Feature selection to classify healthcare data using wrapper method with pso search
Автор: Thinzar Saw, Phyu Hnin Myint
Журнал: International Journal of Information Technology and Computer Science @ijitcs
Статья в выпуске: 9 Vol. 11, 2019 года.
Бесплатный доступ
As a result of the rapid development of technology, data that contain a large number of features are produced from various applications such as biomedical, social media, face recognition, etc. Processing of these data is a challenging task to existing data mining and machine learning algorithms to make the decision. To reduce the size of the data for processing, a feature selection technique is needed. The feature selection is a well-known attribute selection or variable selection. The objective of the feature selection is to minimize the number of attributes contains in the dataset by eliminating the unwanted and repeated attributes to improve the classification accuracy and reduce the computation cost. Although various feature selection methods are proposed, in literature, to classify the healthcare data especially cancer diagnosis, finding an informative feature for medical datasets has still remained a challenging issue in the data mining and machine learning domain. Therefore, this paper presents a feature selection approach with the wrapper method (WFS) using particle swarm optimization (PSO) search to improve the accuracy of healthcare data classification. This work is evaluated on five benchmark medical datasets publicly available from the UCI machine learning repository. The experimental results showed that the WFS-PSO approach produces higher classification accuracy applied to different classification algorithms.
Feature Selection, Particle Swarm Optimization, Healthcare Data Classification, Wrapper Method
Короткий адрес: https://sciup.org/15016385
IDR: 15016385 | DOI: 10.5815/ijitcs.2019.09.04
Текст научной статьи Feature selection to classify healthcare data using wrapper method with pso search
Published Online September 2019 in MECS DOI: 10.5815/ijitcs.2019.09.04
Feature selection (FS), also known as variable selection, is defined as a preprocessing step to eliminate the redundant and irrelevant features for classification to help the good performance result that relates to medical datasets. It can be used to select highly informative features to enhance the accuracy of the classification model. It plays a critical role in classification to remove irrelevant features from the training data. So that the learning algorithm emphases only on these useful training data for analysis and future prediction. In many applications, a feature selection process improves the prediction capability of the classifier. In recent years, many feature selection algorithms have been proposed for classification, that they are different in two key concerns in building an FS approach: subsets generation and evaluation of generated subsets [1].
There are basically two methods for feature selection according to their subset evaluation strategy: filter and wrapper methods. These approaches were performed based on how a feature selection technique combines with the construction of a classification model. In the filter method, an attribute evaluator and search method rank all the attributes in the dataset. The number of attributes selected from the attribute vector can always be defined. After having the rank attributes, the attributes that have lesser ranks are omitted and the predictive accuracy of the classification algorithm can be retained the highest ranks. One problem of the filter method is the weights put by the ranker algorithms are different than those weights by the classification algorithm [2].
In the wrapper method, the subset evaluator is used for creating all possible subsets from the feature vector. Then it uses a classification algorithm such as Naïve Bayes, kNN and C4.5 to make classifiers from the features in each subset. And finally, it considers the subset of features with which the classification algorithm performs the best. To find a subset, a search technique like random search, breadth-first search, hybrid search, a heuristic search is used by the evaluator. Therefore, the possible subsets that evaluated from feature vector depend on these search techniques. Among these search techniques, metaheuristic optimization algorithms or heuristic search is widely used in feature selection in term of computation time. These search methods such as PSO search, Bat search, and Ant search are very efficient and easy to implement. They can also handle large scale data [3].
Particle swarm optimization algorithm becomes the most popular search techniques for the feature selection process in classification. Normally swarm intelligence based FS algorithms perform better than traditional FS techniques mainly for medical datasets [4]. Swarm search algorithms with FS techniques can generate the optimal feature subsets. Each feature subset is evaluated by a fitness function during the feature selection process. In common, prediction accuracy is the important part of the feature selection process for performance evaluation. Therefore, the choice of the best feature subset is the crucial task to perform accurate prediction in healthcare data classification. Hence, this study presents the wrapper-based feature selection approach using particle swarm optimization search algorithms on different healthcare datasets to improve the classification accuracy of classification algorithms.
This research paper is constructed with the following sections. Section 2 presents a literature review related to feature selection for healthcare data classification. The proposed wrapper-based feature selection is clarified in Section 3. In Section 4, the works for the experiments are described and the result for the proposed approach on different datasets is demonstrated in Section 5. Section 6 summarizes the proposed approach and extends for future work.
-
II. Related Works
This section presents the reviews on the background of feature selection algorithms related work on healthcare data classification. Some of the researchers attempt to improve the classification accuracy in high dimensional microarray data. This paper emphasizes on the most closely related FS algorithms in literature since there have been a lot of papers on feature selection.
Thangaraju, P. et al. (2015) analyzed the liver disorder dataset applying particle swarm optimization algorithm combining with the KStar classifier, in two phases to classify the presence of disease or not. A model was proposed to find the chances of the rate of liver diseases based on input attributes using the feature selection approach. The proposed algorithm improved the classification accuracy when compared to existing classification algorithms like Naïve’ Bayes, J48. The results found that the proposed approach was the best suitable algorithm for the classification of liver disorders dataset [5].
Salma, M.U., et al. (2016) created a feature selection approach for medical datasets combine with the clustering technique (Fast K-means) and a stochastic technique (PSO) to select the informative features and to get better results. The feature subset generated from the proposed approach on KDD Cup 2008 Breast cancer dataset produced an accuracy of 99.39% and proficiency of time complexity [6].
Almayyan, et al. (2016) developed a model to predict the diagnosis of the lymphatic disease by using various feature selection techniques: particle swarm optimization (PSO), Information Gain (IG) and Symmetrical Uncertainty (SU) to reduce the variable size. After that, a random forest (RF) classification algorithm is applied to predict the diseases. The result with the selected features obtained from three feature selection methods demonstrated that the proposed model reaches a reasonable improvement in the classification accuracy rate [7].
Rouhi, A, et al (2017) proposed a feature selection (FS) approach based on ensemble method for high dimensional data. In this approach, two meta-heuristic methods binary gravitational search algorithm and binary ant colony optimization algorithm are used on selected features. The results are compared with several state-of-the-art FS methods and the result showed that the effectiveness of the proposed method [8]. In 2018, this author has created a filter based FS approach for microarray data classification which has worked combinations of information gain with improved binary gravitational search algorithm (IBGSA). The classification accuracy, precision, and recall are used for evaluation of subsets generated by the proposed method. The gained results compared with the other methods used for FS in microarray data and confirmed the advantage of the proposed method [9].
Gao, et al. (2017) is utilized the fast correlation-based feature selection (FCBF) method combined with symmetrical uncertainty (SU) to choose the feature subsets for the quality of cancer classification. Then, the accuracy result acquired from the support vector machine classifier is performed as a fitness function adjusting by particle swarm optimization integrates with an artificial bee colony algorithm. The proposed approach is evaluated on 9 cancer datasets. Among them, five datasets with outcome prediction and a protein dataset of ovarian cancer reached the best prediction. The results demonstrated the effectiveness and the robustness of the proposed method in handling various types of data for cancer classification compared to other classification methods [10].
D.sheela et al. (2017) developed an optimal feature selection algorithm using particle swarm optimization (PSO) for high dimensional data sets. The proposed algorithm divided into two parts: preprocessing and PSO based optimization. In the first part, Fuzzy entropy using fuzzy C-means algorithm and symmetrical uncertainty is applied and PSO is used in the second part to select the optimized feature subset. The proposed algorithm is compared with the existing fast correlation-based feature selection algorithm using four different datasets. The performance is evaluated on Sensitivity, specificity, accuracy and selected features for all datasets. The results obtained from the experiment are demonstrated that the proposed hybrid algorithm gives significant and effective results for the feature selection process with respect to the total number of features and classification accuracy [11].
Yiyuan et al. (2018) suggested a confidence based cost-effective feature selection (CCFS) technique using binary PSO. In this approach, two novel points are performed: feature confidence and feature cost. Among these two ideas, feature confidence is employed to update the position of a particle and enhance the performance of the FS. Feature cost is integrated into the scheme of the fitness function. The results through experiments on the Lung Cancer dataset of the proposed method is compared with three different filter methods such as PCFS, InfoGain and CFS and wrapper methods such as BestFirst, GSFS and GSBS methods. The experimental results demonstrated that the proposed CCFS method points to improve the learning accuracy rate and decrease the number of selected features and the total costs of them [12].
Pradana, et al. (2019) implemented an approach combining binary particle swarm optimization (BPSO) and C4.5 decision tree algorithm to detect the cancer diagnosis based on microarray data classification. The objective of this approach is to analyze the prominent feature selection and microarray data classification. Firstly, the decision tree rule model is discretized by K-means algorithms. After that, two phrases: Information Gain (IG)-C4.5 and BPSO-C4.5 are performed in this proposed approach. The accuracy results of these phrases are 54% and 99% respectively. The proposed approach BPSO-C4.5 is able to find the best important feature subset for better performance [13].
In 2019, Amos O Bajeh, et al. presented an experimental study of the influence of particle swarm optimization as a feature selection method on the performance of learning algorithms. SMS spam detection and sentiment analysis datasets were used. On these datasets, PSO is used for the feature selection process. Three classifiers: C4.5 decision tree, k-nearest neighbor and support vector machine were used for the reduced and raw dataset separately. The result of the study indicated that the improvement of classifier performance is case-dependent and some significant improvements are perceived in the sentiment analysis datasets and not in the SMS spam dataset. Additionally, the usage of PSO for feature selection must be done with attention to ensure that performance is actually improved. Though some minimal effect is observed on performance, it suggests that the space complexity of the particle swarm optimization algorithm is reduced while preserving the accuracy of the classifiers [14].
From the literature, it is observed that the feature selection algorithms remove the unnecessary and noisy features from the training data. For this goal, the important features are acquired to develop incredibly classification accuracy. Moreover, feature selection is an intermediate step in the evaluation process for the medical data to increase the accuracy of the classification and increase the efficiency of data mining methods. Therefore, this paper presents a wrapper based feature selection approach using PSO search algorithm to improve the accuracy of the classification task.
-
III. Feature Selection Procedure Using Wrapper Method with PSO Search
In this work, the proposed wrapper-based feature selection approach with particle swarm optimization search strategy is presented. In the wrapper-based approach, the learning algorithm is applied to select the important feature subset from the dataset. The wrapperbased approach can be described in the following procedure.
-
1) Begin
-
2) Read the original dataset with full attributes
D = {A1, A2, A3, …, AN}
N = total number of attributes in D
A = attributes in D
-
3) Choose the attribute subset candidate with the particle swarm optimization search strategy.
-
4) Generate the subset of the attributes using various learning algorithms
-
5) If the subset of selected attributes does not satisfy the condition, go back step 3. Otherwise, go to step 6.
-
6) Output the best selected attributes subset:
D sf = {A 1 , A 2 , A 3 , ..., A n }
-
n = total number of attributes selected from D
-
7) End
This procedure accepts the dataset with full attributes D = {A1, A2, A3, …, AN} and yields the selected attribute set Dsf = {A1, A2, A3, ..., An} is expected as the best output. Firstly, the dataset is read and selecting the attribute candidate subset is made on the dataset using the PSO search. Then the attribute subsets are evaluated using the various classification algorithms. If the evaluated attribute subset satisfied the certain condition then that attribute subset is chosen as the final attribute subset.
In addition, the experiments are applied according to healthcare data classification, since this classification is significant for many patients who need to make a decision. The proposed wrapper based feature selection method for healthcare data classification is a four-phrase procedure. In the first phase, the dataset is read with the initial full set of attributes. The selection of candidate attribute subset with particle swarm optimization search strategy using different classifiers is done in the second phase, while the third phase trains with the various machine learning algorithms that are performed on the selected attribute subset. Finally, the different machine learning algorithms are evaluated on the k-fold crossvalidation technique. The flow diagram of the proposed feature selection approach is shown in Figure 1.
The proposed wrapper-based feature selection method implements iteratively until the best attribute subset has been initiated or a certain number of iterations is achieved by the proposed approach. Then the process ends and the best attribute subset is attained.
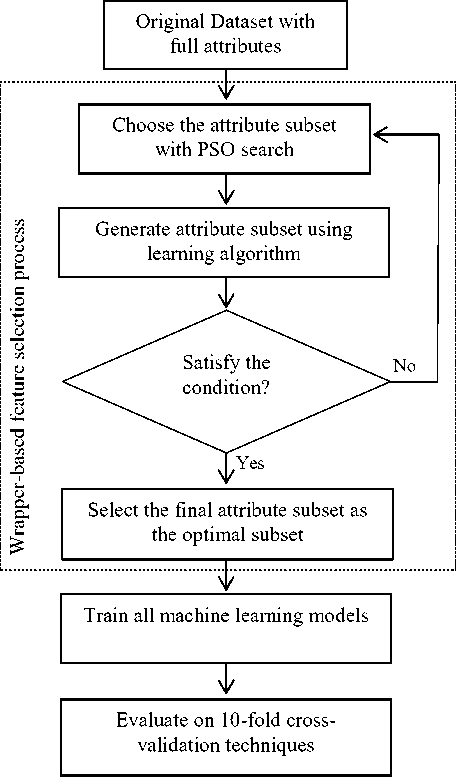
Fig.1. Flow diagram representation for feature selection approach using wrapper method
-
IV. Experimental Work
-
A. Datasets
The Breast Cancer dataset contains 201 instances of one class and 85 instances of another class, where each sample has 9 features, some of which are linear and some are nominal.
The Lung cancer dataset was used to illustrate the power of the optimal discriminant plane even in ill-posed settings. The data described three types of pathological lung cancers. It contains 32 instances and 57 attributes including the predictive attribute which is nominal, taking on integer values 0 to 3. Three types of classes are 9 observations, 13 observations and 10 observations.
The Liver disorder dataset known as BUPA, includes 345 instances and 7 attributes containing the selector
(class label) attribute. The first 5 attributes are all blood tests which are thought to be sensitive to liver disorders that might arise from excessive alcohol consumption. Each instance in the dataset constitutes the record of a single male individual. The presence of liver disorder is determined by the selector attributes.
The Hepatitis dataset consists of 155 samples dispersed between two classes: die with 32 samples and live with 123 samples. There are 19 attributes, 6 attributes are discrete values while 13 attributes are binary. The aim is to predict the presence or absence of the hepatitis virus.
The Diabetes dataset contains 768 samples, where each sample has 8 features which are eight clinical findings. All patients of the dataset are Pima Indian women in whom the youngest one is 21 years old and living near Phoenix, Arizona, USA. The binary target variable can take “0” or “1.” If it takes “1,” it means a positive test for Diabetes, or if it takes “0,” it means a negative test. There are 268 different cases in class “1” and 500 different cases in class “0.”
All above mention datasets are publically available from the University of California Irvine (UCI) machine learning database [15] which is commonly used for classification.
-
B. Experimental Set-up
All of the experiments tested on Windows 7 Professional 64-bit Operating system, Intel(R) Core(TM) i5-3210M CPU @ 2.50 GHz Processor, 4.00 GB RAM and 320 GB Hard disk using the Weka (Witten and Frank, 2000) software [16]. The experiment is conducted on five well-known publically available healthcare datasets collected from the UCI Machine Learning repository. The details of the datasets are presented in Table 1.
Table 1. Summarization of UCI healthcare datasets.
|
Dataset |
No. of Sample |
No. of attributes |
Class |
|
Breast Cancer |
286 |
9 |
2 |
|
Lung Cancer |
32 |
57 |
3 |
|
Liver Disorder |
345 |
7 |
2 |
|
Hepatitis |
155 |
19 |
2 |
|
Diabetes |
768 |
8 |
2 |
-
C. Particle Swarm Optimization (PSO) Parameters
Particle swarm optimization algorithm is a novel metaheuristic optimization algorithm inspired by the natural behavior of the flocking bird in their search process for finding the best particle [17]. The parameters of the PSO algorithm are determined to the default parameters in Weka to certify the correct behavior of it. In this experiments, set individual weight = 0.34, inertia weight = 0.33 and social weight = 0.33, all of these weight equal to 1. The population size is set to 20, the number of generation is determined 20, the number of repetition is 20, and k-fold cross-validation method is used to evaluate the classification accuracy. The selected feature subsets are identified to the various classifier and the accuracy rates were determined.
-
D. Learning Algorithms
In this study, various learning algorithms provided by the WEKA software [16] are used to determine the efficiency of the healthcare datasets for classification by choosing the k-fold cross-validation method. Then compare their evaluation accuracies and give the selected attribute set to the different classification algorithms. In this paper, different classification algorithms are used to validate the feature selection methods specifically the naïve Bayes classifier (NB), k-Nearest Neighbor classifier (kNN), decision tree classifier (C4.5) and Hoeffding Tree classifier (HT) classifier. Moreover, the correlation-based feature selection (CFS) method [18] is used to compare with the proposed method.
-
E. Performance Evaluation
In the experiments, k-fold cross-validation (CV) is performed to estimate the accuracy of various learning algorithms [19]. In this research, the 10-fold crossvalidation is used to predict the classifier’s performance of five healthcare datasets. The classification accuracies for these datasets are measured with the equation (1):
Accuracy = Total number of correctly classifier instances / Total number of instances
-
V. Results and Discussion
The experimental results are summarized in this section. The total number of features (T.F) in the dataset and the number of attributes selected by the proposed approach is shown in Table 2.
Table 2. Number of features selected from the wrapper approaches
|
Dataset |
T.F |
PSO-CFS |
PSO-NB |
PSO-kNN |
PSO-C4.5 |
PSO-HT |
|
Breast Cancer |
9 |
5 |
5 |
2 |
5 |
5 |
|
Lung Cancer |
57 |
7 |
19 |
24 |
23 |
22 |
|
Liver Disorder |
7 |
1 |
3 |
4 |
6 |
5 |
|
Hepatitis |
19 |
10 |
11 |
7 |
2 |
5 |
|
Diabetes |
8 |
4 |
5 |
3 |
4 |
5 |
Table 3 presents the classification accuracy of the NB classifier with the total number of features and different feature selection techniques evaluated on 10 fold crossvalidation. Table 4 illustrates the classification accuracy of the kNN classifier with the total number of features and different feature selection techniques evaluated on 10 fold cross-validation.
Table 5 shows the classification accuracy of the C4.5 classifier with the total number of features and different feature selection techniques evaluated on 10 fold crossvalidation. Table 6 demonstrates the classification accuracy of the HT classifier with the total number of features and different feature selection techniques evaluated on 10 fold cross-validation.
Table 3. Classification accuracy on total no. of features and different feature selection techniques using NB classifier evaluated on 10 fold cross-validation.
|
Dataset |
T.F |
PSO-CFS |
PSO-NB |
PSO-kNN |
PSO-C4.5 |
PSO-HT |
|
Breast Cancer |
71.68 |
72.38 |
75.53 |
72.73 |
72.38 |
75.52 |
|
Lung Cancer |
62.5 |
68.75 |
81.25 |
71.88 |
59.38 |
78.13 |
|
Liver Disorder |
55.36 |
56.52 |
61.16 |
53.04 |
55.36 |
60.87 |
|
Hepatitis |
85.16 |
85.81 |
86.45 |
80.65 |
85.16 |
86.45 |
|
Diabetes |
76.3 |
77.47 |
77.73 |
74.09 |
76.69 |
77.73 |
From Table 3, it is observed that PSO-NB significantly produces better accuracy compared to other methods in terms of accuracy. The PSO-kNN method did not produce significant results in the case of Liver Disorder, Hepatitis and Diabetes datasets. In the Lung Cancer dataset, PSO-C4.5 achieves the lowest accuracy than the accuracy of the total number of features (T.F).
Table 4. Classification accuracy on total no. of features and different feature selection techniques using kNN classifier evaluated on 10 fold cross-validation.
|
Dataset |
T.F |
PSO-CFS |
PSO-NB |
PSO-kNN |
PSO-C4.5 |
PSO-HT |
|
Breast Cancer |
72.38 |
71.68 |
70.28 |
75.17 |
72.03 |
70.28 |
|
Lung Cancer |
53.13 |
68.75 |
71.88 |
84.38 |
75.00 |
71.88 |
|
Liver Disorder |
62.89 |
57.39 |
62.03 |
65.79 |
62.89 |
63.19 |
|
Hepatitis |
80.65 |
82.58 |
83.87 |
85.81 |
80.65 |
82.58 |
|
Diabetes |
70.18 |
68.36 |
66.41 |
71.48 |
68.36 |
66.41 |
In Table 4, it is perceived that PSO-kNN significantly produces better accuracy compared to other methods in terms of accuracy. The PSO-NB, PSO-C4.5 and PSO-HT methods do not produce significant results for Breast Cancer and Diabetes datasets the same as the PSO-CFS method. For Diabetes datasets, it is found that except PSO-kNN, other WFS methods are lower accuracy than the accuracy of the total number of features.
Table 5. Classification accuracy on total no. of features and different feature selection techniques using C4.5 classifier evaluated on 10 fold cross-validation.
|
Dataset |
T.F |
PSO-CFS |
PSO-NB |
PSO-kNN |
PSO-C4.5 |
PSO-HT |
|
Breast Cancer |
75.53 |
73.08 |
73.78 |
75.17 |
76.92 |
73.77 |
|
Lung Cancer |
40.63 |
71.88 |
56.25 |
62.50 |
62.50 |
46.88 |
|
Liver Disorder |
68.69 |
60.87 |
52.46 |
67.54 |
68.69 |
69.28 |
|
Hepatitis |
83.87 |
81.29 |
81.94 |
83.87 |
85.16 |
85.16 |
|
Diabetes |
73.83 |
74.87 |
74.74 |
71.75 |
75.78 |
74.74 |
From Table 5, PSO-C4.5 significantly produces improved accuracy for all datasets compared to other methods and PSO-HT generates more accuracy than that of the total number of features in Lung cancer, Hepatitis and Diabetes datasets. In the Breast Cancer dataset, it is found that other methods are lower accuracy than the T.F not including PSO-C4.5.
In Table 6, PSO-HT significantly yields improved accuracy compared to other methods. PSO-NB and PSO-kNN generate the lowest accuracy than other FS methods on Hepatitis Dataset. It is found that most of the WFS methods are reasonable accuracy with HT classifier on three datasets except Hepatitis and Diabetes datasets.
Table 6. Classification accuracy on total no. of features and different feature selection techniques using HT classifier evaluated on 10 fold cross-validation.
|
Dataset |
T.F |
PSO-CFS |
PSO-NB |
PSO-kNN |
PSO-C4.5 |
PSO-HT |
|
Breast Cancer |
69.93 |
71.33 |
74.83 |
71.33 |
71.68 |
74.83 |
|
Lung Cancer |
43.75 |
59.38 |
62.50 |
68.75 |
75.00 |
75.00 |
|
Liver Disorder |
53.33 |
57.97 |
57.39 |
57.97 |
53.33 |
56.81 |
|
Hepatitis |
80.00 |
81.94 |
79.35 |
79.35 |
84.52 |
87.09 |
|
Diabetes |
76.17 |
77.47 |
77.6 |
73.96 |
76.95 |
77.6 |
In brief, some methods have slightly reduced the accuracy than that of the total number of features (T.F) for all datasets. However, the WFS-PSO produces better accuracy given to the respective classification algorithms. Compare to the PSO-CFS method, it is observed that WFS-PSO with respective learning models produces a well performance in terms of accuracy. Therefore, the wrapper-based feature selection methods using PSO search with their respective learning algorithms produce sensible classification accuracy with the classification algorithms.
-
VI. Conclusion
Feature selection technique is an important phase in data mining and machine learning to increase the performance and decrease the computational cost. It provides an effective way to analyze medical data by reducing irrelevance and redundant data. In this paper, the wrapper-based feature selection approach using particle swarm optimization search for healthcare datasets is presented. The presented wrapper-based feature selection approach using PSO search namely PSO-CFS, PSO-NB, PSO- KNN, PSO-C4.5, and PSO-HT with various classifier produce reasonable accuracy for the respective classification algorithms. Moreover, the results showed that the additional reducing of a feature set can be possible by taking a small drop of the classification amount to improve classification performance and the computational efficiency of the whole classification system. The further extension of this work is to examine the other types of swarm search algorithms instead of particle swarm optimization can be combined with the presented approach. In addition, the presented approach is performed on microarray gene expression data in the future.
Acknowledgments
The authors are thankful to the anonymous reviewers for sharing their valuable comments that definitely improved the quality of the paper and gratitude to people who have contributed towards the development of this manuscript from the planning stage to the finish stage.
Список литературы Feature selection to classify healthcare data using wrapper method with pso search
- Shokouhifar M, Sabet S. A hybrid approach for effective feature selection using neural networks and artificial bee colony optimization [C]. In 3rd international conference on machine vision (ICMV 2010) 2010 Dec (pp. 502-506).
- Yu L, Liu H. Feature selection for high-dimensional data: A fast correlation-based filter solution [C]. In Proceedings of the 20th international conference on machine learning (ICML-03), Washington DC, 2003 (pp. 856-863).
- Mafarja M, Sabar NR. Rank based binary particle swarm optimisation for feature selection in classification [C]. In Proceedings of the 2nd International Conference on Future Networks and Distributed Systems 2018 (ICFNDS’18) June 26-27, 2018, Amman, Jordan, (p. 19). ACM.“doi:10.1145/3231053.3231072”.
- Uzer, M.S., Yilmaz, N. and Inan, O. Feature selection method based on artificial bee colony algorithm and support vector machines for medical datasets classification [J]. The Scientific World Journal, 2013. Article ID 419187, 10 pages.“doi:10.1155/2013/419187”.
- Thangaraju, P. and Mehala, R. Performance analysis of PSO-KStar classifier over liver diseases [J]. International Journal of Advanced Research in Computer Engineering & Technology (IJARCET), 4(7), 2015.
- Salma, M.U., and Doreswamy. PSO based fast K-means algorithm for feature selection from high dimensional medical data set [C]. In 2016 10th International Conference on Intelligent Systems and Control (ISCO) 2016, January (pp. 1-6). IEEE.
- Almayyan, W. Lymph diseases prediction using random forest and particle swarm optimization [J]. Journal of Intelligent Learning Systems and Applications, 2016, January, 8(03), p.51-62.“doi:10.4236/jilsa.2016.83005”.
- Rouhi, A. and Nezamabadi-pour, H. A hybrid feature selection approach based on ensemble method for high-dimensional data [C]. In 2017 2nd Conference on Swarm Intelligence and Evolutionary Computation (CSIEC) (pp. 16-20) March, 2017. IEEE.
- Rouhi, A. and Nezamabadi-pour, H. Filter-based feature selection for microarray data using improved binary gravitational search algorithm [C]. In 2018 3rd Conference on Swarm Intelligence and Evolutionary Computation (CSIEC) (pp. 1-6). March, 2018. IEEE.
- Gao, L., Ye, M. and Wu, C. Cancer classification based on support vector machine optimized by particle swarm optimization and artificial bee colony [J]. Molecules, 2017, 22(12), p.2086.
- D.sheela Jeyarani, Dr.mrs.a.pethalakshmi, and Dr.mrs.k. Jayapriya. Optimal feature selection algorithm for high dimensional data sets using particle swarm optimization [J]. International Journal of Latest Trends in Engineering and Technology (IJLTET), March, 2017, Vol-8, Issue (2) p.200-211. “doi.org/10.21172/1.82.028”.
- Chen, Yiyuan et al. An effective feature selection scheme for healthcare data classification using binary particle swarm optimization [C]. 2018 9th International Conference on Information Technology in Medicine and Education (ITME). IEEE, 2018. “doi:10.1109/ITME.2018.00160”.
- Pradana, A.C. and Aditsania, A. Implementing binary particle swarm optimization and C4.5 decision tree for cancer detection based on microarray data classification [J]. In Journal of Physics: Conference Series 2019, March (Vol. 1192, No. 1, p. 012014). IOP Publishing. “doi:10.1088/1742-6596/1192/1/012014”.
- Bajeh, Amos O., Bukola O. Funso, and Fatima E. Usman-Hamza. Performance Analysis of Particle Swarm Optimization for Feature Selection [J]. FUOYE Journal of Engineering and Technology 4.1 (2019).
- UCI Machine Learning Repository: Available online: http://archive.ics.uci.edu/ml/. Irvine, CA: University of California, School of Information and Computer Science.
- WEKA: Data Mining and Machine Learning Software. Available online: http://www.cs.waikato.ac.nz/ml/weka/
- Kothari, V., Anuradha, J., Shah, S. and Mittal, P. A survey on particle swarm optimization in feature selection [C]. In International Conference on Computing and Communication Systems (pp. 192-201). 2011, December, Springer, Berlin, Heidelberg.
- Hall, M.A. Correlation-based feature selection for machine learning. (Thesis) April, 1999.
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection [J]. In IJCAI 1995, August ,(Vol. 14, No. 2, pp. 1137-1145).


