Enhancing Drug Recommender System for Peptic Ulcer Treatment
Автор: Theresa O. Omodunbi, Grace E. Alilu, Kennedy O. Obohwemu, Rhoda N. Ikono
Журнал: International Journal of Information Technology and Computer Science @ijitcs
Статья в выпуске: 6 Vol. 16, 2024 года.
Бесплатный доступ
Drug Recommender Systems (DRS) streamline prescription process and contribute to better healthcare. Hence, this study developed a DRS that recommends appropriate drug(s) for the treatment of an ailment using Peptic Ulcer Disease (PUD) as a case study. Patients’ and drug data were elicited from MIMIC-IV and Drugs.com, respectively. These data were analysed and used in the design of the DRS model, which was based on the hybrid recommendation approach (combining the clustering algorithm, the Collaborative Filtering approach (CF), and the Knowledge-Based Filtering approach (KBF)). The factors that were considered in recommending appropriate drugs were age, gender, body weight, allergies, and drug interactions. The model designed was implemented in Python programming language with the Flask framework for web development and Visual Studio Code as the Integrated Development Environment. The performance of the system was evaluated using Precision, Recall, Accuracy, Root Mean Squared Error (RMSE) and usability test. The evaluation was carried out in two phases. Firstly, the CF component was evaluated by splitting the dataset from MIMIV-IV into a 70% (60,018) training set and a 30% (25,722) test set. This resulted in a precision score of 85.48%, a recall score of 85.58%, and a RMSE score of 0.74. Secondly, the KBF component was evaluated using 30 different cases. The evaluation for this was computed manually by comparing the recommendation results from the system with those of an expert. This resulted in a precision of 77%, a recall of 83%, an accuracy of 81% and an RMSE of 0.24. The results from the usability test showed a high percentage of performance of the system. The addition of the KBF reduced the error rate between actual recommendations and predicted recommendations. So, the system had a high ability to recommend appropriate drug(s) for PUD.
Drug Recommender System, Peptic Ulcer Disease, Collaborative Filtering, Knowledge-based Filtering, Decision Support Systems
Короткий адрес: https://sciup.org/15019583
IDR: 15019583 | DOI: 10.5815/ijitcs.2024.06.02
Текст научной статьи Enhancing Drug Recommender System for Peptic Ulcer Treatment
Healthcare is a fundamental component of human society, striving to maintain and improve the well-being of individuals. Central to this endeavor is the practice of prescribing drugs to treat a multitude of health conditions. The prescription of appropriate medication is a crucial part of every consultation between patient and healthcare personnel [1], and the goal of physicians in prescribing drugs is to quicken patients' recovery and see them get well.
People fall ill due to factors such as environment, genetics, weather, microbial infections, age, lifestyle choices, etc. These factors cause a number of ailments, one of which is Peptic Ulcer Disease (PUD). To put it simply, a peptic ulcer is a sore on the inner lining of the stomach or duodenum that is caused by peptic acid [2]. The two most common types of PUD are Gastric ulcer and Duodenal ulcer. These names refer to the locations where they are found, which are inside the stomach and the duodenum (upper part of the small intestine), respectively [3]. PUD is one of the most prevalent disorders of the digestive system and affects about 4 million people worldwide each year, with complications occurring in 10–20% of cases [4]. According to Ray-Offor and Opusunju (2020), PUDs are estimated to involve 5–10% of the world’s population [5]. Also, from a study of the global burden of PUD conducted by Xie et al. (2022), it was discovered that in 2019, there were about 8.09 million prevalent cases of PUD [3]. The majority of cases of PUD are caused by Helicobacter Pylori (H. Pylori) infection and Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) [6-8]. Other factors identified by some researchers [3-5] as being associated with an increased risk of PUD are: gastric bypass surgery, alcohol consumption, smoking, starvation, stress, blood group O, and genetic traits. PUD that is untreated or inadequately managed can result in complications, which can in turn lead to the loss of life. The commonly known complications of PUD include perforation in the stomach or intestine, internal bleeding, penetration into a surrounding organ, and obstruction of the gastric outlet that prevents food from leaving the stomach [2,5,6,9,10]. So, PUD needs to be treated early and adequately to prevent it from becoming a complicated case. According to Umegbolu (2022) and the Japanese Society of Gastroenterologists, a number of factors, such as etiology (cause), presence of complications, recurring cases, etc., are considered in prescribing the right treatment for PUD [2,8]. These factors determine the drug(s) to be prescribed for patients.
This process of drug prescription is carried out by physicians who rely on their clinical experience, knowledge, and available guidelines to make informed decisions. However, the increasing number of available drugs, each with varying effectiveness and cost, has made this task increasingly challenging [11]. It has become more difficult for physicians to stay current with the wide range of drugs available, the exponential increase in medical literature, and the development of new drugs nearly on a daily basis [12,13]. Also, when physicians are faced with peculiar cases or unfamiliar cases, they search for information on appropriate medication to prescribe for such cases [14,15]. This can be time-consuming and strenuous, which can lead to complications and the indiscriminate use of drugs or seeking alternative cures by patients [12,13].
In order to assist physicians, this process of drug prescription is now being technologically implemented through the use of Drug Recommender Systems (DRS). A DRS is an information system that recommends appropriate drugs for patients based on their ailment [16,17]. The goal of a DRS is to suggest appropriate drugs to patients based on certain factors, which can be age, type of ailment, allergies, etc. They were developed to support healthcare professionals in performing their duties effectively, not to replace them [18]. In order to recommend appropriate drug(s), DRS employs a number of recommendation approaches, of which the most commonly used are Collaborative Filtering (CF), Content Based (CB), Knowledge Based Filtering (KBF) and the Hybrid recommendation approaches [12,19-21]. The CF approach recommends an item that might interest users based on items that similar users were interested in or items (drugs) that were beneficial to similar users (or patients) [22]. In the CB recommendation approach, items are recommended based on the item or the content or details of an item users were interested in in the past [12]. For instance, if a drug with certain components is effective for a patient, drugs with similar components are recommended for that patient. The KBF approach bases its recommendations on assumptions about a user's preferences and requirements [23]. It relies heavily on existing knowledge, and the necessary knowledges are: knowledge of the users, knowledge of the items (which can be drug), and knowledge of how to match an item with a user's need [12,19,21,23]. This knowledge comes from sources such as answers to a pre-set sequence of questions about users’ preferences, users’ profiles, etc. [24]. An important characteristic of knowledge-based systems is their high level of personalized recommendations [24]. Multiple approaches are combined in the hybrid recommendation approach to enhance performance in terms of recommendation accuracy [19,25,26]. According to [23], the hybrid technique provides recommendations that are more accurate than those from pure approaches such as the CF and CB. It combines a number of techniques to optimize their benefits and reduce their downsides [25,27].
With the aim of assisting healthcare professionals in making informed and personalized drug prescription decisions, this study developed a DRS that recommends the most appropriate drug for a patient by taking factors such as age, gender, body weight, allergies, and drug interactions into consideration. A DRS for PUD will aid in early and appropriate treatment by considering each patient’s features, and in return, it will prevent complications that might arise from delayed treatment. DRS is necessary in a country like Nigeria, which has a ratio of 1:2,500 doctors to patients [28]. This wide ratio results in physicians (especially specialists) not being easily accessible to patients. Patients join endless queues to access or consult a physician, especially in government-owned healthcare facilities. And the time spent in these queues before having access to a physician can worsen an ailment, especially in cases that need urgent attention. Some of these patients result in self-medication, failing to realize that certain factors are taken into consideration in drug prescription, thereby exposing themselves to preventable dangers. So, there is a need for a system that can assist in reducing these issues, directly or indirectly, as the adoption and use of technology in the healthcare sector in Nigeria has transformed the process of care delivery for patients [29,30]. So, a DRS will not only streamline the prescription process but also contribute to better patient care, reduced healthcare costs, and increase patient satisfaction.
This study addressed some gaps in existing study. One of these is that, many DRS failed to take into account underlying medical conditions and allergies in DRS development. [31] recognized this oversight, explicitly acknowledging the exclusion of factors such as allergies and underlying medical issues. Additionally, several other studies omitted these factors without explicit acknowledgment. Investigating the impact of such patient-specific information in
DRS are important. This study also included a user-centric evaluation of the system through a usability test, which was conducted subsequent to users' interactions with the system. While some studies briefly touched on user-based evaluations, there is a lack of comprehensive investigation into the usability, user satisfaction, and effectiveness of the DRS models in real-world scenarios. Understanding how patients and healthcare providers perceive and interact with these systems is essential for successful implementation. In addressing these knowledge gaps this study aims to contribute to the development of more adaptive, user-friendly, and effective DRSs, ultimately improving patient outcomes and satisfaction in healthcare settings.
The next section, which is ‘Methodology’, discusses the methodology used for data collection and analysis and the development of the DRS. The ‘Results’ section describes the results of the system implementation, and the ‘Discussion’ section discusses the implications of the results. The last section, ‘Conclusion’, concludes the paper and gives recommendations for future research.
2. Methodology
This section discusses the methodology used for data collection and analysis, model formulation and implementation of the DRS.
-
2.1. System Design
-
2.2. Dataset Acquisition
-
2.3. Drug Recommender Model Formulation
This study followed the hybrid recommendation approach. This is necessary in order to get the best recommendation and to minimize the weaknesses of the individual recommendation approach.
The system, as depicted in Fig. 1, takes the details of patients (age, gender, weight, existing medical conditions, and current drug being taken) and then checks to determine which cluster the patient falls into. Patients in the database are already grouped into six clusters based on their information using the k-means clustering algorithm. From the cluster, the five closest patients to the new patient in the database are identified using the K Nearest Neighbour (KNN) algorithm, and the drug(s) that were prescribed to those patients are extracted and form the ‘Initial Drug(s) Recommendation’ (IDR). The drugs in the IDR are then used for the knowledge-based filtering of the DRS. This part was implemented using rules. Each drug is checked for allergies and drug-drug and drug-disease interactions based on the drug information in the database. The drug(s) that do not have the tendencies of allergic reactions and interactions then make the ‘Final Drug Recommendation’.
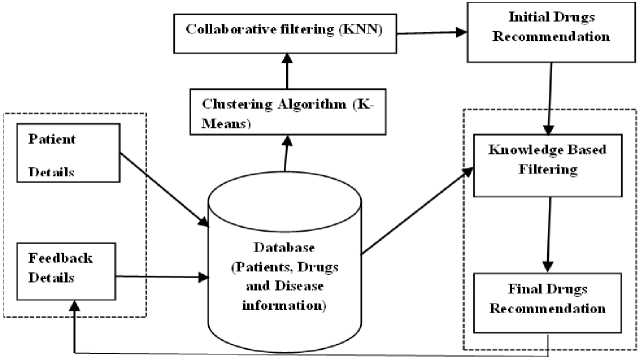
Fig.1. Framework of developed drug recommender system
The data used for this study were in two categories: clinical data and drug data. The clinical datasets were elicited from Medical Information Mart for Intensive Care – IV (MIMIC-IV) [32,33]. MIMIC-IV is a database which contains the de-identified data for patients who were admitted to the BIDMC (Beth Israel Deaconess Medical Center) emergency department or one of the Intensive Care Units (ICU), which were extracted from respective hospital databases. In order to protect patients’ identities, patient identifiers such as name were removed as stipulated by HIPAA (Health Insurance Portability and Accountability Act) and replaced with deidentified integer identifiers (Subject_id). Also, Date and time were randomly pushed into the future using a days-based offset. The MIMIC-IV dataset is divided into two modules: – the hosp and the icu modules. The hosp module was used for this study. In order to gain access to the datasets, the requirements involved getting credentialed, completing CITI’s training on ‘Data or Specimens Only Research’ and signing a ‘data use agreement’. All these were carried out by the authors. The choice of MIMIC-IV dataset is because, it provides access to a wide range of electronic health records, including medications, diagnoses, lab results, and clinical notes. It also has relevant details about patients such as, age, weight, BMI, type of PUD, medical history, etc., and these are some of the important factors considered in PUD drug prescription. In addition to these, MIMIC-IV offers data on a large and diverse patient population, which is essential for developing a robust and generalizable DRS.
After preprocessing, the dataset used for this research consist of 85,740 records of patients with PUD, comprising of 16 variations of PUD. Each record had seven attributes – subject_id, gender, age, ICD code, disease name, drug name, and body weight value. These records made up the data points used in this study. For the drugs Data, a total of five PUD drugs namely; Omeprazole, Pantoprazole, Famotidine, Ranitidine, and Glycopylorrate, were gotten from the MIMIC-IV database. Each of these drugs was checked for information (allergies, drug-to-drug interaction, and drug-to-disease interaction) in the drugs database, Drugs.com [34]. The choice of drugs.com is, Drugs.com is the largest, most widely visited, independent medicine information website available on the Internet [34]. The site provides accurate and independent information on more than 24,000 prescription drugs, over-the-counter medicines and natural products [34]. It offers detailed descriptions of medications, including uses, side effects, interactions, contraindications, warnings and dosage. All these information is necessary for in drug prescription process and some of this information were utilized in the DRS development for this study.
The recommendation phase was based on a hybrid recommendation approach consisting of clustering, collaborative filtering, and knowledge-based filtering. The features from the datasets used for this phase were age, gender, body weight value, name of disease, and drug name. The model formulations for each of the approaches are explained below.
-
A. Clustering
Clustering of data was carried out using the K-means clustering algorithm. The following steps were followed in the clustering phase of the drug recommender model formulation:
Encoding of textual values: The MIMIC-IV dataset used, is a mix of numerical and textual feature values and the K-means algorithm can only be applied on numerical data so, encoding was carried out on the textual values to convert them to numerical values. The textual features were encoded using a one-hot (one-of-K or dummy) encoding scheme. This creates a category for each text and then create a binary column for each category and returns a sparse matrix or dense array (depending on the sparse output parameter). The textual values in the relevant data fields from MIMIC-IV namely, medications, gender and ICD_Codes were converted to numerical values in order to make it easy for clustering to take place. Table 1 shows the result of encoding for the first 10 rows of the dataset.
Standardization/Normalization of Data: When performing clustering on a dataset, it is important to have more normally distributed data because clustering depends on the relative distance of the observations. Standardizing, or normalization, therefore transforms the features of the dataset so that they have a mean of 0 and a standard deviation of 1. The goal is to ensure that all features have the same scale, so they contribute equally to the clustering algorithm because, if the features have different scales, the algorithm may give more importance to features with larger scales, leading to biased results. Normalization of values were carried out before clustering using standard scaler. The standard score of a sample x is calculated as: z = (x - u)/s. Where u is the mean of the training samples and s is the standard deviation of the training samples. A sample of the result gotten after the standard scaling is shown in Table 2.
Optimal Number of Clusters: the elbow method was used to determine the optimal number of clusters to use for this study. This method calculates the Within-Cluster Sum of Squares (WCSS) for various values of K, with lower values generally being better. The plot for this method is shown in Fig. 2. Usually, the point at which the first bend occurs in the plot is picked as the optimal number for the clusters. From figure 2, a bent occurred at point 4 and 6 but 6 was chosen because it has a higher silhouette score of approximately 0.48.
The clustering phase for this study is represented as follows:
-
i. Data Representation: Patient data was represented as data points in a feature space.
Patient 1: X 1 = [age 1 , gender 1 , weight 1 , ICD_code 1 ]
Patient 2: X 2 = [age 2 , gender 2 , weight 2 , ICD_code 2 ]
Patient N: X N = [age N , gender N , weight N , ICD_code N ]
-
ii. Selection of number of Clusters:
The optimal K number of clusters of 6 was chosen using the elbow method.
Centroid 1: `C 1 `
Centroid 2: `C 2 `
Centroid 6: C 6
Table 1. Result of text encoding
|
Medication |
Sex |
Age (Yrs) |
ICD_ code |
Weight (Pounds) |
|
0 |
1 |
34.00 |
3 |
250.300000 |
|
1 |
1 |
62.48 |
11 |
212.415965 |
|
4 |
1 |
57.53 |
12 |
236.880565 |
|
0 |
1 |
62.00 |
11 |
178.200000 |
|
1 |
1 |
60.00 |
12 |
183.866474 |
|
3 |
0 |
77.00 |
12 |
110.254026 |
|
3 |
0 |
67.33 |
12 |
112.667095 |
|
2 |
0 |
22.00 |
11 |
199.800000 |
|
0 |
1 |
34.00 |
3 |
232.397493 |
|
2 |
0 |
66.00 |
5 |
175.800000 |
Table 2. Result of normalization of values
|
0 |
1 |
2 |
3 |
4 |
|
0.009993 |
0.004997 |
0.249836 |
0.059961 |
0.966365 |
|
0.013388 |
0.000000 |
0.254381 |
0.053554 |
0.965527 |
|
0.019938 |
0.000000 |
0.199380 |
0.059814 |
0.977892 |
|
0.015289 |
0.003822 |
0.217861 |
0.045866 |
0.974774 |
|
0.022288 |
0.000000 |
0.572067 |
0.089153 |
0.815043 |
|
0.007392 |
0.002464 |
0.115813 |
0.029569 |
0.992800 |
|
0.019769 |
0.004942 |
0.311368 |
0.059308 |
0.948218 |
|
0.005648 |
0.005648 |
0.333257 |
0.062133 |
0.940752 |
|
0.023588 |
0.005897 |
0.324332 |
0.070763 |
0.942980 |
|
0.000000 |
0.000000 |
0.332117 |
0.061314 |
0.941243 |
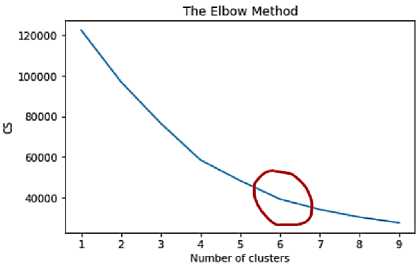
Fig.2. Screenshot of the plot for the elbow method
-
B. Collaborative Filtering
To implement the collaborative filtering phase of this study, the K Nearest Neighbour algorithm was used. The value for K used was 5. The features considered at this stage were gender, age, weight and ICD code of PUD. This is represented as follows:
-
i. Load datasets
The datasets were represented as:
D = {(xi, yi), (x2, y2), ..., (x n , y n )}
Where:
D is the dataset (MIMIC-IV) for the collaborative filtering x represents patient's feature vector (this contains elements for gender, age, weight and ICD code)
y represents the drug(s) prescribed for patient x in the dataset.
-
ii. Input new patient data
For new patients, their features were represented as x_new, where:
x_new[1] is the gender (0 for male, 1 for female).
x_new[2] is the age.
x_new[3] is the weight.
x_new[4] is the ICD code.
-
iii. Calculate the distance between patient x and patient x_new
The similarity measure, Euclidean distance was used in calculating distance between x_new and all data points in D using the formular;
d(x, x_new) = ^(x 1 - x_new 1 ) 2 + (x2 - x_new3) 2 + (x3 - x_new3) 2 + (x 4 - x_new4) 2
where:
d(x, x_new) is the Euclidean distance between two patients x and x_new
-
x 1 and x_new 1 are the values for gender for patients x and x_new respectively
-
x 2 andx_new 2 are the values for age for patients x and x_new respectively
-
x 3 and x_new 3 are the values for weight for patients x and x_new respectively
-
x 4 and x_new 4 are the values for ICD code for the type of PUD for patients x and x_new respectively.
-
iv. Display the drugs prescribed for the nearest neighbours for the value of K = 5.
The results of the above were stored and sort, and the drugs were displayed by selecting the k-nearest neighbours from D based on the smallest distances to x_new.
-
C. Knowledge-based Filtering
This phase of the DRS takes as input, the drugs from the CF phase then, check each drug for interactions and allergies. These drug features (interactions and allergies), were added to the system using the ‘dictionary()’ function. This phase of DRS design for this study is represented as follows:
Notation:
Let D represent the set of all available drugs.
Let P represent the set of patients.
Let A represent known allergies
Let I_d represent the set of known drug-drug interactions.
Let I_dd represent the set of known drug-disease interactions.
Let R represent the set of recommended drugs for a patient.
-
i. Patient Profile:
P = {CD, MC, AR}
Where:
CD = current drugs
MD = Medical Condition
AR = Allergic Reaction
-
ii. Drugs:
D = {di, d2, ..., d5} where di is a drug.
-
iii. Allergy and Drug Interaction Knowledge:
allergic_reaction(drug, allergy)
drug-drug_interactions(drug1, drug2) drug-disease_interactions(drug, disease).
-
iv. Recommendation Rule:
Given a patient's information P and the set of all drugs D, a subset of drugs D was recommended such that:
R = {d i | d i ∈ D and not (allegic(d i , AR) > 0) and not (Interact(d i , CD) > 0) and not (DiseaseInteract(MC, d i ) > 0)} where:
‘di | di £ D’ represents di being element(s) of D allegic(dj, AR) measures allergy between drug di and patient’s allergic reactions
Interact(d i , CD) measures the interaction between the drug d i and the patient's current drugs.
DiseaseInteract(MC, d i ) measures the interaction between the patient's disease and the drug d i .
-
v. Recommendation Process:
For a patient p:
Initialize an empty set Rp for recommended drugs.
Iterate through all drugs in D.
For each drug d:
-
- Check for potential allergic reaction between d and the drugs in AR using A.
-
- Check for potential interactions between d and the drugs in CD using I_d.
-
- Check for potential interactions between d and the patient's medical condition MC using I_dd.
-
- If no interactions are found, add d to Rp.
-
- Return Rp as the set of recommended drugs for patient p.
vi. Output:
2.4. Implementation Environment
3. Results3.1. Implementation Result
For each patient in P, the system outputs their recommended drug set R, which consists of drugs that they are not allergic to and do not interact with their current drugs or medical condition based on the knowledge base.
Visual Studio (VS) Code was the Integrated Development Environment (IDE) used for the implementation of the DRS developed. The back end of the system was designed using Python (version 3.6.3) programming language. Flask, a web framework, which has the ability to integrate results generated by the machine learning algorithms, was used for user interface design.
This section presents the results of the implantation of the DRS and the results of the evaluation of the implemented system.
Fig. 3 shows the interface for inputting the patient’s personal information. The information needed from user include Name, Weight, Age, Gender and ICD Code. Fig. 4 shows the interface for inputting existing medical conditions and current drugs being taken by a patient. These information aids the knowledge-based filtering part of the DRS by enabling the system to recommend only drugs that do not interact with a patient’s existing medical conditions or other drugs being taken by the patient. Fig. 5 shows that one drug (Pantoprazole) was recommended for a male patient, aged 54, with no existing medical condition and who is not taking any other drug.
Fig.3. User interface (patient details)
Fig.4. User interface (drug and existing ailment details)
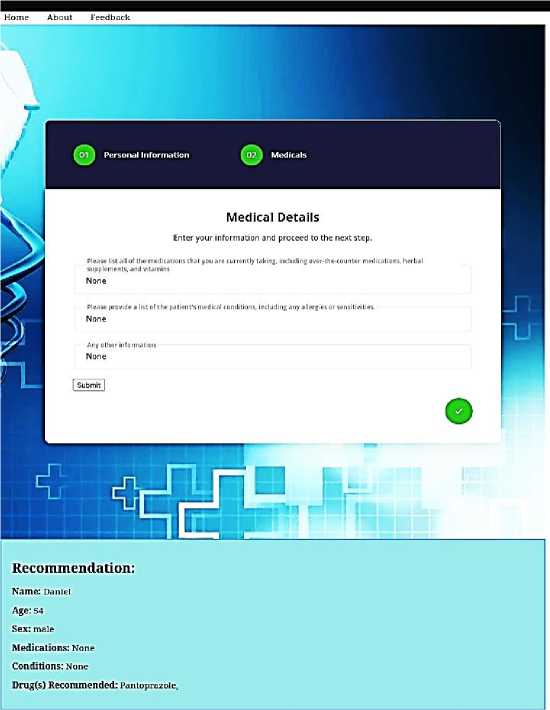
Fig.5. Drug recommendation for a patient
-
3.2. System Evaluation Result
The CF aspect of the system was first evaluated within the system using Precision, Recall and Root Mean Squared Error (RMSE). Then the aspect that involved the Knowledge Based Filtering (KBF) was also evaluated manually using the aforementioned evaluation metrics including accuracy. These evaluation metrics were selected because, evaluating DRS involves assessing how well the system recommends suitable drug(s) for patients based on their features, and these metrics provide insights into DRS’s performance. Also, these are the most widely used evaluation metrics for DRS. The evaluation is separated for each recommendation approach because, both used different datasets. The CF aspect of the DRS used clinical data from MIMIC-IV, while the KBF aspect used the output from the CF in addition to drug information gotten from Drugs.com.
As already stated, the CF aspect of the DRS was first evaluated. Fig. 6. shows a screenshot of the generated evaluation metrics and their values. From the figure, it can be seen that the system had a precision score of 85.48%, a recall score of 85.58%, and a RMSE score of 0.74.
The second part of the evaluation process involved the KBF aspect of the DRS. This evaluated the overall system’s performance. The system was evaluated by considering a total of thirty different cases of patients which were obtained manually in conjunction with an expert who is a public health physician and a resident doctor. The attributes which made up the factors considered for this phase of evaluation were age, gender, weight, ICD code, allergies, current drugs, and existing medical conditions. Considering each of the factors, the expert made recommendations on the most appropriate drug(s) he would prescribe. His recommendations were compared with the drugs that the system recommended after also taking the stated factors into consideration.
Precision Score: 35.48%
Recall Score: 85.58%
Root Mean Squared Error: 0.74
Fig.6. Evaluation scores
Using the expert's and system’s recommendations, the system was evaluated using equations 1, 2, 3 and 4 for precision, recall, accuracy and RMSE respectively. These calculations were carried out manually by using the following formulars:
TP
Precision =
TP+FP
TP
Recall = ~^—
TP+FN
. TP+TN
Accuracy =
TP+TN+FP+FN
Where;
TP = True Positives FP = False Positives
FN = False Negatives
RMSE = J
^ i=1( ^ exp,i ^ drs,i )
n
Where;
n is the total number of drug recommendations.
4. Discussion
X exp is the number of drugs recommended by expert.
X drs is the number of drugs recommended by the system.
The results obtained after the calculation are: 0.77, 0.83, 81% and 0.24 for precision, recall, accuracy and RMSE respectively.
From the overall results, it can be seen that the system has the ability to accurately recommend appropriate drugs for PUD by putting individual and drug factors into consideration. The precision of 0.77 shows that the system can recommend relevant drugs in 77% of the cases. That means that on an average, the expert agrees with 77% of the drugs recommended by the system. Though this value is lower than the values for the CF aspect of the DRS, the score is not bad as this percentage is above average.
For recall, the system had a score of 0.83. This shows that the system has an 83% ability in recommending all relevant drugs in its database after putting some appropriate factors such as gender, age, pre-existing medical condition/state and current drugs into consideration.
From the result for RMSE, it can be seen that the average difference between the system’s recommendations and the expert recommendations is 0.24. This is lower than the score obtained when only the CF was applied. This value shows a low rate of error which can be said to be a good result. The CF aspects performed well in terms of precision and recall but, in terms of the error, the KBF had a very higher performance.
When these values are compared with the DRS developed by Morales et al . (2022) [35], which was hybrid based (Kmeans and KNN), the DRS in this study can be said to have performed well. The DRS in [35] had a RMSE of 0.71 and an accuracy of 61% while the developed DRS had a score of 0.24 for RMSE and 81% for accuracy. It also outperformed the DRSs of [16] which had an accuracy of 80%. [16] is also hybrid based. From these results, it can be inferred that the addition of the KBF enhanced the system’s performance
A usability test was conducted in order to assess the user-friendliness and effectiveness of the DRS was conduction online via Google Forms. 16.7%, 5.5% and 16.7% of these responses were from doctors, pharmacists and other class of medical personnel respectively. The other percentage of responses (61.1%) were from non-medical personnel. 83.3% of the users agreed that the user interface of the DRS was easy to navigate. In terms of suitability of drug(s) recommended by the system after user details were inputted, 88.9% of the respondents agreed that the drug(s) recommended were suitable for the details inputted. In the same vein, in terms of relevance of drug(s) recommended, 88.9% of the users agreed that the drug(s) recommended were relevant. In terms of drugs that should have been recommended but were not, some users stated antibiotics, others herbs, and some, suggested drugs that would be easy to find by patients. Some of these categories of drugs were not recommended because the developed DRS is machine learning based and only drugs contained in the dataset used were recommended. The overall performance of the DRS was also evaluated and 77.8% of the users agreed that the system performed well. This was followed up by checking the likelihood of users in using the system in the future on a scale of 1 to 5. The responses gotten were; 0% for option 1; 27.8% for option 2; 50% for option 3; 16.7% for option 4; and 5.6% for option 5. From all the responses, it can be concluded that the system is easy to use and gives relevant recommendations based on details entered.
From all the evaluation results obtained, it can be concluded that the system has the ability to recommend appropriate drugs for PUD by taking individual and drug factors into consideration.
5. Conclusions
This research presented the development of a DRS using the hybrid recommendation approach. The DRS was developed to aid in the process of making appropriate decisions regarding drugs by putting patient’s unique characteristics into consideration. From the results obtained after evaluation, it can be concluded that the addition of the extra layer of the KBF approach enhanced the performance of the DRS in making more appropriate drug recommendations.
The system’s ability to make more accurate recommendations can be improved by taking the following factors into consideration:
-
• The cost of drugs and the availability of drugs within a patient’s location.
-
• The output of a system should also include information such as;
-
- Instructions and warnings – this are one of the steps in the WHO’s guide to good prescribing.
-
- Serious side effects that patients should look out for.
-
• Also, according to the medical expert consulted, the drug recommendation should include other drugs to tackle the cause of the peptic ulcer. For instance, in drug recommendations for a PUD caused by H. Pylori Infection, there should be an inclusion of an antibiotic to tackle this infection. Also, the presence of coexisting symptoms like heartburn, bloating, bleeding, and pain might require the inclusion of other medications like antacids and anti-flatulents. These were not taken into consideration in this study due to the nature of the datasets used.
In future research, we intend developing a DRS that gives more robust recommendations that will include warnings, potential serious side effects, price of drugs and availability of drugs. We would also make use of diversifying datasets that contains drugs that addresses the cause of a patient’s PUD and coexisting symptoms. More sophisticated machine learning and deep learning algorithms such as Neural networks, would be utilized to enhance the accuracy of drug recommendations. This study used 30 test cases in the final evaluation of the system, for future study, larger and diversifying test cases would be utilized.
In general, the DRS performed well in recommending appropriate drug(s) for patients with peptic ulcer disease.
Conflicts of Interest
There is no conflict of interest among the authors.
Список литературы Enhancing Drug Recommender System for Peptic Ulcer Treatment
- K. Zarour, M. O. Fetni, and S. Belagrouz, “Towards Electronic Prescription System in a Developing Country Cooperative information systems View project ABAH: Agent-Based Architecture for Homecare View project,” pp. 56–67, 2021, [Online]. Available: https://www.researchgate.net/publication/350709082
- E. I. Umegbolu, “Peptic ulcer disease in school children aged 2-11 years in Southeast Nigeria,” Int J Res Med Sci, vol. 10, no. 5, p. 1007, Apr. 2022, doi: 10.18203/2320-6012.ijrms20221169.
- X. Xie, K. Ren, Z. Zhou, C. Dang, and H. Zhang, “The global, regional and national burden of peptic ulcer disease from 1990 to 2019: a population-based study,” BMC Gastroenterol, vol. 22, no. 1, Dec. 2022, doi: 10.1186/s12876-022-02130-2.
- S. Bidokumo Zibima, J. Imawaigha Oniso, K. Belibodei Wasini, J. Chidinma Ogu, and C. Author, “Prevalence Trends and Associated Modifiable Risk Factors of Peptic Ulcer Disease among Students in a University Community South-South Nigeria,” 2020. [Online]. Available: www.ijhsr.org
- E. Ray-Offor and K. A. Opusunju, “Current status of peptic ulcer disease in port harcourt metropolis, nigeria,” Afr Health Sci, vol. 20, no. 3, pp. 1446–1451, Sep. 2020, doi: 10.4314/ahs.v20i3.50.
- M. Ahmed, “Peptic Ulcer Disease,” in Management of Digestive Disorders, IntechOpen, 2019, pp. 1–20. doi: 10.5772/intechopen.8 6652.
- A. Mark Fendrick, R. T. Forsch, R. Van Harrison, J. M. Scheiman, C. J. Standiford, and L. A. Green, “University of Michigan Guidelines for Health System Clinical Care Peptic Ulcer Guideline Team UMMC Guidelines Oversight Team Peptic Ulcer Disease,” May 2005.
- T. Kamada et al., “Evidence-based clinical practice guidelines for peptic ulcer disease 2020,” Journal of Gastroenterology, vol. 56, no. 4. Springer Japan, pp. 303–322, Apr. 01, 2021. doi: 10.1007/s00535-021-01769-0.
- W. Andrew and R. Robert, “Gastric Ulcer,” Stat Pearls. Accessed: Jun. 28, 2024. [Online]. Available: https://www.ncbi.nlm.nih.gov/books/NBK537128/
- L. ME, R.-P. M, P. I, and R. L, “Peptic Ulcer Disease,” Journal of Gastroenterology and Hepatobiliary Disorders, vol. 01, no. 01, Dec. 2015, doi: 10.19104/jghd.2015.105.
- B. Stark, C. Knahl, M. Aydin, and K. Elish, “A Literature Review on Medicine Recommender Systems,” vol. 10, no. 8, pp. 6–13, 2019.
- Q. Zhang, G. Zhang, J. Lu, and D. Wu, “A framework of hybrid recommender system for personalized clinical prescription,” in Proceedings - The 2015 10th International Conference on Intelligent Systems and Knowledge Engineering, ISKE 2015, Institute of Electrical and Electronics Engineers Inc., Jan. 2016, pp. 189–195. doi: 10.1109/ISKE.2015.98.
- S. Garg, “Drug Recommendation System based on Sentiment Analysis of Drug Reviews using Machine Learning,” Mar. 2021, doi: 10.1109/Confluence51648.2021.9377188.
- A. A. R. Kamuhabwa and S. Kisoma, “Factors influencing prescribing practices of medical practitioners in public and private health facilities in dar es salaam, Tanzania,” Tropical Journal of Pharmaceutical Research, vol. 14, no. 11, pp. 2107–2113, Nov. 2015, doi: 10.4314/tjpr.v14i11.22.
- O. R. S. Rao, “Factors influencing Drug Prescription Behavior of Physicians in India,” no. August, 2019.
- J. Ayrine, I. H. Muhammed, and V. Vasudevan, “Medication recommendation system based on clinical documents,” in Proceedings - 2016 International Conference on Information Science, ICIS 2016, Institute of Electrical and Electronics Engineers Inc., Feb. 2017, pp. 180–184. doi: 10.1109/INFOSCI.2016.7845323.
- T. V. N. Rao, A. Unnisa, and K. Sreni, “Medicine Recommendation System Based On Patient Reviews,” vol. 9, no. 02, 2020.
- T. O. Omodunbi, G. E. Alilu, and R. N. Ikono, “Drug Recommender Systems: A Review of State- of-the-Art Algorithms,” in 2022 5th Information Technology Conference for Education and Development (ITED), IEEE XPLORE, 2022, pp. 1–8.
- Y. Bao and X. Jiang, “An intelligent medicine recommender system framework,” in Proceedings of the 2016 IEEE 11th Conference on Industrial Electronics and Applications, ICIEA 2016, Institute of Electrical and Electronics Engineers Inc., Oct. 2016, pp. 1383–1388. doi: 10.1109/ICIEA.2016.7603801.
- Hossain Deloar, Azam Shafiul, Ali Jahan, and Hakilo Sabit, “Drugs Rating Generation and Recommendation from Sentiment Analysis of Drug Reviews using Machine Learning,” in 2020 Emerging Technology in Computing, Communication and Electronics (ETCCE), IEEE XPLORE, 2020, pp. 1–6. doi: 10.1109/ETCCE51779.2020.9350868/20/$31.00.
- V. A. Goyal, D. J. Parmar, N. I. Joshi, and P. K. Champanerkar, “Medicine Recommendation System,” pp. 1658–1662, 2020.
- S. Bhat and K. Aishwarya, “Marketed Pharmaceutical Drugs,” pp. 2107–2111, 2013.
- T. B. Adetoba and A. N. Yekini, “A Comprehensive Study of Recommender Systems Prospects and Challenges,” Int J Sci Eng Res, vol. 6, no. 8, pp. 699–714, 2015, [Online]. Available: http://www.ijser.org
- M. Uta et al., “Knowledge-based recommender systems: overview and research directions,” Frontiers in Big Data, vol. 7. Frontiers Media SA, pp. 1–19, 2024. doi: 10.3389/fdata.2024.1304439.
- P. K. Biswas and S. Liu, “A Hybrid Recommender System for Recommending Smartphones to Prospective Customers,” Expert Syst Appl, vol. 208, pp. 1–23, 2020.
- M. Sharma and S. Mann, “A Survey of Recommender Systems: Approaches and Limitations,” International Journal of Innovations in Engineering and Technology Special Issue-ICAECE, 2013.
- Z. Fayyaz, M. Ebrahimian, D. Nawara, A. Ibrahim, and R. Kashef, “Recommendation systems: Algorithms, challenges, metrics, and business opportunities,” Applied Sciences (Switzerland), vol. 10, no. 21, pp. 1–20, Nov. 2020, doi: 10.3390/app10217748.
- K. Kareem, “As doctors emigrate, Nigerians are left with four doctors to every 10,000 patients,” Dataphyte.
- I. P. Gambo and A. H. Soriyan, “ICT Implementation in the Nigerian Healthcare System,” T Professional, vol. 19, no. 2, pp. 12–15, 2017, doi: 10.1109/MITP.2017.21.
- I. Gambo, E. O. Ayegbusi, O. Abioye, T. Omodunbi, R. Ikono, and K. Olufokunbi, “Design Specification for an M-Health Solution to Improve Antenatal Care,” in Advancing Health Education with Telemedicine, 2021, pp. 41–79. doi: 10.4018/978-1-7998-8783-6.ch003.
- S. Bankhele, A. Mhaske, S. Bhat, and S. V. Shinde, “A Diabetic Healthcare Recommendation System,” International Journal of Computer Applications, vol. 167, no. 5, pp. 14-18, 2017.
- A. Goldberger, L. Amaral, L. Glass, J. Hausdorff, P. C. Ivanov, R. Mark, ... and H. E. Stanley, “PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals”, vol 101, no. 23, 2000, e215–e220.
- A. Johnson, L. Bulgarelli, T. Pollard, S. Horng, L. A. Celi and R. Mark, “MIMIC-IV (version 2.2)”, PhysioNet, 2023, doi: 10.13026/6mm1-ek67.
- Drugs.com, “Drugs & Medications A to Z”, Drugs.com Database, 2023, https://www.drugs.com/drug_information.html.
- L. F. Morales, P. Valdiviezo-Diaz, R. Reátegui, and L. Barba-Guaman, “Drug Recommendation System for Diabetes Using a Collaborative Filtering and Clustering Approach: Development and Performance Evaluation,” Journal of Medical Internet Research, vol. 24 no. 7, pp. 1-9, 2022, https://doi.org/10.2196/37233


