Gene variants that may cause adverse drug reactions in non-Hodgkin’s lymphoma therapy: a genomic and bioinformatics approach
Автор: Gustinanda R., Irham L.M., Supadmi W., Amukti D.P., Adikusuma W., Chong R., Khair R.E., Satria R.D., Wirsahada B.C.
Журнал: Сибирский онкологический журнал @siboncoj
Рубрика: Лабораторные и экспериментальные исследования
Статья в выпуске: 3 т.24, 2025 года.
Бесплатный доступ
Introduction. Chemotherapy remains a cornerstone treatment for most non-Hodgkin lymphoma (NHL) patients, yet it is frequently associated with significant adverse effects that compromise their quality of life. Emerging evidence highlights that genetic factors, particularly single nucleotide polymorphisms (SNPs), play a critical role in determining individual variability in treatment responses and susceptibility to drug-related complications. Aim of this study: to identify SNPs associated with chemotherapy-induced adverse events in NHL through advanced bioinformatics approaches, enabling personalized therapeutic strategies to mitigate risks. Material abd Methods. This study leveraged the PharmGKB database to identify SNPs associated with Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone. SNPs meeting inclusion criteria (Level of Evidence 1A-3, p<0.01) were prioritized for functional impact analysis using PolyPhen-2 scores. Data extraction and computational analysis utilized SNPnexus, HaploReg v4.2, Ensembl Genome Browser (GRCh37), and PharmGKB. The methodology employed a descriptive approach, relying exclusively on secondary data sources. Results. This study identified 11 SNPs that may be important for hematological toxicity, liver damage, and nausea risk. These genes are SLC22A16, GSTP1, NAT2, ATM, ABCB1, CYP2B6, XRCC1, ERCC1, MUTYH PIK3R2, and PNPLA3. In terms of priority and risk, the most significant variants were rs738409 (PNPLA3), rs12210538 (SLC22A16), rs2229109 (ABCB1), and rs56022120 (PIK3R2). The distribution of SNP alleles is more common in European populations than in Asians or Africans. Conclusion. For the first time, we found SNPs that indicate an increase in drug side effects. These SNPs rs738409, rs12210538, rs2229109 and rs56022120 increase the severity of NHL patients during chemotherapy. In order to ensure that these biomarkers can be used in clinical practice and to support the creation of precision medicine strategies, additional clinical validation is needed.
Non-Hodgkin’s lymphoma, adverse drug reactions, SNP, rs738409, rs12210538, rs2229109, rs56022120
Короткий адрес: https://sciup.org/140310574
IDR: 140310574 | УДК: 616-006.441:615.277.3:615.065:575.113 | DOI: 10.21294/1814-4861-2025-24-3-50-64
Текст научной статьи Gene variants that may cause adverse drug reactions in non-Hodgkin’s lymphoma therapy: a genomic and bioinformatics approach
There are numerous subtypes and varieties of non-Hodgkin lymphoma (NHL), which are cancers of the lymphoid cells. Each subtype and variety has its own unique set of biological, clinical, and histological features [1]. With an alarming rise in incidence rates over the past few decades, it has earned its place among the world’s most common malignancies [2]. In the USA, for example, more than 80,350 new cases of NHL are thought to occur annually, making it one of the main reasons why people die from cancer [3]. NHL can present in various age groups, although it is more common in older adults [4]. Treatment of NHL typically involves a multimodal approach, including chemotherapy, radiation therapy, and immunotherapy, each of which has varying degrees of effectiveness and side effect profiles [5]. Although progress has been made in the treatment of NHL, there are still significant challenges associated with resistance to therapy and the emergence of severe side effects, which require further research to understand the factors that influence individual response to treatment [6].
In recent years, studies on pharmacogenetics and pharmacogenomics have provided new insights into how genetic factors influence a patient’s response to NHL therapy. Pharmacogenetics focuses on individual genetic variations that affect drug metabolism, while pharmacogenomics includes the study of the entire genome and how gene expression contributes to response to treatment [7]. Better personalized treatment methods can be achieved by gaining insight into these two areas; this will enable doctors to choose the most effective medications according to a patient’s genetic profile, which in turn reduces the likelihood of adverse effects and increases the likelihood of a cure. Also, pharmacogenetic studies have demonstrated that different medications used to treat NHL have different effects and side effects depending on particular genetic variants. For instance, Conyers et al. (2018) discovered that patients receiving purine analog-based chemotherapy may have increased toxicity if they have variants in the TPMT gene. This provides evidence that tailoring dosages to individual patients’ genotypes has the potential to enhance therapeutic efficacy while decreasing the likelihood of adverse effects [8].
A crucial aspect of this research is the identification of biomarkers that can be used to forecast how a certain therapy will be received, potentially changing the way we treat NHL. These biomarkers not only help in selecting the most effective drugs but also provide insight into disease prognosis and the possible development of resistance to treatment. In addition, pharmacogenetic research can help identify individuals at risk of severe side effects from the therapies used [9]. These side effects, such as myelosuppression, peripheral neuropathy, and immunological reactions, are often a barrier to continuing treatment and can significantly affect patients’ quality of life [10]. With better knowledge of the genetic factors that affect drug tolerability, medical personnel can design safer and more effective treatment plans. This article aims to find the Single Nucleotide Polymorphism (SNP) which were associated with side effects in NHL drugs using bioinformatics-based methods.
Aim of this study: to identify SNPs associated with chemotherapy-induced adverse events in NHL through advanced bioinformatics approaches, enabling personalized therapeutic strategies to mitigate risks.
Material abd Methods
It is important to understand the structure of the human genome and understand the side effects of drugs by identifying genome variants. In this study, we integrated variants associated with drug side effects of NHL cancer using bioinformaticsbased methods. This study was conducted using the PharmGKB (Pharmacogenomics Knowledge Base) database with the keywords «Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisolone» accessed on November 03, 2024. This chemotherapy regimen is based on National Comprehensive Cancer Network (NCCN) patient-resources/guidelines-for-patients/non-hodgkin-lymphoma-resources) guidelines. SNPs were selected based on inclusion criteria, namely Level of Evidence (LOE) 1A-3 (Clinical Annotation), missense variant type, and had a p-value<0.01 as the limit of statistical significance based on genetic association tests listed in the database [5]. Priority analysis was performed using the PolyPhen-2 score results/629178ab/) to determine the level of possible protein damage of each SNP. SNP data was extracted and analyzed using web-based software such as SNPnexus , HaploReg v4.2 haploreg/, Ensembl Genome Browser accessed on November 05, 2024. All analyses were carried out descriptively and based on secondary data.
Results
Genomic Variants Potentially Induce Side Effects of NHL Chemotherapeutic Drugs
Results from studies using the PharmGKB database showed that certain SNPs can raise the risk of adverse effects from the NHL drugs Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisolone (R-CHOP). We identified genetic variant SNPs that have the potential to cause adverse drug events in NHL therapy using genomics and bioinformatics approaches. Table 1. Results from PharmGKB and HaploReg v4.2 present 11 significant SNPs associated with chemotherapy drug toxicity, involving genes such as SLC22A16 , GSTP1 , NAT2 , ATM , ABCB1 , CYP2B6 , XRCC1 , ERCC1 , MUTYH , PIK3R2 , and PNPLA3 . These SNPs showed increased toxicity response based on significant values (p-value<0.01). These genetic variations were spread across different populations, such as European, East Asian, and multiethnic groups. We can learn a lot about how genetic diversity affects the likelihood of side effects from NHL chemotherapy meds from these findings. Although Rituximab was part of the R-CHOP regimen in this study, we did not obtain specific data on Rituximab-induced toxicity from the analyzed database. These limitations highlight the need for further research exploring genetic factors specifically associated with Rituximab-related side effects.
Priority analysis of genes causing adverse reactions to NHL
Gene prioritization analysis using the PolyPhen-2 database (accessed on 5/11/2024) with the aim of
Table 1/Таблица 1
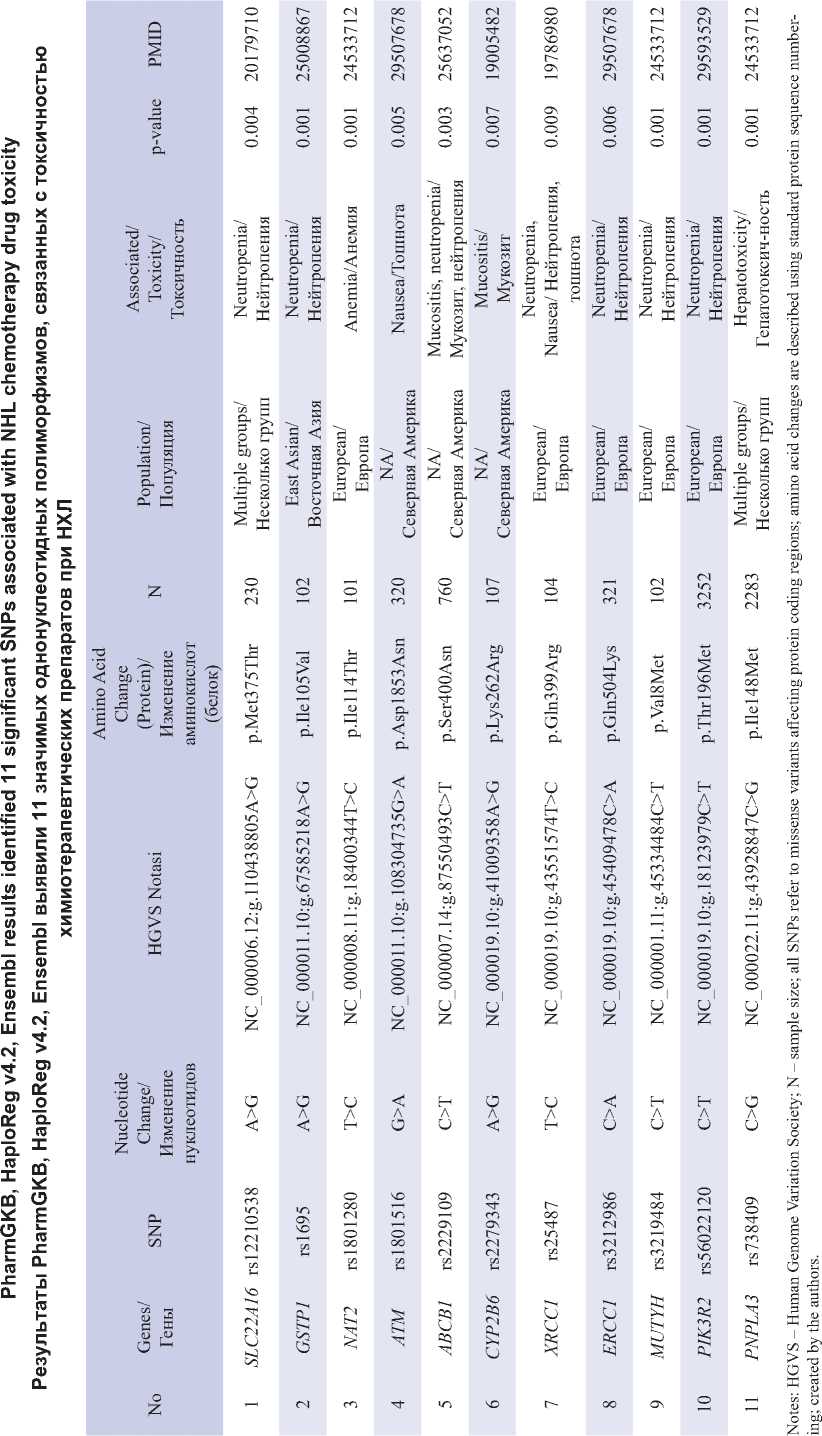
Примечания: HGVS – Human Genome Variation Society; N – размер выборки; все SNP относятся к миссенс-вариациям, влияющим на кодирующие области белков; изменения аминокислот описаны с использованием стандартной нумерации последовательностей белков; таблица составлена авторами.
prioritizing SNPs that have a higher potential risk of side effects of NHL drugs. The SNPs were categorized into three groups: benign, possibly damaging, and probably damaging based on PolyPhen-2 scores. If it is in the score between (0.00 to 0.15), it is benign, then if it is in the score (0.15 to 0.85), it is moderate (possibly damaging), but if it is in the score (0.85 to 1.0), it is very malignant (probably damaging) [11]. So that the gene variant is said to be increasingly malignant if the score is getting closer to one, from this, researchers exclude SNPs that have a potential risk of less than 0.15 (benign).
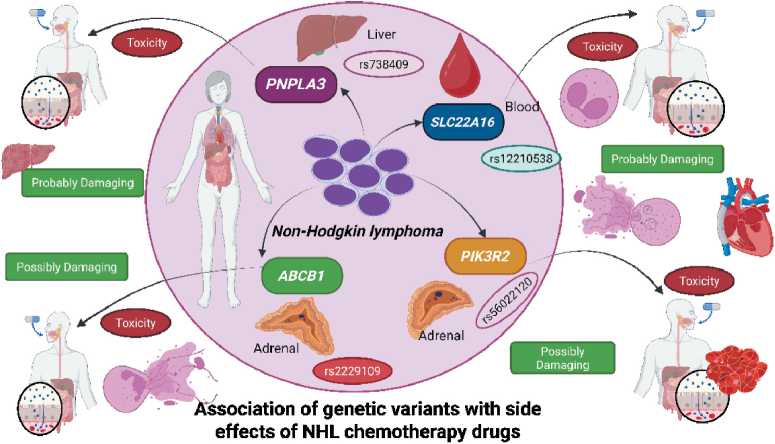
The results of the research from 11 SNPs analyzed with SNPNexus only got 4 variants of the highest data score. Priority analysis targeted the highest ratings of 0.944 and 0.912, two of the eight missense SNP variants rs738409 and rs12210538 being highly dangerous variants. Then, with scores of 0.755 (rs2229109) and 0.505 (rs56022120) are moderate variations, which can be dangerous. So that the 4 SNP data have great potential to cause side effects of NHL chemotherapy drugs. These variations have the potential to be used as biomarkers for the purpose of tracking therapies that may cause side effects of chemotherapy drugs used in NHL and for the purpose of predicting possible side effects [12].
Distribution of genetic variants causing adverse reactions
Table 3 and Figure 1 present the allele frequencies of gene variants associated with adverse drug reactions to NHL therapy across different global populations, including Africa, America, Asia, and Europe. These data were extracted from the 1000 Genomes Project using SNPnexus and Ensembl Genome Browser .The purpose of this mapping is used to see the distribution of susceptibility to side effects of NHL drugs in various world populations. Allele frequencies are different in each population Allele frequencies for each SNP vary across African, American, European, and Southeast Asian populations, as presented in Table 3.
Discussion
SLC22A16
A number of small molecules are transported across the cell membrane by the SLC22A16 gene, which is a member of the SLC22 gene family [11]. SLC22A16 in particular is known as an organic cation transporter. The protein encoded by this gene has an crucial role in the transportation of certain compounds into cells, such as Doxorubicin. Variants in the SLC22A16 gene, which encodes the transporter for doxorubicin, can affect drug toxicity by altering the efficiency of doxorubicin uptake into cells. Loss-of-function variants decrease drug accumulation, reducing toxicity, but can also decrease therapeutic efficacy. On the flip side, doxorubicin accumulation is increased by gain-of-function variants or overexpression, which can lead to oxidative stress on the heart and hematological damage (such as neutropenia). Genetic polymorphisms such as rs12210538 have been linked to individual variation in sensitivity to doxorubicin side effects, making them relevant for pharmacogenomics-based personalized medicine [13].
“Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisolone"
"Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisolone"
"Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisolone"
"Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisolone"
Fig. 1. Prioritized SNPs and their Associated Adverse Drug Effects in NHL Chemotherapy. Notes: created in BioRender by Muhammad Irham, L. (2025).
Рис. 1. Приоритетные однонуклеотидные полиморфизмы и связанные с ними побочные эффекты химиотерапии при НХЛ. Примечание: рисунок выполнен в BioRender Мухаммадом Ирхамом, Л. (2025).
Table 2/Таблица 2
Priority analysis of SNPs associated with adverse effects of NHL and their effect on protein levels Приоритетный анализ однонуклеотидных полиморфизмов, связанных с неблагоприятными эффектами химиотерапии, и их влияние на уровень белка
|
No |
SNP/ Однонуклеотидные полиморфизмы |
Chromosome/ Хромосомы |
Gene/ Гены |
Score/ Счет |
Prediction/ Прогноз |
Consequence/ Последствия |
|
1 |
rs738409 |
22 |
PNPLA3 |
0.944 |
Probably Damaging/ Вероятно повреждающий |
Missense/ Миссенс-мутация |
|
2 |
rs12210538 |
6 |
SLC22A16 |
0.912 |
Probably Damaging/ Вероятно повреждающий |
Missense/ Миссенс-мутация |
|
3 |
rs2229109 |
7 |
ABCB1 |
0.755 |
Possibly Damaging/ Вероятно повреждающий |
Missense/ Миссенс-мутация |
|
4 |
rs56022120 |
19 |
PIK3R2 |
0.505 |
Possibly Damaging/ Вероятно повреждающий |
Missense/ Миссенс-мутация |
Note: created by the authors.
Примечание: таблица составлена авторами.
Table 3/Таблица 3
Allele frequencies in various populations and their potential susceptibility to NHL adverse drug events Частоты аллелей в различных популяциях и их потенциальная восприимчивость к побочным лекарственным эффектам при лечении НХЛ
|
No |
Chr/ Хромосомы |
Gene/ Гены |
SNP/ Однонуклеотидные полиморфизмы |
Allele/Аллели |
Frequency/Частота |
|||||
|
Ref |
Alt |
|||||||||
|
AFR |
AMR |
EAS |
SAS |
EUR |
||||||
|
1 |
22 |
PNPLA3 |
rs738409 |
C |
G |
0.12 |
0.48 |
0.35 |
0.25 |
0.23 |
|
2 |
6 |
SLC22A16 |
rs12210538 |
A |
G |
0.01 |
0.15 |
0.00 |
0.09 |
0.24 |
|
3 |
7 |
ABCB1 |
rs2229109 |
C |
T |
0.00 |
0.02 |
0.00 |
0.01 |
0.03 |
|
4 |
19 |
PIK3R2 |
rs56022120 |
C |
T |
0.00 |
0.09 |
0.04 |
0.06 |
0.01 |
Notes: Chr – chromosome; Ref – reference; Alt – alternative; AFR – Africa; AMR – America; EAS – East Asian; SAS – South Asian; EUR – Europe; created bu the authors.


