Expression of resistance genes in breeding valuable genotypes of pine and fast-growing species of birch and poplar under drought
Автор: Grodetskaya T.A., Evlakov P.M., Fedorovа O.A., Zhuzhukin K.V.
Журнал: Журнал стресс-физиологии и биохимии @jspb
Статья в выпуске: 4 т.20, 2024 года.
Бесплатный доступ
Rationale. This article presents a study of the stress response to the effects of water deficiency in various genotypes of Scots pine, contrasting in seed yield, breeding genotypes of silver and downy birch, hybrids of black, white poplar and intersectional poplar hybrids.
Drought, breeding genotypes, gene expression
Короткий адрес: https://sciup.org/143183441
IDR: 143183441
Текст научной статьи Expression of resistance genes in breeding valuable genotypes of pine and fast-growing species of birch and poplar under drought
In the context of global climate change, long-live plants such as perennial tree species are facing a verse con itions. Large-scale forest mortality will have serious impacts on water an carbon cycles, species istribution an economy (An riantelomanana et al. , 2024). Drought is one of the most etrimental stresses that suppress plant growth an pro uctivity. Among the species threatene by population ecline are fast-growing species such as poplar an birch, which are the main tree species in the broa -leave forests of the mi le zone of Central Russia an are wi ely istribute an use worl wi e. Their ecological role an economic importance cannot be overstate , especially ue to the possibility of their use in pro uction for long-term carbon sequestration (Di Stefano et al. , 2024). Scots pine, one of the most wi esprea tree species worl wi e, is also experiencing massive mortality in a changing climate, the rate of which has only increase over the past two years (Bose et al. , 2024).
Plants have evelope multiple mechanisms to respon to rought stress, inclu ing physiological biochemical an molecular-genetic, complex signaling pathways involving hormones, receptors, protein kinase casca es, transcription factors, an stress-relate protein regulators (Wang et al. , 2017; Yao et al. , 2021). Transcriptomic analysis using microarray technology has i entifie many genes that are in uce by abiotic stress, an these genes have been classifie into two main groups. One group enco es pro ucts that irectly protect plant cells from stress, i.e., functional compoun s.
These pro ucts are likely involve in the metabolic response to stress, protecting cells from stressful influences by removing toxic elements, restoring cellular homeostasis, an possibly restoring normal growth patterns. These inclu e water channel proteins, proline, etoxification enzymes, antifreeze proteins, an late embryogenesis abun ant (LEA) proteins (Shinozaki & Yamaguchi-Shinozaki, 2007; Jan et al. 2013; Rini, 2019).
Pro ucts of another group regulate gene expression an signal trans uction in response to abiotic stress. Previous stu ies, inclu ing genomic ones, have shown that various transcriptional regulatory systems are involve in the in uction of stress-responsive genes. Several groups of cis- an trans-factors are known to be involve in transcription, which is a response to stress. Some of them are controlle by abscisic aci (ABA), while others are not, in icating the involvement of both ABA- epen ent an ABA-in epen ent regulatory systems in the regulation of stress-responsive gene expression. The same group of genes can be in uce by ifferent types of stress, such as rought an col , in icating the existence of cross-pathways between signaling mechanisms (Shinozaki, 2003).
Transcription factors (TFs) are large families of DNA-bin ing proteins that regulate gene expression uring ontogenesis, ifferentiation, an response to various environmental signals (Yanagisawa, 1998). They inclu e families unique to plants, such as AP2/ERF , bZIP , WRKY , MYB , an NAC (Martin & Paz-Ares, 1997). One of the most important families of TFs regulating plant response to stress is APETALA2/ethylene response factors (AP2/ERF). The genes of this family are ivi e into subfamilies that perform ifferent roles in ontogenesis an protection from a verse environmental con itions. TFs control the expression of key genes of secon ary metabolism un er stress by bin ing to regulatory regions in the promoters of target genes. DREB subfamily of transcription factors, specifically bin ing to cis- acting ehy ration-responsive element/C-repeat ( DRE/CRT ), is involve in the response to col an rought (Sakuma, 2006).
The ERF subfamily is involve in the response to abiotic an biotic stress, growth an evelopment, primary an secon ary metabolism, an hormonal signaling (Wang et al , 2017). To ate, sufficient information has been accumulate on the involvement of transcription factors in the mechanisms of plant response to various abiotic stresses. Overexpression of AP2/ERF family of TFs, EFR96 (Wang et al, 2017) an HcTOE3 (Yin et al , 2021) in Arabidopsis contribute to an increase in the expression of Δ-pyrroline-5-carboxylate synthetase (P5CS) an proline content, an greater resistance to rought an col stress. It has been shown that the AP2/ERF family can also control lignan/lignin biosynthesis in Isatis indigotica through the activation of the salicylic aci signaling pathway an the transcriptional regulation of key genes for lignin biosynthesis (Ma et al , 2017), a biopolymer presente in the secon ary walls of tracheal elements an woo fibers an playing an important role in both growth an evelopment an in plant responses to various biotic an abiotic stresses (Cesarino, 2019).
The interest in stu ying gene transcription an its regulation un er the influence of abiotic stress in woo y plants lies in the nee to i entify specific resistance factors an select markers for molecular genetic selection of promising genotypes. In this work, the expression of genes associate with rought resistance was analyze in birch, poplar an pine plants of bree ing value.
MATERIAL AND METHODS
Characteristics of research objects
To assess the rought resistance of plants, we use the bree ing genotype of owny birch ( Betula pubescens Ehrh.) ‘15-1’ an the variety of silver birch ( B. pendula Roth.) ‘Uglyancheskaya-1’ (‘Ug.-1’), poplars of ifferent hybri s an varieties ( Populus L.), belonging to ifferent sections, as well as Scots pine ( Pinus sylvestris L.), iffering in see yiel (Table 1).
The source material for growing see lings of silver birch of the ‘Ug.-1’ variety was prepare in the archive of clones of plus trees, establishe in the territory of the Tellermanovsky forestry enterprise of the Voronezh region. The source material of owny birch an poplar was taken from test crops of the Semiluksky nursery of the Voronezh region. The pine material was selecte from test crops of Scots pine, bre for their pro uctivity, establishe in forest see plantation No. 68 of the Semiluk forest nursery by Efimov Yu.P.
Water Deficiency Experiment Conditions
To simulate rought con itions, the irrigation moisture capacity of birch an poplar samples was re uce to 35±5%, while in the control to 80±5%. Experimental nee le samples were collecte uring the ry perio in June 2023, an control samples were collecte after the restoration of normal precipitation. The samples were fixe by instant freezing in liqui nitrogen an store at -80°C until further analysis.
Determination of expression of resistance genes
RNA was isolate using a stan ar set of NucleoSpin® RNA columns (Macherey-Nagel, Germany) from poplar samples. Mo ifie CTAB metho was use to extract RNA from birch an pine samples (Gro etskaya et al. , 2021).
RNA concentration was etermine on a Qubit 2.0 fluorimeter (Thermo Fisher Scientific, USA) using the manufacturer's stan ar reagents. RNA in an amount of 0.5 μg was selecte for reverse transcription using the AmpliSens kit (Central Research Institute of Epi emiology of Rospotrebna zor, Russia) in accor ance with the manufacturer's recommen ations.
The search for PCR primers was carrie out base on literature ata an using the NCBI (National Center for Biotechnology Information) nucleoti e sequence atabase. For genes whose sequences were not available in the atabase for the esire object, the most conservative fragments i entical in organisms from ifferent families were searche for using the ClustalOmega program (European Bioinformatics Institute EMBL, UK). The selection of primers for poplar, birch an pine resistance genes from the NCBI atabase an sequence fragments selecte base on the analysis through ClustalOmega was performe using the Primer3 program (Estonia). The list of primers for the analysis of rought resistance gene expression is presente in Table 2.
The specificity of primer amplification at the correspon ing temperature was assesse by electrophoresis in 1% agarose gel. Expression was assesse using a Roche Light Cycler 480 II amplifier (Roche, Switzerlan ). A stan ar 5x qPCR–HS SYBR kit (Eurogen, Russia) containing SYBR Green I as an intercalating ye was use for quantitative PCR. The 18S genes for poplar an GAPDH for birch an pine were use as reference genes. The results were calculate using the 2-ΔΔCt metho (Livak & Schmittgen, 2001) using the Light Cycler software (Roche, Switzerlan ) an the MS Excel software package.
Statistical analysis was performe using the stan ar error of the mean (SEM) an Stu ent's t-test in Excel 2010. Differences were consi ere statistically significant at p<0.05.
RESULTS
To analyze the a aptive capacity of promising birch genotypes to the effects of rought, a stu y was con ucte on the expression of the resistance genes bDREB2, bLEA8, bPAL, bNAC19 an bCA. It was foun that the silver birch ‘Ug.- 1’ evelope an a aptive response to rought for all analyze genes. The expression of the genes of owny birch ‘15-1’ i not increase in response to stress, the level of transcripts correspon e to the control (bLEA8) or was re uce (bPAL, bCA, bNAC19 an bDREB2) (Figure 1)
It was reveale that the expression of the analyze genes iffere in poplar genotypes in response to rought (Figure 2). The expression of the pP5CS gene increase un er rought con itions in poplar ‘POK’, ‘E.s.-38’ an ‘Ve uga’ by 1.5-58.3 times. An increase of 1.58.4 times was also reveale for the pCA gene. In black poplar, the transcript level increase only for pCA, while the expression of the pICDH, pDREB2, pNAC034 an pP5CS genes was ecrease relative to the control. Expression of all analyze resistance genes in poplar ‘E.s.-38’ significantly increase relative to the samples un er normal con itions. For the ‘POK’ genotype, an increase in expression was shown for pICDH an pP5CS, while the expression of pNAC034 ecrease relative to the control. Expression of the NAC034 transcription factor genes increase in ‘Ve uga’ an ‘E.s.-38’, while DREB2 respon e positively to rought only in ‘E.s.-38’. For the ‘Ve uga’ genotype, an increase in the expression of pP5CS an pCA was also shown.
The effect of stress in Scots pine genotypes iffering in yiel was stu ie by changes in the expression of the psPAL, psSAMS, psTAT-D an psDREB2 genes (Figure 3).
A 3.3-fol increase in psSAMS gene expression was foun in the me ium-yiel ing genotype No. 4, while psDREB2 expression ecrease 3.1 times. For the low yiel ing genotype No. 23, an increase in psSAMS, psTAT– D, an psDREB2 gene expression was observe by 2.1-2.8 times, while psPAL expression was re uce . For the low-yiel ing genotype No. 22, an increase in pTAT-D expression was etecte , while the level of psPAL, psSAMS, an psDREB2 transcripts i not change relative to the control.
Table 1. Characteristics of genotypes of promising fast-growingwoo yplants
|
N |
Name of genotype |
Invariant number |
Origin |
Author (Bree er) |
|
Birc |
h |
|||
|
4 |
’15-1’ |
20– 17 |
Mother tree 20–17, hybri (B–2 × B . pubescens , pollen mixture) |
Isakov I.Yu. |
|
5 |
‘Ug.-1’* |
– |
B. pendula Roth. |
Kozmin A.V. an others |
|
White poplars with a pyrami al crown shape |
||||
|
1 |
‘Veduga’ |
26– 07 |
P. alba L. × P. bolleana Laurche |
Tsarev A.P. |
|
Intersectional hybri s |
||||
|
2 |
‘Э.с.-38’* |
49 |
P. deltoides Marsh. × P. balsamifera L. |
Veresin M.M. |
|
Black poplars with a pyrami al crown shape |
||||
|
3 |
‘POK’ * |
91 |
P. piramidalis Ros. × P. nigra L. |
Albensky A.V. |
|
Black poplars with a sprea ing crown shape |
||||
|
4 |
Black poplar (‘osokor‘) |
P. nigra L. |
species |
|
|
Pine |
||||
|
5 |
Me ium-yiel ing No.4 |
4 |
P. sylvestris L. |
Efimov Yu.P. |
|
6 |
Low-yiel ing №22 |
22 |
||
|
7 |
Low-yiel ing №23 |
23 |
||
* 'Ug.-1' – 'Uglyancheskaya-1'; 'E.s.-38' – 'Elitesee ling-38';'POK' –'Pyrami al-osokoreviyKamyshinsky
Table 2. Primer sequences for stress resistance genes
Gene Sequence (5′→3′)
Birch (Pääkkönen et al. , 1998)
F: AGGCAGAGAACATGGGGAAA bDREB2
R: GAAAGTTGAGGCGAGCGTAA
F: AATGACTTTGACATGGGCGT bLEA8
R: TATCCCAAACTGCAGAGCCA
F: CTGTGGCTGCAACGGTTT bPAL
R: TCAATTTGAGGTCCGAGCCA
F: CAAGCTCAGGGTCAGACTCA bNAC19
R: ACACGTTCGAGTTCTGCCTA
F: ACATAGTCGTTGTGGTGGGA bCA
R: TGACAAGTTCACTGCCTCCT
F: CAGCCGAAGATGTCAATGCA bGAPDH
R: GGCCACTTGTTTGCTACCAA
Poplar (Chen et al. ,2009; Lan et al. , 2013; Wu et al. , 2015)
F: ACTCGGCATTACAGGGTTCA pICDH
R: GACTCCACAGCTCCGATACA
F: TCTATGGCCTGCACTGTTGA pP5CS
R: TGCTTATTCCGACCTCTGCA
F: TGTATGCTCGTATGCTCGT pDREB2
R: TCCTCATACACGCAGACCTC
F: GTGTATTTCGACACGTCAGATTCT pNAC034
R: ATACATGAACATGTCCTGAAGCG
F: TGCGAGAAGGAGGCTGTTAA pCA
R: GCCGGCTACAGTTTCACATT
F: GGCTCTGCCCGTTGCTCT p18S
R: CGTCACCCGTCACCACCA
Pine (Sun et al. , 2022)
F: GAG GGA ATT TCC AGG GCA CA psPAL
R: GAT CTC GGC CCC TTT CAG TC
F: ACT GCA AAG TGC TGG TT psSAMS
R: ATG GGT CAG TGG CAT AAG
F: TGG ATG TTC CTT AAA GAC AGT GG psTAT– D
R: TCT CAC AGT ATG GAG CGT CTG
F: GGT TCC TGT GCT CGT CTC AAT CTG PmAP2/ERF11
R: GGT ACT AGC GGC TGA GGC ATT AAA G
F: GGA CAG TGG AAG CAT CAT psGAPDH
R: AAC CGA ATA CAG CAA CAG A
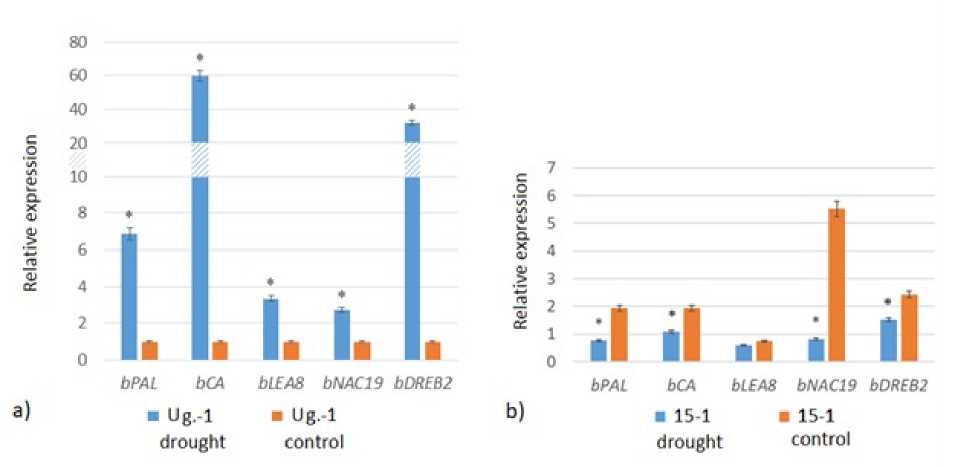
Figure 1. Expression of rought resistance genes in birch: a) ‘Ug.-1’ an b) ‘15– 1’. * significant change in expression is in icate
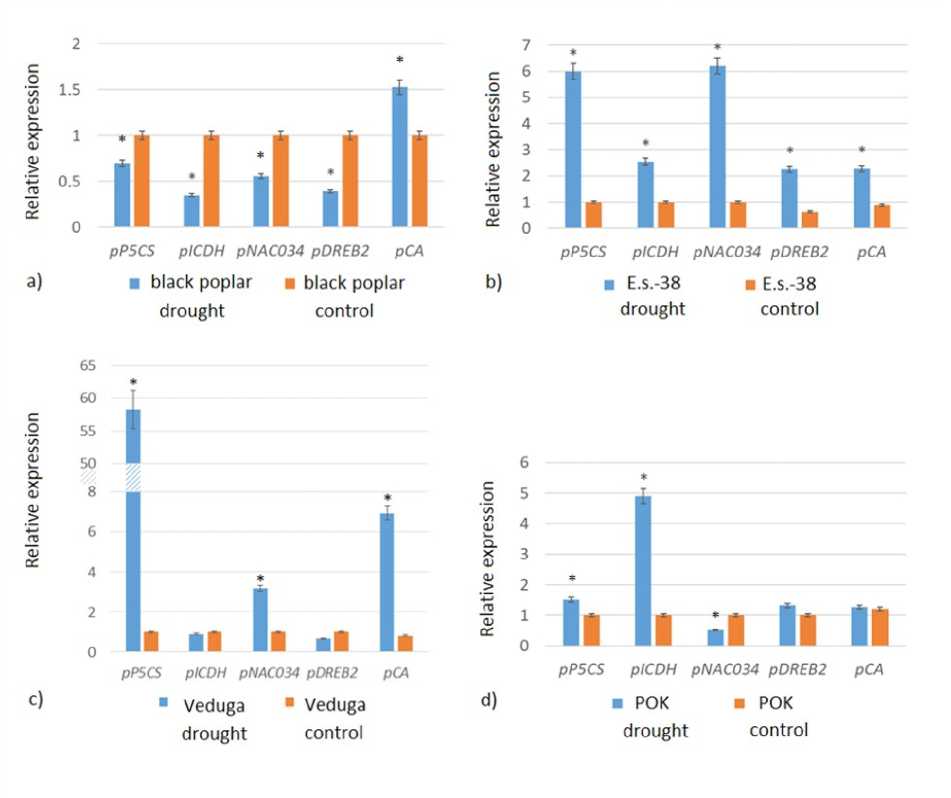
Figure 2. Expression of rought resistance genes in ifferent poplar genotypes: a) black poplar, b) ‘E.s.-38’, c) ‘POK’, ) ‘Ve uga’. * significant change in expression is note
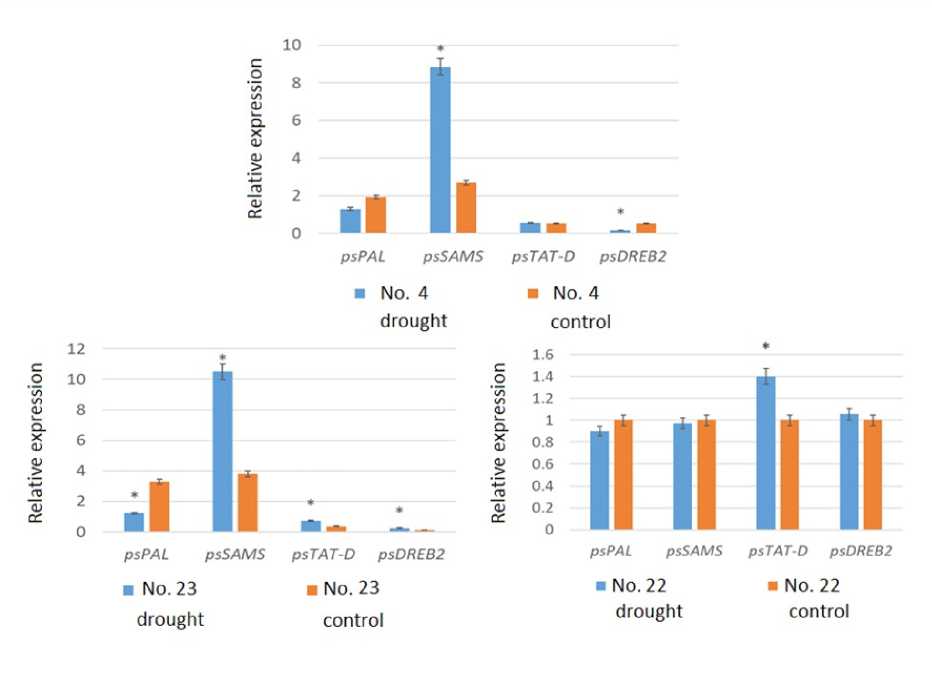
Figure 3 . Expression of roughtresistance genes in pine genotypes with contrastingsee pro uctivity
Abiotic stress
SAM
SAM.
ABAI
NAC
AP2/ERBP
NAC1
P5CS
PvPsbP
PvCA
LEA
ICDH
Transcription factors
SINAC10 НсТОЕЗ
ERF9(
DREB2 PtrMYB94 PvMYBlOl
MY В OsMYB30
ABA-independent
ABA-dependent
PAL
PAL a-ketoglutarate
Rubisco
Proline
NADPH
Osmoprotection
Lignin biosynthesis
Phenylalanine ammonia lyase
Oxidative stress reduction
--^--
Carbonic anhydrase
Photosystem II operation Photosynthesis
__M_
Functional protein
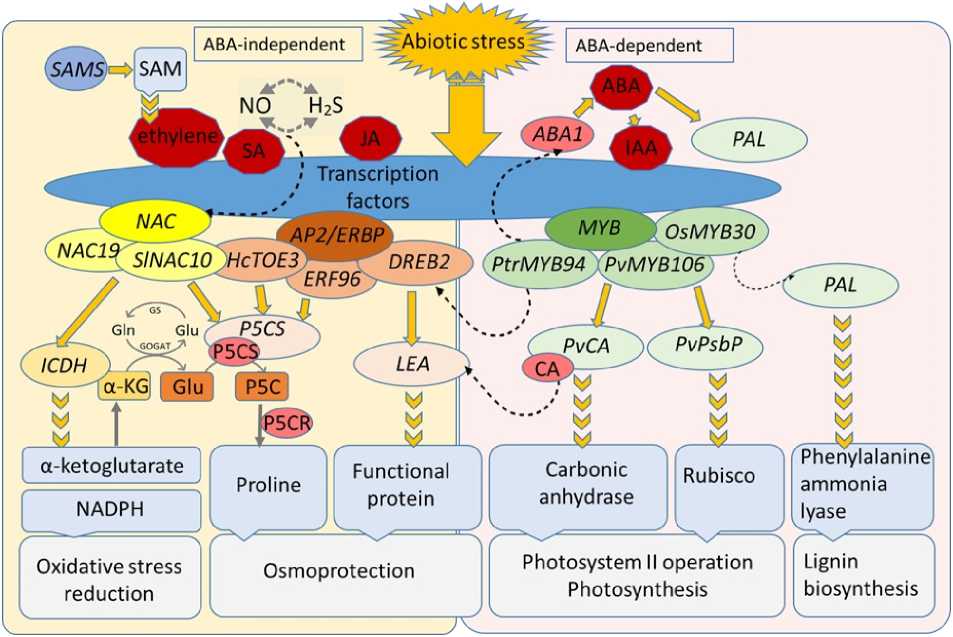
Figure 4 . Plant key genes of secon ary metabolism transcription regulation un er abiotic stress (author’s own composition).
DISCUSSION
The mechanism of a aptation to abiotic stress, such as rought, through the expression of transcription factors an the genes they control in plants can be represente as a iagram (Figure 4). A aptation to stress involves abscisic aci (ABA)- epen ent TF families, such as the ABA-responsive element/ABRE-bin ing factors ( ABRE/ABF ), WRKY , Nuclear Factor Y families, an ABA-in epen ent AP2/ERF an NAC ( NAM , ATAF1/2 , an CUC2 ) families (Yao et al. , 2021). Other authors attribute TFs of the AP2/ERBP subfamily ( AP2/ERF ) an NAC to be regulate by ABA, as well as cytokinins an jasmonate (JA) (Liu & Zhang, 2017; Khan et al ., 2019). One of the most important AP2/ERBP subfamilies is the DREB superfamily, whose members are involve in the mechanisms of response to rought, salinity, an low-temperature stress, with DREB2 pre ominantly associate with rought an salinity. The expression of DREB an , in particular, DREB2 genes was increase un er rought stress in Broussonetia papyrifera (Sun et al. , 2014) an Eucalyptus sp., (Nguyen et al. , 2017), Malus sp. (Zhao et al. , 2012), rought-sensitive an rought-tolerant coffee genotypes (Torres et al. , 2019). Overexpression of DREB2 promote rought tolerance in Robinia pseudoacacia (Xiu et al. , 2016).
In almon , rought-, col -, an light-relate stress-responsive elements an hormone-responsive cis- regulatory elements have been i entifie within the promoter regions of the PnaDREB family of proteins (Qian et al. , 2023). The Arabi opsis DREB2A orthologs in euphrates poplar have been shown to be involve in rought response (Yao et al. , 2021). In Pinus massoniana Lamb., nine PmAP2/ERF genes were constitutively expresse upon rought stress, an other genes showe up- or own-regulation in response to rought stress, which also epen e on the tissue stu ie (Sun et al. , 2022). NAC transcription factors are one of the largest families of transcription factors in plants an play an important role in their evelopment an response to a verse factors. The expression of SlNAC10 an NAC19 genes was in uce by rought, salinity an ABA (Du et al. , 2022; Khan et al ., 2019).
NAC19 expression showe an almost 150-fol increase upon exposure to S-nitrosocysteine, a nitric oxi e (NO) onor that plays a signaling role in stress (Khan et al. , 2019). In the present stu y, it was reveale that NAC an /or DREB2 transcription factors are involve in rought a aptation of birch genotypes ‘U.g.-1’, poplar ‘E.s.-38’ an ‘Ve uga’ an pine No. 23.
Phenylalanine ammonia lyase (PAL) is the first etermine enzyme of the phenylpropane metabolic pathway, involve in phenolic metabolism an playing a crucial role in plant growth an evelopment (Zhang et al., 2022; Fe uraev et al., 2020). PAL enco es the first enzyme of the lignin synthesis pathway, an extremely important in icator of abiotic stress, inclu ing salinity, waterlogging, rought, temperature fluctuations an heavy metal exposure (Han et al., 2022). High levels of PAL gene expression were foun in the roots of rought-tolerant wheat genotypes compare to rought-sensitive genotypes (Rasool et al., 2021). Increase PAL expression was observe un er rought stress in peony (Li et al., 2020), poplar hybri P. tremula × P. alba (Ahme et al., 2021). In Populus cathayana, an increase in the expression of the PAL isoform was etecte uring an experiment on the effect of rought, while the other isoform was activate only in plants from a rought-resistant population, but not in poplar growing in a well-watere population (Xiao et al., 2009). In hibiscus, the effect of rought suppresse the expression of the PAL gene, while the effect of salinity an abscisic aci provoke its activation (Jeong et al., 2012). The con ucte stu y shows that bPAL is involve in the response to rought in silver birch. In pine, psPAL was re uce in the low-yiel ing genotype No. 23 or i not iffer significantly from the control in genotypes No. 4 an No. 22 un er natural rought con itions, in icating the absence of a significant role of the PAL isoform in question in regulating the rought stress response in Scots pine. Plants overcome osmotic stress cause by rought through the synthesis of a wi e range of metabolites an functional proteins - ehy rins an late embryogenesis proteins (LEA) (Yao et al., 2021). It was reveale that the expression of the bLEA8 gene increase significantly in silver birch in response to rought, while in owny birch it i not iffer from the level of the control variant, which in icates the participation of LEA8 in the evelopment of the stress response in birch, on the one han , an the presence of genotypic ifferences in the activity of representatives of the late embryogenesis protein family in the two birch species un er rought con itions, on the other.
Carbonic anhy rase (CA) is a common protein in most photosynthetic organisms. In plants, CA isoforms belong to three ifferent families (α, β, an γ) an mainly catalyze the reversible conversion of CO2 to HCO3–. Ensuring the equilibrium of CA in this reaction is necessary to maintain the CO2 concentration for the function of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). CA re uces the resistance to CO2 iffusion insi e mesophyll cells by facilitating the transport of CO2 in both gas an liqui phases. Furthermore, physical an /or biochemical interactions between CA an other membrane-boun compartments, such as aquaporins, are propose to trigger the CO2 sensing response through stomatal movement (Momayyezi et al., 2020). Accor ing to current un erstan ing (Çevik et al., 2023], mil to mo erate rought stress stimulates carbonic anhy rase activity, an the main isoform involve in the stress response is β–CA. Severe rought causes a ecrease in CA activity, which may be ue to low intracellular CO2 levels (Momayyezi et al., 2020). Activation of Rubisco an carbonic anhy rase, as well as other proteins associate with carbon an nitrogen metabolism, coul contribute to the rought tolerance of cassava by increasing the efficiency of carbohy rate conversion an protecting the plant from oxi ative stress (Chang et al., 2019). CA activity generally increases un er mil to mo erate stress severity, in icating that CA is likely to be involve in a aptation to rought, high salinity, heat, bright light, free carbon eficiency, an bicarbonate excess (Polishchuk, 2021). In the stu y, bCA expression increase almost 60 times relative to the control in silver birch, while in owny birch, bCA expression ten e to ecrease, which may be ue to their genotypic ifferences. (Kotra e et al., 2019) showe that β–CA expression iffere in two oak species, in icating their species-specific characteristics.
S-a enosylmethionine (SAM) is wi ely involve in plant stress response an growth regulation, ethylene an polyamine biosynthesis, transmethylation an transsulfuration. This coenzyme is enco e by a group of genes, some of which showe an increase in expression in response to rought stress, which was more significant in rought-tolerant cotton than in rought-sensitive cotton (Sun et al., 2022). An increase in SAMS gene expression was also observe in tomato in response to rought an floo ing (Hei ari et al., 2020), an in cotton un er rought an salinity con itions (Kilwake et al., 2023). Un er rought con itions, the expression of lignin biosynthesis-relate genes such as SAMS can change significantly, suggesting that lignin content is an effective in icator for assessing rought tolerance (Han et al., 2022). The stu y foun that the psSAMS gene may be an important in icator of rought stress in Scots pine, which a apts to rought con itions, among other pathways, through lignin synthesis.
The Tat– D nuclease gene ( TAT– D ) is associate with programme cell eath (apoptosis), which is in uce by exposure to unfavorable factors, such as oxi ative, salt stress an UV irra iation (Qiu et al ., 2005). The effect of osmotic stress on the expression of the TAT– D gene was stu ie in embryogenic cell cultures of Scots pine. It was shown that the gene expression level i not change throughout the experiment (Muilu-Mäkelä et al. , 2015). The stu y reveale that the TAT– D gene was significantly activate in pine as a result of controlle rought exposure un er greenhouse con itions, as well as in the low-yiel ing genotype No. 23, while the change in expression in genotype No. 22 was insignificant, an in the me ium-yiel ing genotype No. 4 this in icator remaine at the control level. The obtaine results may in icate a negative impact of rought on sensitive pine genotypes, which was expresse in an increase in the expression of TAT–D , which is involve in the evelopment of the programme cell eath mechanism.
CONCLUSION
Thus, the key mechanisms involve in the evelopment of the response to rought were the biosynthesis of phenolic compoun s an lignin (increase expression of PAL an SAMS in birch an pine), proline an osmoprotectant proteins (activation of P5CS an LEA8 in poplar). The synthesis of proline an ehy rin proteins in ifferent tree species can be controlle by the ERF an NAC transcription factor families, which is known from previous stu ies an is confirme by the results of the present work, since the expression of the DREB2, NAC034 an NAC19 genes increase simultaneously with the increase in the level of LEA8 an P5CS transcripts. A aptation of photosynthesis also apparently plays an important role in rought in birch an poplar, which was expresse in an increase in the expression of the carbonic anhy rase gene CA, which probably contribute to maintaining the supply of CO2 for the normal functioning of Rubisco un er mo erate stress. The negative impact of stress on the stu ie tree species was also reveale in the course of the work, which was expresse in a ecrease in the expression of ICDH, which ensures the functioning of nitrogen metabolism an the supply of 2-oxoglutarate for the synthesis of amino aci s. An increase in the expression of TAT- D, which is involve in the evelopment of apoptosis, in sensitive pine genotypes (low-yiel ing pine genotype No. 23 un er greenhouse experiment con itions) also in icate a negative impact of rought. In owny birch ‘15-1’ an black poplar, the expression of all analyze genes was re uce , which in icates a negative impact of rought on these genotypes.
The stu ie genes are part of large gene families, the stu y of the expression of representatives of which uring rought will help to more eeply reveal the mechanisms of a aptation in tree species. The significant increase in pCA expression shown in the present work in icates an important role of CA in the evelopment of a response to rought an the a aptation of the photosynthetic apparatus to stress con itions, however, the specific mechanism of this process, especially in woo y plants, has not been establishe an requires further stu y.
ACKNOWLEDGEMENTS
The stu y was supporte by the Ministry of Science an Higher E ucation of the Russian Fe eration (state assignment No. 1023013000020-6-4.1.2 “Selection of economically valuable an climate-resistant tree crops with high biological pro uctivity an carbon sequestration potential, taking into account regional soil an climatic features for the implementation of forest climate projects (FZUR-2023-0002)”).
CONFLICTS OF INTEREST
The authors eclare that they have no potential conflicts of interest.
Список литературы Expression of resistance genes in breeding valuable genotypes of pine and fast-growing species of birch and poplar under drought
- Ahmed, U., Rao, M. J., Qi, C. and Xie, Q. (2021). Expression profiling of flavonoid biosynthesis genes and secondary metabolites accumulation in Populus under drought stress. Molecules, 26(18), 5546.
- Andriantelomanana, T., Ameglio, T., Delzon, S., Cochard, H. and Herbette, S. (2024). Unpacking the point of no return under drought in poplar: insight from stem diameter variation. New Phytologist, 242(2), 466-478.
- Bose, A. K., Gessler, A., Buntgen, U. and Rigling, A. (2024). Tamm review: Drought-induced Scots pine mortality-trends, contributing factors, and mechanisms. Forest Ecology and Management, 561, 121873.
- Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Current opinion in biotechnology, 56, 209-214.
- Cevik, S., Kurt, O., Guzel Deger, A. Y. §. i. N., Kocacinar, F. and Binzet, R. (2023). Beta-carbonic anhydrase gene expression levels change depending on the drought severity in both the leaves and roots of Arabidopsis thaliana. Turkish Journal of Botany, 47(6), 541-555.
- Chang, L., Wang, L., Peng, C., Tong, Z. and Wang, D. (2019). The chloroplast proteome response to drought stress in cassava leaves. Plant Physiology and Biochemistry, 142, 351-362.
- Chen, J., Xia, X. and Yin, W. (2009). Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica. Biochemical and Biophysical Research Communications, 378(3), 483-487.
- Corpas, F. J. and Palma, J. M. (2023). Functions of NO Di Stefano, V., Di Domenico, G., Menta, M., Pontuale, E., Bianchini, L. and Colantoni, A. (2024). Comparison between Different Mechanization Systems: Economic Sustainability of Harvesting Poplar Plantations in Italy. Forests, 15(3), 397.
- Du, M., Ding, G. and Cai, Q. (2018). The transcriptomic responses of Pinus massoniana to drought stress. Forests, 9(6), 326.
- Du, X., Su, M., Jiao, Y., Xu, S. and Song, J. (2022). A transcription factor SlNAC10 gene of Suaeda liaotungensis regulates proline synthesis and enhances salt and drought tolerance. International Journal of Molecular Sciences, 23(17), 9625.
- Feduraev, P., Skrypnik, L., Riabova, A., Pungin, A. and Tokupova, E. (2020). Phenylalanine and tyrosine as exogenous precursors of wheat (Triticum aestivum L.) secondary metabolism through PAL-associated pathways. Plants, 9(4), 476.
- Gongalves, A. Z. and Mercier, H. (2021). Transcriptomic and biochemical analysis reveal integrative pathways between carbon and nitrogen metabolism in Guzmania monostachia (Bromeliaceae) under drought. Frontiers in Plant Science, 12, 715289.
- Grodetskaya, T., Fedorova, O., and Evlakov, P. (2021, October). Optimized method for RNA extraction from leaves of forest tree species. In IOP Conference Series: Earth and Environmental Science (Vol. 875, No. 1, p. 012008). IOP Publishing.
- Han, X., Zhao, Y., Chen, Y., Xu, J. and Jiang, C. (2022). Lignin biosynthesis and accumulation in response to abiotic stresses in woody plants. Forestry Research, 2(1).
- Heidari, P., Mazloomi, F., Nussbaumer, T. and Barcaccia, G. (2020). Insights into the SAM synthetase gene family and its roles in tomato seedlings under abiotic stresses and hormone treatments. Plants, 9(5), 586.
- Jan, A. T., Singhal, P. and Haq, Q. M. R. (2013). Plant abiotic stress: deciphering remedial strategies for emerging problem. Journal of Plant Interactions, 8(2), 97-108.
- Jeong, M. J., Choi, B. S., Bae, D. W., Shin, S. C. and Park, S. U. (2012). Differential expression of kenaf phenylalanine ammonia-lyase (PAL) ortholog during developmental stages and in response to abiotic stresses. Plant Omics, 5(4), 392-399.
- Khan, M., Imran, Q. M., Shahid, M., Mun, B. G. and Lee, S. U. (2019). Nitric oxide-induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana. BMC Plant Biology, 19, 1-19.
- Kilwake, J. W., Umer, M. J., Wei, Y., Mehari, T. G. and Magwanga, R. O. (2023). Genome-Wide characterization of the SAMS gene family in cotton unveils the putative role of GhSAMS2 in enhancing abiotic stress tolerance. Agronomy, 13(2), 612.
- Kotrade, P., Sehr, E. M. and Bruggemann, W. (2019). Expression profiles of 12 drought responsive genes in two European (deciduous) oak species during a two-year drought experiment with consecutive drought periods. Plant Gene, 19, 100193.
- Lan, T., Gao, J. and Zeng, Q. Y. (2013). Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree genetics & genomes, 9, 253-264.
- Li, T., Wang, R., Zhao, D. and Tao, J. (2020). Effects of drought stress on physiological responses and gene expression changes in herbaceous peony (Paeonia lactiflora Pall.). Plant signaling & behavior, 15(5), 1746034.
- Liu, C. and Zhang, T. (2017). Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC genomics, 18, 1-16.
- Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-AACT method. methods, 25(4), 402-408.
- Ma, R., Xiao, Y., Lv, Z., Tan, H. and Chen, R. (2017). AP2/ERF transcription factor, Ii049, positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Frontiers in Plant Science, 8, 1361.
- Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends in Genetics, 13(2), 67-73.
- Momayyezi, M., McKown, A. D., Bell, S. C. and Guy, R. D. (2020). Emerging roles for carbonic anhydrase in mesophyll conductance and photosynthesis. The Plant Journal, 101(4), 831844.
- Muilu-Makela, R., Vuosku, J., Hamberg, L., Latva-Maenpaa, H. and Haggman, H. (2015). Osmotic stress affects polyamine homeostasis and phenolic content in proembryogenic liquid cell cultures of Scots pine. Plant Cell, Tissue and Organ Culture (PCTOC), 122, 709-726.
- Nguyen, H. C., Cao, P. B., San Clemente, H., Ployet, R. and Mounet, F. (2017). Special trends in CBF and DREB2 groups in Eucalyptus gunnii vs Eucalyptus grandis suggest that CBF are master players in the trade-off between growth and stress resistance. Physiologia plantarum, 159(4), 445467.
- Paakkonen, E., Seppanen, S., Holopainen, T., Kokko, H. and Karenlampi, S. (1998). Induction of genes for the stress proteins PR-10 and PAL in relation to growth, visible injuries and stomatal conductance in birch (Betula pendula) clones exposed to ozone and/or drought. New Phytologist, 138(2), 295-305.
- Polishchuk, O. V. (2021). Stress-related changes in the expression and activity of plant carbonic anhydrases. Planta, 253(2), 58.
- Qian, C., Li, L., Guo, H., Zhu, G. and Yang, N. (2023). Genome-Wide analysis of DREB family genes and characterization of cold stress responses in the woody plant Prunus nana. Genes, 14(4), 811.
- Qiu, J., Yoon, J. H. and Shen, B. (2005). Search for apoptotic nucleases in yeast: role of Tat-D nuclease in apoptotic DNA degradation. Journal of Biological Chemistry, 280(15), 15370-15379.
- Rasool, F., Uzair, M., Naeem, M. K., Rehman, N. and Afroz, A. (2021). Phenylalanine ammonia-lyase (PAL) genes family in wheat (Triticum aestivum L.): Genome-wide characterization and expression profiling. Agronomy, 11(12), 2511.
- Rini, D. (2019). Sequence variation of DREB2 gene as a potential molecular marker for identifying resistant plants toward drought stress. Nusantara Bioscience, 11(1), 35-43
- Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K. and Yamaguchi-Shinozaki, K. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell, 18(5), 1292-1309.
- Shinozaki, K. and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. Journal of experimental botany, 58(2), 221-227.
- Shinozaki, K., Yamaguchi-Shinozaki, K. and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Current opinion in plant biology, 6(5), 410-417.
- Sun, J., Peng, X., Fan, W., Tang, M. and Liu, J. (2014). Functional analysis of BpDREB2 gene involved in salt and drought response from a woody plant Broussonetia papyrifera. Gene, 535(2), 140-149.
- Sun, S., Liang, X., Chen, H., Hu, L. and Yang, Z. (2022). Identification of AP2/ERF transcription factor family genes and expression patterns in response to drought stress in Pinus massoniana. Forests, 13(9), 1430.
- Tikhomirova, T. S., Krutovsky, K. V., and Shestibratov, K. A. (2022). Molecular traits for adaptation to drought and salt stress in birch, oak and poplar species. Forests, 14(1), 7.
- Torres, L. F., Reichel, T., Déchamp, E., de Aquino, S. O. and Duarte, K. E. (2019). Expression of DREB-like genes in Coffea canephora and C. arabica subjected to various types of abiotic stress. Tropical Plant Biology, 12, 98-116.
- Wang, X., Hou, C., Zheng, K., Li, Q., Chen, S. and Wang, S. (2017). Overexpression of ERF96, a small ethylene response factor gene, enhances salt tolerance in Arabidopsis. Biologia Plantarum, 61, 693-701.
- Wang, X., Hou, C., Zheng, K., Li, Q., Chen, S. and Wang, S. (2017). Overexpression of ERF96, a small ethylene response factor gene, enhances salt tolerance in Arabidopsis. Biologia Plantarum, 61, 693-701.
- Wu, H. L., Li, L., Cheng, Z. C., Ge, W. and Gao, J. (2015). Cloning and stress response analysis of the PeDREB2A and PeDREB1A genes in moso bamboo (Phyllostachys edulis). Genetics and Molecular Research, 14(3), 10206-10223.
- Xiao, X., Yang, F., Zhang, S., Korpelainen, H. and Li, C. (2009). Physiological and proteomic responses of two contrasting Populus cathayana populations to drought stress. Physiologia plantarum, 136(2), 150-168.
- Xiong, C., Zhao, S., Yu, X., Sun, Y. and Li, H. (2020). Yellowhorn drought-induced transcription factor XsWRKY20 acts as a positive regulator in drought stress through ROS homeostasis and ABA signaling pathway. Plant Physiology and Biochemistry, 155, 187-195.
- Xiu, Y., Iqbal, A., Zhu, C., Wu, G. and Chang, Y. (2016). Improvement and transcriptome analysis of root architecture by overexpression of Fraxinus pennsylvanica DREB2A transcription factor in Robinia pseudoacacia L.'Idaho'. Plant biotechnology journal, 14(6), 1456-1469.
- Yanagisawa, S. (1998). Transcription factors in plants: physiological functions and regulation of expression. Journal of Plant Research, 111, 363371.
- Yao, T., Zhang, J., Xie, M., Yuan, G., Tschaplinski, T. J., Muchero, W. and Chen, J. G. (2021). Transcriptional regulation of drought response in Arabidopsis and woody plants. Frontiers in plant science, 11, 572137.
- Yin, F., Zeng, Y., Ji, J., Wang, P., Zhang, Y. and Li, W. (2021). The halophyte Halostachys caspica AP2/ERF transcription factor HcTOE3 positively regulates freezing tolerance in Arabidopsis. Frontiers in Plant Science, 12, 638788.
- Zhang, C., Yao, X., Ren, H., Wang, K. and Chang, J. (2022). Genome-wide identification and characterization of the phenylalanine ammonia-lyase gene family in pecan (Carya illinoinensis). Scientia Horticulturae, 295, 110800.
- Zhao, T., Liang, D., Wang, P., Liu, J., and Ma, F. (2012). Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Molecular genetics and genomics, 287, 423-436.


