Genetic aberrations in multipe myeloma
Автор: Blau O., Bullinger L., Blau I.W.
Журнал: Вестник гематологии @bulletin-of-hematology
Рубрика: Передовые статьи
Статья в выпуске: 2 т.17, 2021 года.
Бесплатный доступ
Multiple myeloma (MM) is a plasma cell malignancy characterized by complex cytogenetic and molecular genetic aberrations. Genomic analysis shows a variety of gene mutations, aneuploidies, segmental copynumber changes, translocations that are extremely heterogeneous, and more numerous than other hematological malignancies. It is known that the development of MM is preceded by pre-malignant stages, and therefore it represents a well-defined model of disease progression, suitable for studies of clonal evolution and heterogeneity. Here we review at the main genetic abnormalities in patients with MM, involvement in pathogenesis, and prognostic value
Короткий адрес: https://sciup.org/170175820
IDR: 170175820
Текст научной статьи Genetic aberrations in multipe myeloma
Multiple myeloma (MM) is an incurable disease characterized by the clonal proliferation of bone marrow (BM) plasma cells (PC) that secrete a monoclonal immunoglobulin [1]. Despite the introduction of novel drugs, it remains an incurable disease as most patients eventually relapse. Although second and subsequent remissions can be achieved with further therapy, MM usually appears more aggressive after each relapse, culminating in treatment-resistant dis-ease with short survival [2, 3].
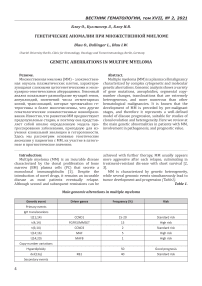
MM is characterized by genetic heterogeneity, while several genomic events simultaneously lead to tumor development and progression (Table1).
Table 1.
Main genomic alterations in multiple myeloma
|
Genetic event |
Driver genes |
Frequency (%) |
Risk |
|
Primary events |
|||
|
IgH translocations |
|||
|
t(11;14) |
CCND1 |
15-20 |
Standard risk |
|
t(4;14) |
FGFR3/MMSET |
15 |
High risk |
|
t(6;14) |
CCND3 |
2 |
Standard risk |
|
t(14;16) |
MAF |
5 |
High risk |
|
t(14;20) |
MAFB |
1 |
High risk |
|
Copy-number variations |
|||
|
Hyperdiploidy |
50 |
Good prognosis |
|
|
del(13q) |
RB1 |
40 |
Standard risk |
|
Secondary events |
|
Chromosome gains |
|||
|
1q |
40 |
High risk |
|
|
8q |
MYC |
15 |
Standard risk |
|
11q |
CCND1 |
15 |
Standard risk |
|
Chromosome losses |
|||
|
1p |
30 |
High risk |
|
|
17p |
TP53 |
10 |
High risk |
|
MYC rearrangements |
MYC |
15 |
High risk |
|
Somatic mutations |
|||
|
MAPK pathway |
KRAS, NRAS, BRAF |
45 |
Standard risk |
|
NF-κB pathway |
CYLD, TRAF3, LBT, NIK |
15 |
Standard risk |
|
RNA metabolism |
DIS3, FAM46C |
15 |
Standard risk |
|
DNA-repair pathway |
TP53, ATM, ATR |
10 |
High risk |
|
Plasma cell differentiation |
IRF4, PRDM1 |
10 |
Standard risk |
Numerous genetic abnormalities are major prognostic factors in MM. Molecular cytogenetic methods such as G-band karyotyping, fluorescence in situ hybridization (FISH), and compara-tive genomic hybridization (CGH) combined with more advanced genetic techniques, encom-passing single nucleotide polymorphism (SNP) arrays and, more recently, nextgeneration se-quencing (NGS), allow the identification of numerous recurrent chromosomal and genetic alterations in MM [4, 5]. These changes can be divided into three types: chromosomal translo-cations, copy number abnormalities and point mutations. The prognostic value of many of genetic anomalies has been analyzed in several studies with large numbers of patients [6, 7].
Over the past decade, the results of large-scale studies of whole exome sequencing have pro-vided new insights into clonal heterogeneity and disease evolution. Moreover, the repeated coincidence of genomic events helps to analyze the genomic complexity underlying tumor progression. The development of MM is considered as a multistep process. Most MM patients progress from an asymptomatic premalignant stage, monoclonal gammopathy of undetermined significance (MGUS) [2, 3]. How quickly progression occurs is largely determined by genetic aberrations. Translocations involving immunoglobulin heavy chain (IgH) genes or hyperdip-loidy are known to be frequent initiating events that deregulate normal PC behavior, leading to the development of MGUS [8-10]. Further mutational burden leads to the intermediate stage of smoldering multiple myeloma (SMM). The main cytogenetic type of the disease in-fluences the rate of progression. Thus, patients with translocation t(4; 14), deletion of 17p, and gain (1q) have a higher risk of progression from SMM to MM [8, 10, 11]. Since it has been observed that MGUS / SMM patients usually already have genetic abnormalities and do not show any clinical symptoms of MM, it seems that the initial genetic defects alone are not enough to induce MM transformation [12].
MM is a clinical model of disease continuity between MGUS, SMM, MM, and plasmacell leukemia (PCL) provides a unique framework for understanding the genomic hier-archy and clonal evolution. Genetic abnormalities found in the MGUS stage are likely to be the primary events involved in tumor development. In contrast, the aberrations present in stages of MM that were absent in MGUS are likely secondary events leading to tumor pro-gression. Recent applications of NGS have shown that MM patients exhibit complex muta-tional landscapes. In addition, there intraclonal genetic heterogeneity exist in the main tumor clone [10, 13-18].
Since intraclonal heterogeneity was observed at all stages of MM, it can be assumed that the progression of disease may be mediated by competition between subclones and the growth of the fittest of these subclones. Recently, a study based on the use of whole exome sequencing has shown evidence of intraclonal heterogeneity and subclonality from the earliest stages of MGUS / SMM, where most of the transformed subclonal populations involved in progression to MM are already present at diagnosis. There was no significant difference in subclonality characteristics at the initial asymptomatic stage of MGUS and intermediate stage of SMM, suggesting that severe subclonal remodeling is also not the only phenomenon associated with the transition between asymptomatic stages [10].
Chromosomal Translocations
Somatic hypermutation and class-switching recombination are two key features of normal B cell development. During early differentiation of B-cells in the BM, after contact with antigens, immunoglobulin (Ig) genes segments are rearranged to form the primary Ig rep-ertoire. These are physiological mechanisms of affinity maturation in the germinal center, but they are also involved in the pathogenesis of MM [19].
The vast majority of chromosomal translocations affect chromosome 14, namely the IGH lo-cus at 14q32, which is one of the most actively transcribed genes in PC. As a result of transla-tions, the partner genes are brought under the control of the IGH enhancer, and the expression of certain of these oncogenes is activated. This event gives a selective advantage to subclones carrying these translocations. IGH translocations are considered triggering events and are therefore called primary translocations. They include five chromosomal loci, 11q13, 6p21, 4p16, 16q23, and 20q11, which contain the oncogenes CCND1, CCND3, FGFR3 / MMSET, MAF, and MAFB, respectively [4, 7].
Translocation t(11;14)
The t(11;14)(q13;q32) is the most common translocation in MM with a reported fre-quency of 15–20% based on FISH and conventional cytogenetic analyses [4]. The breakpoints on 14q32 fall within either the JH region or the switch region. As a result of the translocation, cyclin D1 (CCND1) is juxtaposed to the powerful IgH 3’ enhancer on chromosome 14, its expression is dysregulated, as indicated by gene expression profiling [20]. Traditionally, MM patients with t(11;14) have been categorized as standard risk [3]. However, some recent stud-ies indicate that in the era of novel agents t(11;14) is associated with shorter survival, acting as a marker of intermediate risk. These data also suggest that high-dose chemotherapy benefits patients with t(11;14) [22]. Recently it has been shown that MM patients with t(11; 14) have a standard risk when they receive induction therapy with new agents in combination with high dose of melphalan auto stem cell transplantation (ASCT) [23].
Additional genetic aberrations may also play a role in disease risk. Patients with a t(11;14) alone have better overall survival (OS) compared to patients with a t(11;14) translocation in combination with any other assessed aberrations. The combination of t(11;14) with multiple amplifications 1q, del(1p), and del(IgH) had the most negative effect on OS [24].
A particularly optimistic factor in determining the risks of MM patients with t(11;14) is the fact that there is a specific therapy for this type of myeloma. Venetoclax, a BCL-2 inhibitor, induces cell death, especially in those with t(11;14) who express high levels of BCL-2 com-pared to BCL-XL and MCL-1 [25].
Translocation t(4;14)
This translocation is observed in about 15% of MM patients, more often in young peo-ple [7]. This aberration can be identify using FISH analysis, but they cannot be detected by karyotyping techniques. In the IgH locus, the break-points occur in the switch region and dis-sociate the intronic enhancer from the 3’ enhancer [4]. The translocation t(4;14) results in the simultaneous overexpression of two genes, FGFR3
(fibroblast growth factor receptor 3) and the multiple myeloma SET domain gene (MMSET) on der(4) [4, 7].
Both FGFR3 and MMSET genes are not normally expressed on PC but are overexpressed as a result of the t(4;14). It was shown that only 75% of the MM with t(4;14) display a simultane-ous overexpression of both genes. In the remaining 25% of cases, only MMSET is activated, and the absence of FGFR3 expression in most cases is associated with the loss of the FGFR3 gene on the der(14) [26]. Furthermore, the poor prognosis of t(4; 14) persists irrespective of FGFR3 expression. These data suggest that MMSET may be a critical transforming event in MM with t(4; 14). In some cases (10%), displaced FGFR3 contains activating mutations that may be involved in the progression of MM [26].
FGFR3 is one of the high-affinity tyrosine kinase receptors for the FGF ligand family. These proteins play a role in several important cellular processes, including regulation of cell growth and division, cell type determination, angiogenesis, and embryo development[27].
Numerous data prove that MMSET is an oncogene and plays an important role in the devel-opment of MM. MMSET encodes histone-3-lysine 36 (H3K36) methyltransferase, and its overactivation in MM has been shown to affect the expression of many genes [28]. Histones are the stage of a variety of post-translational modifications that ultimately regulate gene tran-scription. Lysine methylation is one of the characteristic processes of post-translational histone modifications in the regulation of the structure and function of chromatin. It is suggested that the principal physiologic activity of MMSET is demethylation of H3K36, and in the process rules out generation of H3K36me3, H4K20me2, and several other putative methyl products of MMSET [29]. Signaling pathway analysis indicated that MMSET could regulate cell death and the p53 pathway (e.g., BAX, BCL2, and caspase 6), the cell cycle (cyclin E2, E2F2, TP53, INP1, and CDC25A), genes for DNA repair (ATM, E2F2, and GADD45A), and integrin-mediated signaling (CDC42 and integrin alpha-L) [30, 31].
The t(4;14) translocation is associated with unfavorable prognosis and patients bearing it were classified in the high-risk category [3]. Interestingly, despite the poor prognosis associated with t(4;14), early treatment of these patients with the proteasome inhibitor bortezomib results in a survival improvement [7, 14]. Clinical testing of selumetinib (MEK1/2 inhibitor) did not affect treatment outcomes for MM patients, which stoped the development of this drug.
Translocation t(6;14)
The rare translocation t(6;14) is present in only 3% of patients with MM. This chromosomal rearrangement leads to fusion of cyclin D3 (CCND3) with IGH enhancers. It was shown that the t(6;14)(p21;q32) translocation leads to overexpression of cyclin D3. The initial oncogenic event for this type of MM is the primary translocation of Ig, which disrupts the regulation of cyclin D3, providing a proliferative stimulus to PC. By microarray analyses high levels of cyclin D3 mRNA have been shown in cases with t(6;14) detected by FISH [32]. Although the low prevalence of t(6;14) precludes the estimation of survival, this translocation is included in the standard-risk category [3].
Translocations t(14;16) and t(14;20)
Translocations t(14;16) and t(14;20) are the least common class of primary IgH translocation. They observed in less than 5% of MM patients [33]. Translocations deregulate the MAF and MAFB genes, respectively. Both genes belong to the MAF family. MAF genes family are transcription factors. MAFB is a basic leucine zipper transcription factor that plays an important role in the regulation of lineagespecific hematopoiesis. Overexpression of MAFB in MM induces proliferation and protects cells from drug-induced apoptosis, conferring resistance. Increased levels of MAF induce upregulation of cyclin D2 through its transactivation function, resulting in an accelerated rate of division and DNA synthesis [34]. The overexpression of MAF is observed in half of MM cases. This proportion is much more frequent than was expected from the low prevalence of the t(14;16) [4]. It is shown, that the oncoprotein MAF increases the expression of integrin β7, an adhesion molecule that heterodimerises with integrin to bind to Ecadherin on the surface of BM stroma cells. This finding suggests that MAF enhances the adhesion of myeloma cells to BM stroma through the integrin αΕβ7/E-cadherin pathway [35].
It was noted, that the distance between the enhancer and the oncogene MAF in t(14;16) is considerably longer than in other IgH translocations. Taking into account such a long distance it is still an open question whether or not IgH may act as enhancer for MAF in this translocation [36].
Translocations t(14;16) and t(14;20) are considered high-risk cytogenetic factors [3, 7]. Some studies reported that MEK inhibition induces apoptosis of MAF-expressing MM cells and blocks survival signals provided by the microenvironment [37]. The blockade of FOS activity is also toxic for MM cells harboring t(4;14), which is associated with the MAF upregulation in myeloma cells with MMSET translocations [7].
MYC aberration
MYC has long been recognized as one of the most frequently deregulated oncogenes in human cancer. Structural variants of MYC are common in B cell malignant neoplasms, including MM. Rearrangements involving the MYC proto-oncogene on the long arm of chromosome 8 (8q24.1) are common cytogenetic abnormalities detected by FISH in approximately 15% of newly diagnosed MM patients [38]. The structural variants of MYC are distributed over at least two broad regions and serve to amplify or transpose large enhancers to drive MYC expression. Interestingly, almost all MYC translocations are also accompanied by copy number changes, with most of them showing large duplicated sequences at both break points of the translocation [34]. Types of MYC-mediated genomic instability include single nucleotide substitutions and double-stranded breaks resulting from induction of reactive oxygen species, gene amplification, and generation of extrachromosomal elements, as well as numerical chromosomal defects resulting from aberrant DNA synthesis and defects in genes, checkpoint of the mitotic spindle. These aberrations result in overexpression of MYC, which is a prerequisite for the creation of important behavioral characteristics of the tumor, such as invasiveness, metastasis, and the acquisition of resistance to chemotherapy. Recently there has been a clear understanding of how this contributes to transformation. Due to its role as a transcription factor, MYC alters the expression of hundreds of target genes, many of which are themselves oncogenes or tumor suppressors [39-43]. Since structural variants of MYC are less common in MGUS than in MM, this suggests that MYC changes contribute to disease progression. The activation of MYC is a key event in the progression from MGUS and smoldering myeloma (SMM) to symptomatic myeloma [44].
Prognostic value of MYC rearrangements has not yet been established; at this time, they are not included in risk stratification systems for newly diagnosed MM. While some studies have found MYC rearrangements to be associated with inferior outcomes, other studies failed to show prognostic significance [45, 46].
Copy number abnormalities
Most MM cases are aneuploid, in which there are frequent gains and losses of complete chromosomes or chromosome arms. According to the ploidy status, MM is usually categorized in hyperdiploid (H-MM) and non-hyperdiploid (NH-MM) MM. The H-MM group, which accounts for 50–60% of all MM cases, is characterized by the presence of trisomies that typically affect the odd chromosomes [47].
The NH-MM group includes hypodiploid (up to 44/45 chromosomes), pseudodiploid (44/45 to 46/47), and near-tetraploid (more than 74) cases. NH-MM is frequently characterized by the loss of chromosomes 13, 14, 16, and 22. Hyperhaploid karyotypes, as a result of the loss of nearly a haploid set of chromosomes, have also been found in MM. The most frequent monosomie or deletions present in hyperhaploid cases affect 17p, 1p, 13q, and 16q [48, 49]. The NH-MM is characterized by a very high prevalence of IGH translocations. Similarly, monosomy or deletion of chromosome 13 occurs predominantly in NH-MM [50, 51].
Hyperdiploidy
Hyperdiploidy is the other common type of initiating genetic event in MM. The high proportion of chromosomal imbalances observed in MM is a clear sign of underlying genomic instability. Unlike translocations, it is very difficult to trace the oncogenic effects of hyperdiploidy due to the aneuploidy of numerous chromosomes. Hyperdiploidy is hypothesized to occur during rapid germinal center proliferation that results in chromosome segregation errors [34]. Both IgH translocations and hyperdiploidy were found to be clonal aberrations at all stages of gammopathy, which is consistent with their basic role in the MM pathogenesis [4].
Notably, the H-MM is associated with a low incidence of structural chromosomal abnormalities [50]. Hyperdiploidy, in itself, seems to be an early event in MM evolution since it has been described in MGUS [52].
Hyperdiploidy, is almost mutually exclusive with IgH translocations, tends to have a better prognosis than IgH-translocated MM [34, 53]. Several studies have shown that hyperdiploid patients have better response rates to treatment and longer survival than patients with other aneuploidies H-MM rarely results in extramedullary disease or PCL [54]. On the contrary, hyperhaploid karyotypes are associated with an adverse prognosis, even worse than that of the hypodyploid group, with an estimated 20–25% survival after five years, despite intensive treatments based on PIs, IMIDs, and tandem ASCT [49].
Deletion of 13q
Among the numerical abnormalities, monosomy 13 is the most common; it is detected in about 45% of cases. Indeed, deletions of chromosome 13 identified by interphase FISH usually represent monosomy 13 and only occasionally represent an interstitial deletion, accounting for only 15% of cases [55, 56]. Del(13q) was described in MM for the first time in 1995 and has consistently been related to adverse prognosis [57].
This deletion lose the RB1 gen. The RB protein is a well-studied tumor suppressor. A multifunctional protein regulates a number of critical cellular activities, including cell cycle development, response to DNA damage, checkpoint activation, and differentiation [58]. Depending on phosphorylation, RB either inhibits or activates the G1 / S cell cycle transition [59]. Recently was demonstrated that RB interacts with nuclear factor κB (NF-κB) [58]. NF-κB is a transcription factor that is highly implicated in cancer. It can be activated by proinflammatory cytokines [60]. Recently, RB has been shown to specifically bind to p65 protein from NF-κB/Rel family [58]. A recent CRISPR/Cas9-based screening identifies the NF-κB pathway as one of the key mechanisms that promote cancer cell escape from immune attack of T cells [61]. RB deletion seems to increase PDL1 (Programmed death-1) expression, mediated by the NFkB pathway.
Deletion of 17p, TP53 Mutation
Deletion of 17p13 is found from 5 to 12% in MM patients [62, 63]. Its incidence increases with a progression of disease [64]. Importantly, that these percentages can vary from study to study depending on the chosen cut-off value. The 20% threshold is the most widely used and recommended by the European Myeloma Network (EMN) [65, 66]. Additionally, it has been shown that an MLPA cut-off value of <0.75 is equivalent to 20% by iFISH. Both are suitable molecular methods for detecting del(17p13), that can be readily applied in standard diagnostic laboratories [66].
Deletion (17p13) has consistently been associated with shorter progression free survival (PFS) OS in patients with MM [46, 62, 67]. Detailed analysis of TP53 aberrations in a large study supports the independent predictive value of monoallelic TP53 changes [66].
This deletion entails the loss of the TP53 gene, which is a key suppressor gene that organizes multiple functions associated with cell cycle control and DNA damage response [68]. In spite of the infrequent presence of TP53 mutations in MM, the TP53 gene is mutated in about half of patients who harbor del(17p), giving rise to its biallelic inactivation [69]. Patients with biallelic inactivation of TP53 have an aggressive clinical course and poor prognosis [66, 70]. The TP53 mutation frequency increases in the late stages of the disease, which indicates its important role in the progression of MM [70]. These results have also been confirmed by massive parallel sequencing [71].
TP53 mutations are distributed mainly in coding exons with a strong predominance for exons 4–9, which encode the DNA-binding domain of the protein. In fact, 95% of the TP53 mutations stand in the core DNA-binding domain [68]. TP53 mutations can be classified in two categories: contact and structural mutations. Contact mutations affects residues involved directly in DNA-contacts without altering p53 folding, and perturb the transcriptional function of p53 protein. In contrast, structural mutations lead to destabilization of the local structure of p53 core domain [68, 72]. In all types of human cancers, the missense TP53 mutations have been detected predominantly in 6 hotspot residues located within the DNA-binding domain (residues R175, G245, R248, R249, R273, and R282) [68, 72, 73].
Gain of 1q
The long arm of chromosome 1 is gained (three copies) or amplified (more than three copies) in nearly 50% of MM patients, these percentages increase as the disease progress [62, 74]. Gain or amplification of chromosome arm 1q21 occurs in a subclone of the tumor as a secondary genomic event and is more amplified as the tumor progresses and a risk factor for the progression from smoldering multiple myeloma to MM [75].
Trisomy of chromosome 1, tandem duplications of 1q21, and jumping whole-arm translocations of 1q are some of the ways in which gain of 1q21 occurs, suggesting that these aberrations are heterogeneous at the genomic level [76]. Genes located in the 1q21 amplicon, including CKS1B, PSMD4, IL6R, ADAR, MCL1, and others, are associated with tumor proliferation and/or drug sensitivity because of upregulation of the expressed genes resulting from the increased gene dosage in MM cells with trisomy of chromosome 1 [75]. To date, the relevant genes on 1q21 remain unclear. Putative targets of this amplification include CKS1B and PMSD4 genes, mediating cell cycle progression and resistance to bortezomib, respectively.
A common feature shared by CKS1B and PMSD4 is that both of their expressions are highly sensitive to copy number variation of 1q21, and higher expression levels adversely affect clinical outcome, suggesting the existence of a gene-dosage effect that is of biological and prognostic significance [77-79]. Other genes mapped at 1q, such as MUC1, MCL1, ANP32E, BCL9, PSMD4, and PDZK1, have been proposed as candidate participants in myelomagenesis.
Several studies have shown that a 1q21 gain is a significant and independent factor in poor prognosis [80-82]. Although there are also a number of studies that have not been able to confirm this. The prognostic value depends on the treatment regimens, cohorts, and newly identified prognostic markers. Nevertheless, numerous publications suggest that at diagnosis, trisomy 1q seems to be an independent adverse prognosis marker for OS, even in the era when new drugs are used [46, 62, 80, 83].
Deletions of 1p
Deletions of 1p, del(1p), have been observed in up to 30% of MM patients [84], their fréquence increases to 60% in PCL [85], indicating that del(1p) may be related to clonal evolution [86].
Several minimally altered regions on 1p have been identified, including 1p32.3, 1p31.3, 1p22.1-1p21.3 and 1p12, were the FAM46C, CDC14A, MTF2, and CDKN2C genes are located [84, 86, 87]. Previously, the recurrent homozygous deletions and down expression of CDKN2C and FAF1 has already been described in homozygous 1p32.3 losses [86, 88]. In addition, recurrent homozygous deletions of FAM46C at 1p12 was identified as a gene with potential pathogenic relevance [87]. The next generation sequencing of 38 myeloma samples found FAM46C to be frequently mutated, highlighting the potential significance of this gene [12].
Chromosome 1p deletion is associated with poor outcome in myeloma patients [86]. In particular, del(1p) is associated with shorter survival in transplant-eligible patients, according to data from both the MRC Myeloma IX trial (United Kingdom) and the IFM (Institut Francophone du Myélome, France) group [84, 89].
Mutations
Whole-genome and whole-exome sequencing by NGS of thousands of MM samples has led to the detection of around 60 exonic mutations per patient [14]. The frequency of muta-tions in MM is higher than in acute leukemia, but much lower than in solid tumors, which tend to have hundreds of mutations [90]. In contrast with other hematological malignancies, there is no universal, unique, and specific mutation in MM, although many recurrently mutated genes have been detected [7]. In fact, about 250 mutated genes have been described in MM [90], about 60 of which are considered driver genes [91]. Most mutations are single nucleotide variants with consequences for the structure of the final protein. Mutations are present at both the clonal and sub-clonal levels, and increase as the disease progression. Various studies have investigated the mutational landscape of MM at diagnosis [92] and relapse [93, 94]. Some even compared the landscapes on both occasions by using paired samples [95, 96]. Even though there are many mutated genes, only a few of them are mutated in more than 5% of patients [97]. However, many of the mutated genes belong to key pathways that are usually dysregulated in MM: KRAS (20–25%), NRAS (20–25%), TP53 (8–15%), DIS3 (11%), FAM46C (11%), BRAF (6–15%), TRAF3 (3– 6%), ROBO1 (2–5%), EGR1 (4–6%), SP140 (5–7%), and FAT3 (4–7%) [98].
It seems that deregulation of RAS / MAPK, the NF-κB pathway, and apoptotic response plays a central role in the development of MM. However, the RAS / MAPK or NF-κB muta-tion was not predictive in this clinical trial [92].
The most significant predictive mutational marker in this study was TP53. The combination of TP53 mutation with additional mutations in ATM or ATR was found in 17% of cases with a significantly poor outcome. Besides ATM and ATR, ZFHX4 has been identified as a frequent-ly mutated gene. These genes are related apoptotic response to DNA damage DNA-Damage-Repair pathway [92].
Conclusion
Over the past decade, major improvements have been observed in the outcome of MM patients. This progress is mainly related to the availability of novel drugs. However, despite these improvements, it is clear that a huge heterogeneity persists in patient outcome. One of the most important predictors of prognosis for patients with MM is the detection of recurring genetic abnormalities. Pronounced genetic heterogeneity is the main feature of MM. This heterogeneity is mainly the result of biologic variations observed at the individual level.
It has been clearly shown that most tumor plasma cells share a common pool of mutations, but they may distinguish in several subclonal aberrations. The major clone at the time of diagnosis may be different from the major clone seen at the first relapse, which may also be different from the subclones seen at later relapses. There is probably competition between clones in the bone marrow niche for survival. Bone marrow niche characteristics (such as nutriment accessi-bility or hypoxia) may influence to the selection the clone with the best fitness. In addition, mutations itself can generate resistant clones. The different proliferative capacity of subclones can also be involved in selection. Finally, chemotherapy can also play an important role in the destruction of the most sensitive cells, but in the selection of more resistant ones, as well as for drugs that have mutagenic effects, with a direct effect on tumor cells.
The primary genomic events in MM are the acquisition of hyperdiploidy or translocations affecting the IGH genes; these events are mutually exclusive. Secondary genomic events in-clude other chromosomal translocations, copy number variations, loss or addition of chromo-somes. Recently, whole-exome sequencing studies have revealed numerous genomic muta-tions. Genomic events underlying MM affect multiple signalling pathways including the MYC, NF-κB, and MAPK pathways, plasma-cell differentiation, cell-cycle regulation or DNA-damage repair. The development of specific and, in particular, target therapy based on this knowledge opens up great opportunities in the future. However, typing of multiple genet-ic lesions can now identify high-risk patients, allowing stratification of the treatment.
Список литературы Genetic aberrations in multipe myeloma
- Joshua, D.E., et al., Biology and therapy of multiple myeloma. Med J Aust, 2019. 210(8): p. 375-380.
- Kumar, S.K., et al., Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group stu-dy. Leukemia, 2012. 26(1): p. 149-57.
- Rajkumar, S.V., Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. American Journal of Hematology, 2020. 95(5): p. 548-567.
- Gutiérrez, N.C., R. García-Sanz, and J.F. San Miguel, Molecular biology of myeloma. Clin Transl Oncol, 2007. 9(10): p. 618-24.
- Ryland, G.L., et al., Novel genomic findings in multiple myeloma identified through routine diagnostic sequencing. J Clin Pathol, 2018. 71(10): p. 895-899.
- Palumbo, A., et al., Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J Clin Oncol, 2015. 33(26): p. 2863-9.
- Cardona-Benavides, I.J., C. de Ramón, and N.C. Gutiérrez, Genetic Abnormalities in Multiple Myeloma: Prognostic and Therapeutic Implications. Cells, 2021. 10(2).
- Walker, B.A., et al., Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia, 2014. 28(2): p. 384-390.
- Landgren, O., et al., Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood, 2009. 113(22): p. 5412-7.
- Dutta, A.K., et al., Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia, 2019. 33(2): p. 457-468.
- Kyle, R.A., et al., Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N Engl J Med, 2007. 356(25): p. 2582-90.
- Chapman, M.A., et al., Initial genome sequencing and analysis of multiple myeloma. Nature, 2011. 471(7339): p. 467-72.
- Dutta, A.K., et al., Cutting edge genomics reveal new insights into tumour development, disease progression and therapeutic impacts in multiple myeloma. Br J Haematol, 2017. 178(2): p. 196-208.
- Manier, S., et al., Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol, 2017. 14(2): p. 100-113.
- Szalat, R. and N.C. Munshi, Genomic heterogeneity in multiple myeloma. Curr Opin Genet Dev, 2015. 30: p. 56-65.
- Brioli, A., et al., The impact of intra-clonal heterogeneity on the treatment of multiple myeloma. Br J Haematol, 2014. 165(4): p. 441-54.
- Morgan, G.J., B.A. Walker, and F.E. Davies, The genetic architecture of multiple myeloma. Nat Rev Cancer, 2012. 12(5): p. 335-48.
- Greaves, M. and C.C. Maley, Clonal evolution in cancer. Nature, 2012. 481(7381): p. 306-13.
- González, D., et al., Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood, 2007. 110(9): p. 3112-21.
- Zhan, F., et al., Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood, 2002. 99(5): p. 1745-57.
- Baldin, V., et al., Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev, 1993. 7(5): p. 812-21.
- Lakshman, A., et al., Natural history of t(11;14) multiple myeloma. Leukemia, 2018. 32(1): p. 131-138.
- Gao, W., et al., What Multiple Myeloma With t(11;14) Should Be Classified Into in Novel Agent Era: Standard or Intermediate Risk? Front Oncol, 2020. 10: p. 538126.
- Leiba, M., et al., Translocation t(11;14) in newly diagnosed patients with multiple myeloma: Is it always favorable? Genes Chromosomes Cancer, 2016. 55(9): p. 710-8.
- Kumar, S., et al., Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood, 2017. 130(22): p. 2401-2409.
- Pawlyn, C. and G.J. Morgan, Evolutionary biology of high-risk multiple myeloma. Nat Rev Cancer, 2017. 17(9): p. 543-556.
- Kalff, A. and A. Spencer, The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: prognostic implications and current clinical strategies. Blood Cancer Journal, 2012. 2(9): p. e89-e89.
- Brito, J.L., et al., MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica, 2009. 94(1): p. 78-86.
- Xie, Z. and W.J. Chng, MMSET: role and therapeutic opportunities in multiple myeloma. Biomed Res Int, 2014. 2014: p. 636514.
- Martinez-Garcia, E., et al., The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood, 2011. 117(1): p. 211-20.
- Min, D.J., et al., MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia, 2013. 27(3): p. 686-94.
- Shaughnessy, J., et al., Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood, 2001. 98(1): p. 217-223.
- Castaneda, O. and R. Baz, Multiple Myeloma Genomics - A Concise Review. Acta Med Acad, 2019. 48(1): p. 57-67.
- Barwick, B.G., et al., Cell of Origin and Genetic Alterations in the Pathogenesis of Multiple Myeloma. Front Immunol, 2019. 10: p. 1121.
- Hurt, E.M., et al., Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell, 2004. 5(2): p. 191-9.
- Bergsagel, P.L. and W.M. Kuehl, Chromosome translocations in multiple myeloma. Oncogene, 2001. 20(40): p. 5611-22.
- Annunziata, C.M., et al., A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood, 2011. 117(8): p. 2396-404.
- Avet-Loiseau, H., et al., Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood, 2001. 98(10): p. 3082-6.
- Smadbeck, J., et al., Mate pair sequencing outperforms fluorescence in situ hybridization in the genomic characterization of multiple myeloma. Blood Cancer J, 2019. 9(12): p. 103.
- van Riggelen, J., A. Yetil, and D.W. Felsher, MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer, 2010. 10(4): p. 301-9.
- Prochownik, E.V., c-Myc: linking transformation and genomic instability. Curr Mol Med, 2008. 8(6): p. 446-58.
- Hoffman, B. and D.A. Liebermann, Apoptotic signaling by c-MYC. Oncogene, 2008. 27(50): p. 6462-72.
- Misund, K., et al., MYC dysregulation in the progression of multiple myeloma. Leukemia, 2020. 34(1): p. 322-326.
- Chng, W.J., et al., Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia, 2011. 25(6): p. 1026-35.
- Walker, B.A., et al., Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J, 2014. 4(3): p. e191.
- Avet-Loiseau, H., et al., Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood, 2007. 109(8): p. 3489-95.
- Chng, W.J., R.P. Ketterling, and R. Fonseca, Analysis of genetic abnormalities provides insights into genetic evolution of hyperdiploid myeloma. Genes Chromosomes Cancer, 2006. 45(12): p. 1111-20.
- Hoctor, V.T. and L.J. Campbell, Hyperhaploid plasma cell myeloma. Cancer Genet, 2012. 205(7-8): p. 414-8.
- Sawyer, J.R., et al., Hyperhaploidy is a novel high-risk cytogenetic subgroup in multiple myeloma. Leukemia, 2017. 31(3): p. 637-644.
- Smadja, N.V., et al., Further cytogenetic characterization of multiple myeloma confirms that 14q32 translocations are a very rare event in hyperdiploid cases. Genes Chromosomes Cancer, 2003. 38(3): p. 234-9.
- Mateo, G., et al., Genetic abnormalities and patterns of antigenic expression in multiple myeloma. Clin Cancer Res, 2005. 11(10): p. 3661-7.
- Chng, W.J., et al., A validated FISH trisomy index demonstrates the hyperdiploid and nonhyperdiploid dichotomy in MGUS. Blood, 2005. 106(6): p. 2156-61.
- Avet-Loiseau, H., et al., Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol, 2009. 27(27): p. 4585-90.
- Besse, L., et al., Cytogenetics in multiple myeloma patients progressing into extramedullary disease. Eur J Haematol, 2016. 97(1): p. 93-100.
- Fonseca, R., et al., Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia, 2001. 15(6): p. 981-6.
- Avet-Louseau, H., et al., Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br J Haematol, 2000. 111(4): p. 1116-7.
- Tricot, G., et al., Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other kary-otype abnormalities. Blood, 1995. 86(11): p. 4250-6.
- Jin, X., et al., Phosphorylated RB Promotes Cancer Immunity by Inhibiting NF-κB Activation and PD-L1 Expression. Mol Cell, 2019. 73(1): p. 22-35.e6.
- Burkhart, D.L. and J. Sage, Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nature Reviews Cancer, 2008. 8(9): p. 671-682.
- Taniguchi, K. and M. Karin, NF-κB, inflammation, immunity and cancer: coming of age. Nature Reviews Immunology, 2018. 18(5): p. 309-324.
- Manguso, R.T., et al., In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature, 2017. 547(7664): p. 413-418.
- Avet-Loiseau, H., et al., Long-term analysis of the IFM 99 trials for myeloma: cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J Clin Oncol, 2012. 30(16): p. 1949-52.
- Hu, B., et al., High-risk myeloma and minimal residual disease postautologous-HSCT predict worse outcomes. Leuk Lymphoma, 2019. 60(2): p. 442-452.
- Tiedemann, R.E., et al., Genetic aberrations and survival in plasma cell leukemia. Leukemia, 2008. 22(5): p. 1044-52.
- Merz, M., et al., Baseline characteristics, chromosomal alterations, and treatment affecting prognosis of deletion 17p in newly diagnosed myeloma. Am J Hematol, 2016. 91(11): p. E473-e477.
- Shah, V., et al., Molecular Characterisation of TP53 Aberrations in 1,777 Myeloma Trial Patients. Blood, 2017. 130(Supplement 1): p. 4331-4331.
- Xiong, W., et al., An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood, 2008. 112(10): p. 4235-46.
- Herrero, A.B., et al., Molecular Mechanisms of p53 Deregulation in Cancer: An Overview in Multiple Myeloma. International Journal of Molecular Sciences, 2016. 17(12): p. 2003.
- Thakurta, A., et al., High subclonal fraction of 17p deletion is associated with poor prognosis in multiple myeloma. Blood, 2019. 133(11): p. 1217-1221.
- Chin, M., et al., Prevalence and timing of TP53 mutations in del(17p) myeloma and effect on survival. Blood Cancer Journal, 2017. 7(9): p. e610-e610.
- Lionetti, M., et al., Molecular spectrum of TP53 mutations in plasma cell dyscrasias by next generation sequencing: an Italian cohort study and overview of the literature. Oncotarget, 2016. 7(16): p. 21353-61.
- Joerger, A.C. and A.R. Fersht, Structural Biology of the Tumor Suppressor p53 and Cancer‐Associated Mutants, in Advances in Cancer Research. 2007, Academic Press. p. 1-23.
- Rivlin, N., et al., Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes & Cancer, 2011. 2(4): p. 466-474.
- An, G., et al., Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica, 2014. 99(2): p. 353-9.
- Hanamura, I., Gain/Amplification of Chromosome Arm 1q21 in Multiple Myeloma. Cancers (Basel), 2021. 13(2).
- Sawyer, J.R., et al., Evidence for a novel mechanism for gene amplification in multiple myeloma: 1q12 pericentromeric heterochromatin mediates breakage-fusion-bridge cycles of a 1q12 approximately 23 amplicon. Br J Haematol, 2009. 147(4): p. 484- 94.
- Fonseca, R., et al., International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia, 2009. 23(12): p. 2210-21.
- Shaughnessy, J.D., Jr., et al., Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myelo-ma treated with Total Therapy 3. Blood, 2011. 118(13): p. 3512-24.
- Zhan, F., et al., CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood, 2007. 109(11): p. 4995-5001.
- Fonseca, R., et al., Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia, 2006. 20(11): p. 2034-40.
- Hanamura, I., et al., Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood, 2006. 108(5): p. 1724-32.
- Shaughnessy, J.D., Jr., et al., Testing standard and genetic parameters in 220 patients with multiple myeloma with complete data sets: superiority of molecular genetics. Br J Haematol, 2007. 137(6): p. 530-6.
- Shah, V., et al., Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia, 2018. 32(1): p. 102-110.
- Boyd, K.D., et al., Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res, 2011. 17(24): p. 7776-84.
- Li, F., et al., Identification of characteristic and prognostic values of chromosome 1p abnormality by multi-gene fluorescence in situ hybridization in multiple myeloma. Leukemia, 2016. 30(5): p. 1197-201.
- Leone, P.E., et al., Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res, 2008. 14(19): p. 6033-41.
- Walker, B.A., et al., A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood, 2010. 116(15): p. e56-65.
- Walker, B.A., et al., Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood, 2011. 117(2): p. 553-562.
- Hebraud, B., et al., Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: the IFM experience on 1195 patients. Leukemia, 2014. 28(3): p. 675-9.
- Corre, J., et al., Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia, 2018. 32(12): p. 2636-2647. 91. Maura, F., et al., Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun, 2019. 10(1): p. 3835.
- Walker, B.A., et al., Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J Clin Oncol, 2015. 33(33): p. 3911-20.
- Lohr, J.G., et al., Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell, 2014. 25(1): p. 91-101.
- Bolli, N., et al., Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature Communications, 2014. 5(1): p. 2997.
- John, R.J., et al., Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica, 2019. 104(7): p. 1440-1450.
- Weinhold, N., et al., Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood, 2016. 128(13): p. 1735-1744.
- Lionetti, M. and A. Neri, Utilizing next-generation sequencing in the management of multiple myeloma. Expert Rev Mol Diagn, 2017. 17(7): p. 653-663.
- Kumar, S.K. and S.V. Rajkumar, The multiple myelomas — current concepts in cytogenetic classification and therapy. Nature Reviews Clinical Oncology, 2018. 15(7): p. 409-421.