Feature Selection based on Hybrid Binary Cuckoo Search and Rough Set Theory in Classification for Nominal Datasets
Автор: Ahmed F. Alia, Adel Taweel
Журнал: International Journal of Information Technology and Computer Science(IJITCS) @ijitcs
Статья в выпуске: 4 Vol. 9, 2017 года.
Бесплатный доступ
Feature Selection (FS) is an important process to find the minimal subset of features from the original data by removing the redundant and irrelevant features. It aims to improve the efficiency of classification algorithms. Rough set theory (RST) is one of the effective approaches to feature selection, but it uses complete search to search for all subsets of features and dependency to evaluate these subsets. However, the complete search is expensive and may not be feasible for large data due to its high cost. Therefore, meta-heuristics algorithms, especially Nature Inspired Algorithms, have been widely used to replace the reduction part in RST. This paper develops a new algorithm for Feature Selection based on hybrid Binary Cuckoo Search and rough set theory for classification on nominal datasets. The developed algorithm is evaluated on five nominal datasets from the UCI repository, against a number of similar NIAs algorithms. The results show that our algorithm achieves better FS compared to two known NIAs in a lesser number of iterations, without significantly reducing the classification accuracy.
Feature Selection, Rough Set Theory, Cuckoo Search, Binary Cuckoo Search, Nature Inspired Algorithms, Meta-heuristic Algorithms
Короткий адрес: https://sciup.org/15012639
IDR: 15012639
Текст научной статьи Feature Selection based on Hybrid Binary Cuckoo Search and Rough Set Theory in Classification for Nominal Datasets
Published Online April 2017 in MECS
The rapid growth of the number of features in structured datasets in classification causes a major problem, known as “curse of dimensionality”, that reduces the classification accuracy, increases the classification model complexity and increases the computational time, thus the need for Feature Selection (FS) [1] [2]. Koller and Sahami [3] define FS as selecting "a subset of features for improving the classification performance or reducing the complexity of the model without significantly decreasing the classification accuracy of the classifier built using only the selected features ".
Good FS approaches are capable of minimizing the number of selected features without significantly reducing the classification accuracy of all features [1] [4]. In general, the search strategy, which is used to select the candidate feature subsets, and the objective function, which is used to evaluate these candidate subsets, are the two main steps employed in any FS approach [1] [5].
Based on the objective function, existing FS approaches are categorized into two categories: filter approaches and wrapper approaches [1] [6][60]. Filter approaches select the feature subset independently from the classification algorithms using statistical characteristics, such as dependency degree [7] and information measurement [8] of the data to evaluate features of a subset [6][58]. But the wrapper approaches include a classification algorithm as part of the objective function to evaluate the selected feature subsets [2]. Filter approaches are often recognized as faster and more generic than wrapper approaches [2] [6].
The RST approach, proposed in the early 1980s, is one of the effective approaches to feature selection [9] [10] [11]. Its main concept is to search or generate all possible feature subsets and select the one with maximum dependency and a minimum number of features. However, its search strategy (complete search) is very expensive [10]. Several researchers have shown that meta-heuristic algorithms are more efficient for searching [12]. They have been shown to provide an efficient solution for an optimization problem. FS is effectively an optimization problem, since it needs less number of assumptions to find the near optimal feature subset [2] [5] [12]. Thus, many approaches combine meta-heuristic algorithms and RST to solve the FS with higher accuracy and lesser cost.
NIAs are an efficient type of meta-heuristic algorithms [13]. They process multiple candidate solutions concurrently, and they are developed based on characteristics of biological systems [13]. They are widely used for improving the search strategy in FS, because they are easy to implement and incorporate mechanisms to avoid getting trapped in local optima [54] [59][61][65]. These algorithms are also able to find best/optimal solution in a reasonable time with efficient convergence [13][14][65]. Ant Colony Optimization (ACO) [15], Particle Swarm Optimization (PSO) [16], Genetic algorithm (GA) [17], Artificial Bee Colony (ABC) [18], and Cuckoo Search (CS) are known examples of NIAs [65]. Binary CS (BCS) is a binary version of the CS, in which the search space is modelled as a binary string [19].
The ACO algorithm uses graph representation and needs more parameters than others, which makes it much more complex and more expensive compared to other NIAs [20] [16] [15] [18]. PSO, ABC, and BCS use binary representation, which makes their implementation easier and less expensive. The convergence of ABC and BCS is more efficient compared to ACO and PSO because ABC and BCS use a hybrid search mechanism. But BCS is easy to implement and has a more efficient convergence compared to ABC, it uses lesser number of parameters in its local search which makes it one of the fastest algorithms [14] [21][65].
There are several studies in filter FS for classification that combine NIA with Rough Set Theory Dependency Degree (RSTDD). [22] [23] [24][63] employ ACO with RSTDD, [25] [26] [27][62] combine PSO and RSTDD, while [28] [29] [30] [64] use ABC and RSTDD. But according to our knowledge, none of the existing FS approaches use BCS [19] [31] [32][55][56] and RSTDD. Thus this paper proposes a new classification filter FS approach for nominal datasets that combines between BCS and RSTDD. It minimizes the number of selected features (i.e. achieve significant Size Reduction % (SR %)) without significantly reducing the classification accuracy. This approach has developed an algorithm referred to as Feature Selection based on hybrid Binary Cuckoo Search and rough set theory in classification for nominal datasets (FS-BCS).
The remainder of this paper is organized as follows. Section 2 describes RST, CS and BCS. In section 3, details the proposed algorithm FS-BCS. The followed evaluation methodology is described in section 4. Section 5 presents the results and discussion. The conclusion is discussed in section 6. The last section presents the made assumptions and future work.
-
II. RST and Binary Cuckoo Search
RST and binary cuckoo search are the key concepts that our approach bases itself on, these are described briefly below.
-
A. Rough Set Theory
RST was developed, by Z Pawlak, in the early 1982s [9] as a mathematical tool that deals with classificatory analysis of data table. The advantages of RST are as follows [33] [11]: First, it provides efficient methods for finding hidden patterns in data. Second, it allows to reduce original data without additional information about data. Third, it is easy to understand. Fourth, it allows to evaluate the significance of data using data alone.
RSTDD and positive region are two important issues in data analysis to discover the dependency between the feature subsets and class labels. Positive region (POSp(Q)) contains all objects that can be classified into classes of Q using information in P. The RSTDD can be defined in equation (1) [10] [33] [34].
Y p =
| posp ( Q ) | | U |
Where |U| is the total number of objects, |POSp(Q)| is the number of objects in a positive region, and yp (Q) is the dependency between feature subset p and classes Q.
-
B. The Principle of Cuckoo Search
CS is a new and powerful NIA algorithm that was developed by Yang and Deb in 2009 [20]. CS is a search algorithm inspired by the breeding behavior of cuckoos and L’evy flight behavior of some birds and fruit flies which is a special case of random walks [20] [19] [35]. The reproduction strategy for Cuckoo is aggressive. Cuckoos use the nests of other host birds to lay their eggs in and rely on these birds for hosting the egg. Sometimes, the other host birds discover these strange eggs and they either throw these strange eggs or abandon their nest and build a new one. Cuckoos lay eggs that look like the pattern and color of the native eggs to reduce the probability of discovering them. If the egg of the cuckoo hatches first, it gets all the food that is provided by its host bird [20] [36].
Algorithmically, each nest represents a solution, CS aims to replace the "not so good" solution (nest) with a new one that is better. CS starts to generate the population of nests randomly, then in each iteration, CS uses hybrid search to update the population of nests as follows: The nests that have the lowest quality are updated randomly (global search) [20] [36]. And the remaining nests are updated using local search that uses L’evy flight via the equation (2) and equation (3) [19]:
x t + 1 = x t + a О L ’ evy ( 2 ) . (2)
Where xt+1 is a new candidate solution (nest), α is the step size (α>0) scaling factor of the problem, in most cases, we can use α=1. ʘ Means entry-wise multiplications, and λ: L’evy distribution coefficient (0 < λ ≤ 3). Random step length (L’evy (λ)) is calculated from power low by equation (3):
L ’evy uu = s "x . (3)
Where s is step size. L'evy flight is a semi-random search that moves in search space in diverse step lengths depending on the current location to find a good location. L'evy flight makes the CS more efficient in exploring the search space.
-
C. Binary Cuckoo Search
BCS is proposed by [19] in 2013. It is a binary version of the CS, in which the search space is modelled as a
Classification for Nominal Datasets binary n-bit string, where n is the number of features [19]. BCS represents each nest as a binary vector, where each 1 corresponds to a selected feature and 0 otherwise. This means each nest represents a candidate solution, and each egg represents a feature. The initialization strategy is generating an initial population of n nests randomly by initializing each nest with a vector of binary value. Then according to a probability pa ϵ [0, 1]), it selects the worst nests and updating them using L'evy flight. Finally, BCS stops when the number of iterations reaches the maximum predefined by the user, and produces the best global nest. Algorithm 1 explains how BCS work.
In order to build binary vectors in local search, it employs equation (4) and equation (5) [19]:
t1
-
5 ( x ( i . j ) ) =----------------. (4)
-
1 + e - x t ( j
1, 5 ( x t ( i , j ) ) > °
0, otherwise
(features) at nest i in iteration t.
-
III. The Proposed Algorithm
Feature selection based on hybrid binary cuckoo search and rough set theory in classification for nominal datasets (FS-BCS) uses BCS as a search technique to generate candidate feature subsets, and it uses objective function based on RSTDD to evaluate these candidates' features subsets to guide the BCS to reach best feature subset. This objective function is proposed by Jensen et al. [30] that selects the minimum number of selected features and maximum classification accuracy, it is shown in equation (6). The proposed FS-BCS is presented in Algorithm 1.
Objective Fuh.(R)=/r(D) *1C | 1R 1 (6)|C |
Where |C| is the number of conditional (total-class) features, |R| is the number of selected features, D is class, and Y r (D) is the dependency degree between feature subset (selected features) and class label.
Where о belongs to [0, 1], x^p stands for new eggs js
Input
-
1: Number of nests N. // Each nest represent candidate feature subset
-
2: Probability p a ϵ [0, 1].
-
3: Maximum number of iteration T, initial iteration t=0.
Output
-
4: Best global feature subset g_best.
Initialization (Initialize the population of nests and best global feature subset g_best).
-
5: for i=1 to N do
-
6: Generate Initial population of N candidate feature subsets x i randomly.
-
7: Sort the x i descending according to value of objective function.
-
8: Evaluate x i using equation (6).
-
9: Best local feature subset=top candidate feature subset.
-
10: g_best=best local feature subset.
-
11: end for
Updating the population several time ( T times)
-
12: Repeat
-
13: Abandon a fraction p a of worse candidate feature subsets and updating them using L´evy flight.
-
14: Evaluate each candidate feature subset in population
-
15: Sort the population of nests descending according to value of objective function.
-
16: Best local solution= top candidate feature subset in the population.
-
17: If g_best< best local solution
-
18: g_best=local best solution.
-
19: end if.
-
20: t=t+1.
-
21: Until t
-
22: Produce the global best solution (g_best).
Algorithm 1. FS-BCS algorithm
-
IV. Evaluation
There are a number of potential ways to evaluate our approach. However, the most commonly used and perhaps provides comparative results on its efficiency is one that uses existing datasets, which have known features, and then compares our approach to a set of other competitive approaches. These are described below.
-
A. Datasets Selection
To evaluate the performance of FS-BCS, a group of experiments have been run on five nominal datasets with different number of features (from 9 to 36), different number of objects (from 366 to 8124) and different number of classes (from 2 to 19). These nominal datasets are randomly selected from a set that has very little missing values from the University of California at Irvine, known as the UCI data repository of machine learning database [37]. Table 1 shows the selected datasets and their characteristics.
Table 1. Datasets
|
Dataset |
Features |
Objects |
Classes |
|
Breast-W |
9 |
699 |
2 |
|
Mushroom |
22 |
8124 |
2 |
|
Dermatology |
34 |
366 |
6 |
|
Soybean(large) |
35 |
683 |
19 |
|
Chess |
36 |
3196 |
2 |
The following is a brief description of the datasets.
Breast-W. It is obtained from the university of Wisconsin hospitals, it has 9 features, and each one has ten distinct values. There are 699 objects, 458 objects classified into benign class, and the remaining objects are classified into malignant class.
Mushroom. It contains records drawn from the Audubon society field guide to North American mushrooms. This dataset has 22 features with different number of distinct values (from 2 to 12). Also, it has 8124 objects that are classified roughly into two classes.
Dermatology. This dataset has 34 features which have distinct values from 2 to 4. 366 objects are classified into six classes, 61 objects for first class, 112 objects for second class, 72 objects for third class, 52 for fourth class, 49 for fifth class, and 20 objects for sixth class.
Soybean (large). The task is to diagnose diseases in soybean plants. There are 304 objects which are classified into 19 classes.
Chess (King-Rook vs. King-Pawn). It has 36 features which have 2 distinct values. This dataset has 3196 objects, 1669objects belong to White can win class, and remaining objects belong to White cannot win class.
-
B. Evaluation Method
The indirect approach [2][57] is used to evaluate the developed algorithm, three comparisons are used in the experiments. First, “before and after comparison” is employed, which measures the classification accuracy of all available features and of the selected features. Secondly, our approach is evaluated against the Genetic algorithm [17] with Correlation based on Feature Selection (CFS) [38]. Thirdly, our approach is evaluated against PSO [39] with CFS [38]. Decision Tree (DT) [40] and Naive Bayes (NB) [41], which are two different classification algorithms (types), are used to measure the classification accuracy for all approaches that are used in the experiments. DT and NB are from the top ten data mining algorithms, and do not need complex initial parameters [42]. These are evaluated against three factors, their number of selected features, classification accuracy and number of iterations.
All implementations are run on a personal computer running Windows 8.1 with (i7), 3.0 GHZ processor and 16 GB memory. FS-BCS is implemented with PHP. The Genetic algorithm with CFS and PSO are known filter FS approaches, and are already implemented in the Weka tool [43]. The Genetic and PSO algorithms are selected, they are common NIAs for FS, and they are implemented in the Weka tool. CFS is an efficient objective function for FS, because it measures the redundant and relevant features in each candidate feature subset by evaluating the correlation between each feature and the class labels and between each pair of features using mutual information [38].
All parameters in these approaches are selected according to default parameters in the Weka tool, some of them are shown as follows: population size is 20, maximum number of iterations is 20, and the size of k is 10 objects in K-fold cross-validation which will be mentioned later in this section.
Each dataset was divided into two datasets randomly, training set, and a learning and test set. A training set that has about 70% of the dataset objects, and a learning and test set has about 30% of the dataset objects [44]. The training set is used by FS approaches to achieve features reduction. The learning and test set is used to build the classification model and estimates the performance of classification. We use K-fold cross-validation [45], also implemented in the Weka tool, to split the learning and test set into two disjoint sets to build the model and estimate the classification performance using DT and NB. When an object belongs to the test set, its class is hidden from the built classification model based on the learning set only.
In this work, we found, running the FS-BCS five times over a training set was sufficient to obtain good results, running the experiments longer than five times did not provide additional value. The best run, with the best classification accuracy, is selected.
-
V. Results and Discussion
In these experiments, classification Accuracy (Acc) is used to evaluate the classification performance, and Size Reduction percentage (SR %) is used to evaluate the percentage of removed features compared to all available features (or Subset Size (SS), which is the number of
Classification for Nominal Datasets selected features). In table 2, all means all features in a dataset. The best approach is the one that achieves maximum SR% without significantly reducing the classification accuracy compared to the total number of features in the least number of iterations [1]. Differences in accuracy is considered significant when it is more than 5% [46], and it is considered the same when it is less than 1% [47].
Table 2 shows that FS-BCS achieves better SR% without significantly reducing the classification accuracy in lesser number of iterations compared to the Genetic and PSO approaches. The main reason for these results is that BCS’s convergence is more efficient and faster compared to convergence of the PSO and Genetic approaches, because BCS uses efficient and simple hybrid mechanism [20] [19]. To be more specific, the local search in the BCS's hybrid mechanism plays the main role in the efficiency and speed of its convergence, because it uses levy flight technique to maximize the guarantee and speed of convergence [35]. The following subsections discuss the experiments results in more details in reference to SR%, "before and after FS" and a number of iterations.
-
A. Size Reduction Percentage and Classification Accuracy
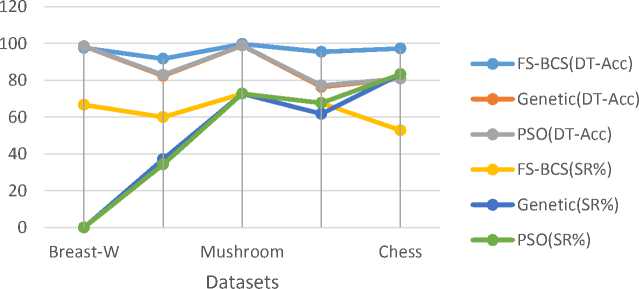
Table 2 and fig. 1 show the results of the comparisons between the FS-BCS, PSO and Genetic approaches.
The results show that FS-BCS achieves better FS for four datasets and the same for one dataset compared to the Genetic and PSO. In Breast-W dataset, FS-BCS achieves significant SR% (66.7%) compared to PSO and genetic (SR%=0), while the classification accuracy, according to DT and NB, is the same in the three approaches. FS-BCS achieves better SR% and classification accuracy according to DT and NB for Soybean (large). In Mushroom dataset, three approaches achieved the same FS (SR% and classification accuracy). But FS-BCS is capable to achieve better SR% and classification accuracy (DT and NB) in the dermatology dataset compared to the Genetic approach. Also in this dataset, FS-BCS achieves the same SR% and better classification accuracy (DT and NB) compared to PSO approach. The Genetic and PSO approaches achieve better SR% and significant reduction in classification accuracy (DT and NB) compared to FS-BCS, which achieves SR% without significant DT classification accuracy and improvement NB classification accuracy.
Table 2. Results of FS-BCS, genetic and PSO approaches
|
Dataset |
Method |
Size |
SR% |
DT Acc. |
NBAcc. |
|
Breast-W |
All |
9 |
93.9 |
96.1 |
|
|
FS-BCS |
3 |
66.7 |
97.6 |
98.1 |
|
|
Genetic |
9 |
0 |
98.5 |
98.1 |
|
|
PSO |
9 |
0 |
98.5 |
98.1 |
|
|
Mushroom |
All |
22 |
100 |
95.8 |
|
|
FS-BCS |
6 |
72.7 |
99.7 |
98.1 |
|
|
Genetic |
6 |
72.7 |
98.8 |
98.7 |
|
|
PSO |
6 |
72.7 |
98.8 |
98.8 |
|
|
Soybean (large) |
All |
35 |
92.3 |
92 |
|
|
FS-BCS |
14 |
60 |
91.7 |
90.7 |
|
|
Genetic |
22 |
37.1 |
82.4 |
85.3 |
|
|
PSO |
23 |
34.2 |
82.9 |
84.8 |
|
|
Dermatology |
All |
34 |
93.9 |
97.2 |
|
|
FS-BCS |
11 |
67.6 |
95.4 |
94.5 |
|
|
Genetic |
13 |
61. 6 |
76.3 |
86.3 |
|
|
PSO |
11 |
67.6 |
77.2 |
78.1 |
|
|
Chess |
All |
36 |
99.4 |
87.8 |
|
|
FS-BCS |
17 |
52.7 |
97.3 |
90.8 |
|
|
Genetic |
6 |
83.3 |
80.9 |
78.6 |
|
|
PSO |
6 |
80.9 |
81.2 |
83.3 |
All: Original Datasets. Size: Number of features. SR%: Percentage of size reduction against all features Acc: Accuracy.
CD сю
CD
CD
Datasets
—•—FS-BCS(NB-Acc)
—•—Genetic(NB-Acc)
—•—PSO(NB-Acc)
—•— FS-BCS(SR%)
—•—Genetic(SR%)
—•— PSO(SR%)
Fig.1. Comparisons between FS-BCS, genetic and PSO based on SR% and Classification Accuracy.
On average, in all datasets that are used in our experiment, FS-BCS achieves about 64% SR% and improves the DT and NB classification accuracy compared to the Genetic and PSO approaches, which achieve 50.9% and 51.6% SR% respectively
-
B. Classification Accuracy “before and after” FS-BCS, Genetic and PSO.
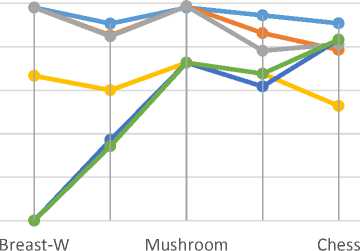
The results in Table 2 and fig. 2 show the classification accuracy according to DT and NB classification algorithms before and after running the FS-BCS, Genetic and PSO.
These results show that FS-BCS improves the classification accuracy-DT in breast-W and Dermatology datasets, as well as in the Soybean (large) and mushroom, while in the chess dataset, there is insignificant DT reduction. But the genetic and PSO achieve significant DT reduction in three datasets. According to NB classification algorithm, FS-BCS improves the NB in three datasets, and it achieves slight reduction, in NB, in two datasets, while the Genetic and PSO achieve significant reduction.
These results show that FS-BCS improves the classification accuracy-DT in breast-W and Dermatology datasets, as well as in the Soybean (large) and mushroom, while in the chess dataset, there is insignificant DT reduction. But the Genetic and PSO achieve significant DT reduction in three datasets. According to NB classification algorithm, FS-BCS improves the NB in three datasets, and it achieves slight reduction in NB in two datasets, while genetic and PSO achieve significant reduction.
Finally, according to DT and NB classification algorithms, FS-BCS succeeded to avoid significant reduction in classification accuracy for all the five datasets, while the Genetic and PSO failed to. Since FS-BCS is a general filter FS approach, it is efficient for DT and NB, which are different types of classification algorithms. According to [1], when a filter FS approach is efficient for DT and NB, it can be generalised for different types of classification algorithms, thus the results of FS-BCS.
Classification for Nominal Datasets
Q
го
о го
го
со
го
ого
го
IIII
Breast-W
Mushroom
Datasets
Chess
IIII
Breast-W
Mushroom
Chess
-
■ Before FS
-
■ After FS-BCS
-
■ After Genetic
-
■ After PSO
-
■ Before FS
-
■ After FS-BCS
-
■ After Genetic
-
■ After PSO
Datasets
Fig.2. Classification Accuracy Before and After FS
-
C. Analysis based on the number of iterations.
Table 3 and fig. 3 show the number of iterations needed to find the best solution for FS-BCS, genetic and PSO approaches. As shown, FS-BCS requires the least number of iterations compared to genetic and PSO in all datasets to reach the best solution. FS-BCS was able to find the best solution in 25% and 34% (these percentages are the average of all datasets) from the number of iterations in the Genetic and PSO respectively.
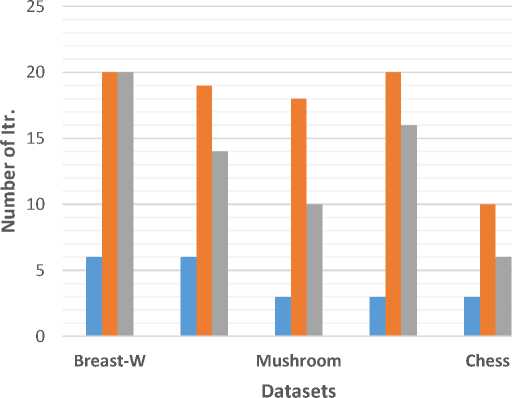
Fig.3. C omparisons between FS-BCS, genetic and PSO based on number of iterations
■ FS-BCS ■ Genetic ■ PSO
Table 3. Number of iterations of FS-BCS, genetic and PSO approaches
|
Dataset |
FS-BCS |
Genetic |
PSO |
|
Breast-W |
6 |
20 |
20 |
|
Mushroom |
6 |
19 |
14 |
|
Soybean (large) |
3 |
18 |
10 |
|
Dermatology |
3 |
20 |
16 |
|
Chess |
3 |
10 |
6 |
-
VI. Conclusion
BCS has quick and efficient convergence, less complexity, easier to implement, and fewer parameters compared to other NIAs such as ACO, ABC and PSO, and thus is increasingly used in many algorithms as a search technique. Similarly, RSTDD is used in many filter FS approaches as an objective function, for its relatively cheaper, easier to implement characteristics, and does not need any preliminary or additional information.
This paper proposed a new algorithm, FS-BCS, as a classification filter FS approach for nominal datasets using BCS and RSTDD to achieve significant SR% without significantly reducing the classification accuracy compared to the total number of features in lesser number of iterations. According to our knowledge, FS-BCS is the first approach that combines the BCS with RSTDD.
The experimental results show that FS-BCS achieved better FS in four datasets, and same FS in the fifth dataset compared to known filter FS approaches (PSO and Genetic) in the Weka tool implementations.
On average, FS-BCS achieved significantly higher SR% and improved the DT and NB classification accuracy compared to the Genetic and PSO approaches. Also our approach succeeded to maximize the SR% without significantly reducing the classification accuracy compared to the total number of features for all datasets, while PSO failed to do so in two datasets and Genetic failed in three datasets. Our approach also took lesser number of iterations to reach the best solution compared to PSO and genetic approaches. On average our approach needed significantly lesser number of iterations than those needed in Genetic and PSO.
Although these results show that BCS with RSTDD has faster and more efficient convergence compared to PSO and Genetic approaches, the authors appreciate further experiments would needed to evaluate the scalability of the proposed approach on large high dimensional datasets.
Список литературы Feature Selection based on Hybrid Binary Cuckoo Search and Rough Set Theory in Classification for Nominal Datasets
- H. Liu and H. Motoda, Feature selection for knowledge discovery and data mining, vol. 454, Springer Science & Business Media, 2012.
- I. A. Gheyas and L. S. Smith, "Feature subset selection in large dimensionality domains," Pattern recognition, vol. 43, no. 1, pp. 5-13, 2010.
- D. Koller and M. Sahami, "Toward optimal feature selection," 1996.
- N. Kwak and C.-H. Choi, "Input feature selection for classification problems," IEEE Transactions on Neural Networks, vol. 13, no. 1, pp. 143-159, 2002.
- M. Dash and H. Liu, "Feature selection for classification," Intelligent data analysis, vol. 1, no. 3, pp. 131-156, 1997.
- O. Maimon and L. Rokach, "Introduction to knowledge discovery and data mining," in Data mining and knowledge discovery handbook, Springer, 2009, pp. 1-15.
- L. Yu and H. Liu, "Efficient feature selection via analysis of relevance and redundancy," Journal of machine learning research, vol. 5, no. Oct, pp. 1205-1224, 2004.
- P. A. Estvez, M. Tesmer, C. A. Perez and J. M. Zurada, "Normalized mutual information feature selection," IEEE Transactions on Neural Networks, vol. 20, no. 2, pp. 189-201, 2009.
- Z. Pawlak, "Rough sets," International Journal of Computer & Information Sciences, vol. 11, no. 5, pp. 341-356, 1982.
- R. Jensen and Q. Shen, "Semantics-preserving dimensionality reduction: rough and fuzzy-rough-based approaches," IEEE Transactions on knowledge and data engineering, vol. 16, no. 12, pp. 1457-1471, 2004.
- Z. Pawlak, Rough sets: Theoretical aspects of reasoning about data, vol. 9, Springer Science & Business Media, 2012.
- H. Zhao, A. P. Sinha and W. Ge, "Effects of feature construction on classification performance: An empirical study in bank failure prediction," Expert Systems with Applications, vol. 36, no. 2, pp. 2633-2644, 2009.
- I. Fister Jr, X.-S. Yang, I. Fister, J. Brest and D. Fister, "A brief review of nature-inspired algorithms for optimization," arXiv preprint arXiv:1307.4186, 2013.
- Z. Beheshti and S. Shamsudding, "A review of population-based meta-heuristic algorithms," Int. J. Adv. Soft Comput. Appl, vol. 5, no. 1, pp. 1-35, 2013.
- M. Dorigo, V. Maniezzo and A. Colorni, "Ant system: optimization by a colony of cooperating agents," IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), vol. 26, no. 1, pp. 29-41, 1996.
- R. Poli, J. Kennedy and T. Blackwell, "Particle swarm optimization," Swarm intelligence, vol. 1, no. 1, pp. 33-57, 2007.
- D. Goldberg, "Genetic algorithms in search, optimization, and machine learning. Reading, MA: Addison-Wesley; 1989," Objective Function Value [Eq.(18)].
- B. Basturk and D. Karaboga, "An artificial bee colony (ABC) algorithm for numeric function optimization," in IEEE swarm intelligence symposium, 2006.
- D. Rodrigues, L. A. Pereira, T. Almeida, J. P. Papa, A. Souza, C. C. Ramos and X.-S. Yang, "BCS: A Binary Cuckoo Search algorithm for feature selection," in 2013 IEEE International Symposium on Circuits and Systems (ISCAS2013), 2013.
- X.-S. Yang and S. Deb, "Cuckoo search via L{\'e}vy flights," in Nature & Biologically Inspired Computing, 2009. NaBIC 2009. World Congress on, 2009.
- S. Kamat and A. Karegowda, "A brief survey on cuckoo search applications," Int. J. Innovative Res. Comput. Commun. Eng, vol. 2, no. 2, 2014.
- L. Ke, Z. Feng and Z. Ren, "An efficient ant colony optimization approach to attribute reduction in rough set theory," Pattern Recognition Letters, vol. 29, no. 9, pp. 1351-1357, 2008.
- R. Jensen and Q. Shen, "Finding rough set reducts with ant colony optimization," in Proceedings of the 2003 UK workshop on computational intelligence, 2003.
- M. Mafarja and D. Eleyan, "Ant colony optimization based feature selection in rough set theory," International Journal of Computer Science and Electronics Engineering (IJCSEE) Volume, vol. 1, no. 2, 2013.
- X. Wang, J. Yang, X. Teng, W. Xia and R. Jensen, "Feature selection based on rough sets and particle swarm optimization," Pattern Recognition Letters, vol. 28, no. 4, pp. 459-471, 2007.
- H. Shen, S. Yang and J. Liu, "An attribute reduction of rough set based on PSO," in International Conference on Rough Sets and Knowledge Technology, 2010.
- H. H. Inbarani, A. T. Azar and G. Jothi, "Supervised hybrid feature selection based on PSO and rough sets for medical diagnosis," Computer methods and programs in biomedicine, vol. 113, no. 1, pp. 175-185, 2014.
- Y. Hu, L. Ding, D. Xie and S. Wang, "A Novel Discrete Artificial Bee Colony Algorithm for Rough Set-based Feature Selection.," International Journal of Advancements in Computing Technology, vol. 4, no. 6, 2012.
- N. Suguna and K. Thanushkodi, "A novel rough set reduct algorithm for medical domain based on bee colony optimization," arXiv preprint arXiv:1006.4540, 2010.
- N. Suguna and K. G. Thanushkodi, "An independent rough set approach hybrid with artificial bee colony algorithm for dimensionality reduction," American Journal of Applied Sciences, vol. 8, no. 3, p. 261, 2011.
- M. Moghadasian and S. P. Hosseini, "Binary Cuckoo Optimization Algorithm for Feature Selection in High-Dimensional Datasets," in International Conference on Innovative Engineering Technologies (ICIET’2014), 2014.
- M. Sudha and S. Selvarajan, "Feature Selection Based on Enhanced Cuckoo Search for Breast Cancer Classification in Mammogram Image," Circuits and Systems, vol. 7, no. 04, p. 327, 2016.
- Z. Pawlak, "Some issues on rough sets," in Transactions on Rough Sets I, Springer, 2004, pp. 1-58.
- S. Rissino and G. Lambert-Torres, "Rough set theory--fundamental concepts, principals, data extraction, and applications," Data mining and knowledge discovery in real life applications, p. 438, 2009.
- X.-S. Yang and S. Deb, "Engineering optimisation by cuckoo search," International Journal of Mathematical Modelling and Numerical Optimisation, vol. 1, no. 4, pp. 330-343, 2010.
- J. Jona and N. Nagaveni, "Ant-cuckoo colony optimization for feature selection in digital mammogram," Pakistan Journal of Biological Sciences, vol. 17, no. 2, p. 266, 2014.
- C. Blake and C. J. Merz, "Repository of machine learning databases," 1998.
- M. A. Hall, "Correlation-based feature selection for machine learning," 1999.
- A. Moraglio, C. Di Chio and R. Poli, "Geometric particle swarm optimisation," in European conference on genetic programming, 2007.
- S. L. Salzberg, "C4. 5: Programs for machine learning by j. ross quinlan. morgan kaufmann publishers, inc., 1993," Machine Learning, vol. 16, no. 3, pp. 235-240, 1994.
- G. H. John and P. Langley, "Estimating continuous distributions in Bayesian classifiers," in Proceedings of the Eleventh conference on Uncertainty in artificial intelligence, 1995.
- X. Wu, V. Kumar, J. R. Quinlan, J. Ghosh, Q. Yang, H. Motoda, G. J. McLachlan, A. Ng, B. Liu, S. Y. Philip and others, "Top 10 algorithms in data mining," Knowledge and information systems, vol. 14, no. 1, pp. 1-37, 2008.
- G. Holmes, A. Donkin and I. H. Witten, "Weka: A machine learning workbench," in Intelligent Information Systems, 1994. Proceedings of the 1994 Second Australian and New Zealand Conference on, 1994.
- K. K. Dobbin and R. M. Simon, "Optimally splitting cases for training and testing high dimensional classifiers," BMC medical genomics, vol. 4, no. 1, p. 1, 2011.
- J. D. Rodriguez, A. Perez and J. A. Lozano, "Sensitivity analysis of k-fold cross validation in prediction error estimation," IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 32, no. 3, pp. 569-575, 2010.
- N. Parthalain, Q. Shen and R. Jensen, "A distance measure approach to exploring the rough set boundary region for attribute reduction," IEEE Transactions on Knowledge and Data Engineering, vol. 22, no. 3, pp. 305-317, 2010.
- B. Xue, M. Zhang and W. N. Browne, "Particle swarm optimisation for feature selection in classification: Novel initialisation and updating mechanisms," Applied Soft Computing, vol. 18, pp. 261-276, 2014.
- G. Lang, Q. Li and L. Guo, "Discernibility matrix simplification with new attribute dependency functions for incomplete information systems," Knowledge and information systems, vol. 37, no. 3, pp. 611-638, 2013.
- E.-G. Talbi, Metaheuristics: from design to implementation, vol. 74, John Wiley & Sons, 2009.
- D. Henderson, S. H. Jacobson and A. W. Johnson, "The theory and practice of simulated annealing," in Handbook of metaheuristics, Springer, 2003, pp. 287-319.
- R. Ruiz, J. C. Riquelme and J. S. Aguilar-Ruiz, "Fast feature ranking algorithm," in International Conference on Knowledge-Based and Intelligent Information and Engineering Systems, 2003.
- I. Guyon and A. Elisseeff, "An introduction to variable and feature selection," Journal of machine learning research, vol. 3, no. Mar, pp. 1157-1182, 2003.
- K. Kira and L. A. Rendell, "The feature selection problem: Traditional methods and a new algorithm," in AAAI, 1992.
- P. Shrivastava, A. Shukla, P. Vepakomma, N. Bhansali and K. Verma, "A survey of nature-inspired algorithms for feature selection to identify parkinson's disease," Computer Methods and Programs in Biomedicine, 2016.
- S. A. Medjahed, T. A. Saadi, A. Benyettou and M. Ouali, "Binary Cuckoo Search algorithm for band selection in hyperspectral image classification," IAENG International Journal of Computer Science, vol. 42, no. 3, 2015.
- M. Sudha and S. Selvarajan, "Feature Selection Based on Enhanced Cuckoo Search for Breast Cancer Classification in Mammogram Image," Circuits and Systems, vol. 7, no. 04, p. 327, 2016.
- C. Freeman, D. Kuli{\'c} and O. Basir, "An evaluation of classifier-specific filter measure performance for feature selection," Pattern Recognition, vol. 48, no. 5, pp. 1812-1826, 2015.
- E. Ghumare, M. Schrooten, R. Vandenberghe and P. Dupont, "Comparison of different Kalman filter approaches in deriving time varying connectivity from EEG data," in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2015.
- R. Diao and Q. Shen, "Nature inspired feature selection meta-heuristics," Artificial Intelligence Review, vol. 44, no. 3, pp. 311-340, 2015.
- S. Goswami and A. Chakrabarti, "Feature selection: A practitioner view," International Journal of Information Technology and Computer Science (IJITCS), vol. 6, no. 11, p. 66, 2014.
- R. Parimala and R. Nallaswamy, "Feature selection using a novel particle swarm optimization and It’s variants," International Journal of Information Technology and Computer Science (IJITCS), vol. 4, no. 5, p. 16, 2012.
- X. Jian, L. Fu, H. H. Tao and W. Haiwei, "Rough reduction algorithm for reduction of metagenomic DNA digital signature," in Control and Decision Conference (CCDC), 2016 Chinese, 2016.
- P. R. K. Varma, V. V. Kumari and S. S. Kumar, "A novel rough set attribute reduction based on ant colony optimisation," International Journal of Intelligent Systems Technologies and Applications, vol. 14, no. 3-4, pp. 330-353, 2015.
- S. Chebrolu and S. G. Sanjeevi, "Attribute reduction on real-valued data in rough set theory using hybrid artificial bee colony: extended FTSBPSD algorithm," Soft Computing, pp. 1-27, 2016.
- A. Alia and A. Taweel, "Hybrid Nature Inspired Algorithms and Rough Set Theory in Feature Selection for Classification: A Review," International Journal of Innovative Research in Computer and Communication Engineering, vol. 3, p. 7, 2016.


