the bacteria are positive for the ability to ferment mannitol. Catalase testThe test use distinction between Staphylococci and streptococci, a definitive classification can be made on the basis of the presence or absenc of the enzyme catalase. Staphylococci contain this enzyme, streptococci do not. Motility testing is very useful in the differentiation and identification of bacteria. Staphylococcus aureus is not motility .
Identification of S. aureus Isolates via VITEK2 System
VITEK 2 System is the following version of the gold standard of bacterial diagnosis and represents a new colorimetric technic. Procedure: All the subsequent measures were accomplished according to the producer’s protocol (Biomerieux) (Lee et al ., 2018):
-
1. In a test tube 3 ml of normal saline was applied, then adding colony by using loop.
-
2. The test tube was placed into a Dens Check device for modification of colony to McFarland standard solution (1.5×108 cell / ml)
-
3. The inoculum then implanted in the container and a sample detection number introduced into the software program by barcode.
-
4. Then read using barcode of VITEK 2 card formulas, was put on card through production and card was therefore attached to ID of sample.
-
5. The container was implanted in the filler element, after the cards were full, moved the container to incubator system for reading.
The following processes were accomplished by the instrument; the temperature of incubation is controlled by the instrument. The visual interpretation of the cards is persistently monitors and passes test 110 | Cardiometry | Issue 30. February 2024
outcomes to the device for assessment. When the assessment period is ended, the machine spontaneously expelled the cards into a leftover bin.
Antibiotic Susceptibility Testing
(Disk Diffusion Method)
Antibiotic Susceptibility Test was done according to (Bauer et al , (1966) by using a pure culture of previously identified bacterial organism and antibiotic disks listed in table [3-5]
-
1. With a sterile wire loop, the tips of 4-5 isolated colonies of the organism to test tube containing 5 ml of sterile normal saline in a cell density equivalent to turbidity of McFarland tube No. (0.5) which approximately equal to bacterial cells density of 1.5x108 cells/ml
-
2. A sterile cotton swab was dipped into the standardized bacterial suspension. The excess fluid was removed by rotating the swab firmly against the inside of the tube above fluid level. The swab was then streaked onto the dried surface of a Muller-Hinton Agar plate in 2 different planes to obtain an even distribution of the inoculums.
-
3. The plate lids were replaced and the inoculated plates were allowed to remain on a flat and level surface undisturbed for 3-5 min to allow absorption of excess moisture.
-
4. With the sterile forceps, the selected discs were placed on the inoculated plate and pressed gently into the agar. Within 15 min the inoculated plates were incubated at 37 ºC for 18 -24 hr in an inverted position.
-
5. After incubation, the diameters of the complete inhibition zone were noted and measured using reflected light and a ruler. The end point, measured to the nearest millimeter, was taken as the area showing no visible growth.
-
6. The results interpreted according to (CLSI, 2023), the critical diameters and to the leaflet of antibiotics manufactures.
Molecular Study virulence genes (MecA) genes. PCR technique was done.
Extraction of DNA
The S. aureus isolates after being cultured on mannitol salt agar and Brain Heart Infusion agar. The Genomic DNA Extraction Kit (Favorgen® Genomic DNA Mini Kit) was used for DNA extraction by manufacturer s protocol, as follow:
-
1. Transfer the appropriate number of bacterial cell (up to 1x 10^9) to a 1.5ml microcentrifuge tube (not provided) and centrifuge at speed 18,000 x g for 1 min. Discard the supernatant
-
2. Add 200 μl of lysozyme buffer (20 mg/ml lysozyme; 20 mM Tris-HCl; 2 mM EDTA: 1% Triton X-100, pH 8.0; prepare fresh lysozyme buffer immediately prior to use) and resuspend the pellet by vortex.
-
3. Incubate for 10 minutes at room temperature. During incubation, invert the tube every 2-3 min.
-
4. Add 200 μl of FABG Buffer to the sample and vortex for 5 seconds.
-
5. Incubate for 10 min at 70 °C until the sample lysate is clear. During incubation, invert the tube every 3 min.
-
6. Add 200 ul of ethanol (96-100%) to the sample and vortex for 10 sec, then placed a FABG column to a collection tube then transfer the sample mixture to FABG column and centrifuge at speed 18,000xg for 1 min.
-
7. Add 400 μl of W1 Buffer to the FABG Column and centrifuge for 30 sec at speed 18,000 x g. Discard the flow-through and place the FABG Column back to the Collection Tube.
-
8. Add 600 μl of Wash Buffer to the FABG Column and centrifuge for 30 sec at speed 18,000 x g. Discard the flow-through and place the FABG Column back to the Collection Tube.
-
9. Centrifuge for additional 3 min at speed 18,000xg to dry the column
-
10. then transfer the dry FABG column to a new 1.5 ml microcentrifuge tube.
-
11. Added 100 μl of Preheated Elution Buffer or TE to the membrane center of FABG Column.
-
12. Incubated the FAGB Column at 37 °C for 10 min in an incubator.
-
13. Centrifuged for 1 minute at full speed at 18,000 xg to elute the DNA.
-
14. Stored the DNA fragment at -20°C.
Conditions of PCR Program
|
Gene |
Temperature(°C) /Time |
Cycles number |
||||
|
Initial Denaturation |
Condition of one cycle |
Final Extension |
||||
|
Denaturation |
Annealing |
Extension |
||||
|
MecA |
95/5min |
95/30sec |
58/30sec |
72/1min |
72/5min |
35cycle |
RESULTS AND DISCUSSION
Bacteria were collected from August 2023 to February 2024 from different hospitals in Najaf Governorate and at different ages. Bacterial isolates were diagnosed by routine examination for the purpose of diagnosing Staphylococcus aureus bacteria, for example, Gram staining, as well as cultivation on menthol salt medium, and chemical tests. On this basis, the bacteria were diagnosed as Staphylococcus aureus bacteria. After that, we made a confirmatory diagnosis using the Vitic-2 device. Also, the results appeared identical to the routine examination and it was Staphylococcus aureus bacteria.
-
S. aureus related staphylococcal infection is one of the most common types of hospital-acquired infections, which requires selective and effective treatment in clinical practice [6].
From Figure No(1-2) we notice that the sensitivity rate of bacteria to the antibiotics used in the study was different if there was a difference in their sensitivity and resistance to the antibiotics used in the study, which included thirteen antibiotics. If the highest sensitivity rate was observed, it was from the share of the antibiotic Vancomycin, which reached 85%. While the antibiotics that followed, we note that they were insensitive to bacterial isolates,
From the figure ( 1– 1), we notice that the rate of bacterial resistance to different antibiotics was very high depending on the types of antibiotics. It was noted that the highest resistance rate reached 100% for the antibiotic Tetracycline, as well as the antibiotic Azithromycin. The resistance rate was also 100% for all bacterial isolates, as well as the antibiotic Erythromycin. The resistance rate of bacterial isolates was 100%, followed by the antibiotic Rifampin. The resistance rate of bacterial isolates was 100%, as well as the antibiotic Oxacillin. The resistance rate of bacterial isolates was 100%, as well as the antibiotic Cefoxitin. The resistance rate of bacterial isolates was 100%. While other types of antibiotics showed the least resistance, for example, to the antibiotic Clindamycin, the rate of resistance to bacterial isolates was 95%. Likewise, the next lowest rate of resistance was to the antibiotic Levoflox-acine, which had a resistance rate of 80%. Likewise, the next lowest rate of resistance was to the antibiotic Trimethoprim, where the rate was Resistance to bacterial isolates was 75%. It was also followed by the lowest rate of resistance to the antibiotic Chloramphenicol, where the rate of resistance to bacterial isolates was
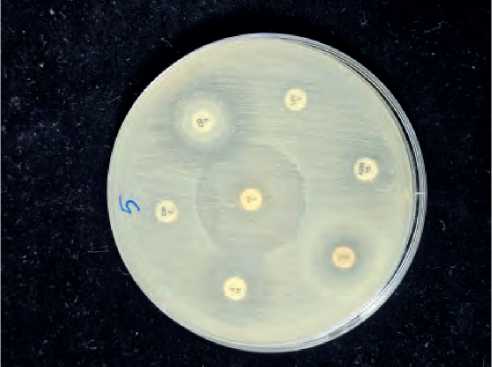
Figure (1-1): Detection resistance of various antibiotic S. aureus isolates (No. of isolate 5).
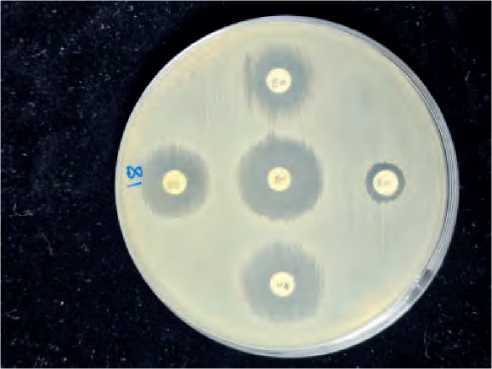
Figure (1-2): Detection sensitivie of various antibiotic S. aureus isolates (No. of isolate 18).
60%. It was followed by the lowest rate of resistance to the antibiotic Vancomycin, where the rate of resistance to bacterial isolates reached 12%, which is the lowest rate among all types of antibiotics used in the study.
This difference in sensitivity and resistance of bacterial isolates can be explained by the different places where the bacteria are collected. Therefore, the extent of their sensitivity and resistance was different due to the presence or absence of resistance genes or phenotypic factors, which made them different in terms of resistance and sensitivity. From the Table [1 – 4].
Table (1-4)
Frequency of Antibiotic Susceptibility of 80 S aureus Isolates
|
Type of Antibiotic |
Sensitive (%) |
Resistance(%) |
|
Vancomycin |
68 (85%) |
12 (15%) |
|
Ciprofloxacin |
0 (0%) |
64 (80%) |
|
Tetracycline |
0 (0%) |
80 (100%) |
|
Azithromycin |
0 (0%) |
80 (100%) |
|
Erythromycin |
0 (0%) |
80 (100%) |
|
Minocycline |
0 (0%) |
72 (90%) |
|
Levofloxacine |
0 (0%) |
64 (80%) |
|
Clindamycin |
0 (0%) |
76 (95%) |
|
Trimethoprim |
0 (0%) |
60 (75%) |
|
Chloramphenicol |
0 (0%) |
48 (60%) |
|
Rifampin |
0 (0%) |
80 (100%) |
|
Oxacillin |
0 (0%) |
80 (100%) |
|
Cefoxitin |
0 (0%) |
80 (100%) |
М 12 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
3000 bp
500 bp

100 bp
Figure (1-3): The PCR product of MecA gene in 1.5% agarose gel electrophoresis, voltage (80 V), time (60 minute) and 5 μL of PCR product loaded in each well. Lane M: DNA Ladder (100bp – 300bP) Lanes PCR product (positive case band 855 bp).
The current study included the molecular detection of genes in 20 Staphylococcus. aureus isolates responsible for the resistance of a large group of important drugs widely used in the country against the S. aureus isolates, which is the MecA. The PCR results showed total compatibility with the phenotypic results of bacteria. The MecA gene ratio reached (45%) of what could be adopted as a positive indication of the presence and spread of the Mec-A gene, figure (1-3). while [11]isolates (%55) were lacking the mec A gene. Figure (1-3) shows gel electrophoresis of PCR amplification products of mec A gene.
REFRENCES
-
1. Zetola N, et al. Community acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 2005; 9: 978-84.
-
2. Bauer AW, et al. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966; 45(4): 493–6.
-
3. Brea D, et al. Altered Growth, Pigmentation, and Antimicrobial Susceptibility Properties of Staphylo-coccusaureus Due to Loss of the Major Cold Shock Gene cspB Antimicrob. Agents Chemother. 2010; 54(6): 2283-90.
-
4. Peacock SJ, Silva DI, Lowy FD. What determines nasal carriage of Staphylococcus aureus? Trends. Microbiol. 2001; 9(12): 605-10.
-
5. VonEiff C, et al. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 2001; 344: 11-6.
-
6. Qiu L, et al. Gelatinase-responsive release of an antibacterial photodynamic peptide against Staphylococcus. aureus. Biomaterials Science. 2021.
-
7. Lee JYH, et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermid-is. Nat Microbiol. 2018; 3: 1175-85.
-
8. CLSI, CLSI M100-ED30: Performance Standards for Antimicrobial Susceptibility Testing, 30th Edition (2023).
Список литературы Detection MEC a gene and susceptibity some various antibiotic of Staphlococcus aureus
- Zetola N, et al. Community acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 2005; 9: 978-84.
- Bauer AW, et al. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966; 45(4): 493-6.
- Brea D, et al. Altered Growth, Pigmentation, and Antimicrobial Susceptibility Properties of Staphylo-coccusaureus Due to Loss of the Major Cold Shock Gene cspB Antimicrob. Agents Chemother. 2010; 54(6): 2283-90.
- Peacock SJ, Silva DI, Lowy FD. What determines nasal carriage of Staphylococcus aureus? Trends. Mi-crobiol. 2001; 9(12): 605-10. EDN: EJUQNF
- VonEiff C, et al. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 2001; 344: 11-6.
- Qiu L, et al. Gelatinase-responsive release of an antibacterial photodynamic peptide against Staphylococcus. aureus. Biomaterials Science. 2.
- Lee JYH, et al. Global spread of three multi-drug-resistant lineages of Staphylococcus epidermid-is. Nat Microbiol. 2018; 3: 1175-85. EDN: VJEMTY
- CLSI, CLSI M100-ED30: Performance Standards for Antimicrobial Susceptibility Testing, 30th Edition (2023).


