Клонирование и экспрессия гена эндонуклеазы I бактериофага Т7
Автор: Машков Олег Игоревич, Чубукова Ольга Вячеславовна
Журнал: Известия Самарского научного центра Российской академии наук @izvestiya-ssc
Рубрика: Генетика
Статья в выпуске: 5-3 т.13, 2011 года.
Бесплатный доступ
В настоящей статье сообщается о клонировании гена эндонуклезы I бактериофага Т7 в экспрессирующей векторной конструкции, а также о создании бактериального штамма-продуцента данного фермента.
Короткий адрес: https://sciup.org/148200461
IDR: 148200461 | УДК: 03.4.1.1
Текст научной статьи Клонирование и экспрессия гена эндонуклеазы I бактериофага Т7
Генетическая рекомбинация является одним из фундаментальных генетических процессов. Она необходима при внедрении вирусной ДНК в геном клетки-хозяина [16], поддержании общей стабильности генома [4] посредством рекомбинант-зависимой репарации ДНК [14], при повторном запуске или расщеплении остановленных реплика-ционных вилок [3]. Рекомбинационный процесс является основой непостоянства генома, это источник новых комбинаций существующих единиц наследственности [8, 9]. В ходе рекомбинации образуется промежуточный субстрат – структура Холлидея – четырёхнитевой перекрест [7], который впоследствии подвергается ферментативному разрешению [6, 9, 12, 17]. Многие перекрест-расщепляющие ферменты можно отнести к одному из двух суперсемейств белков – интеграз или нуклеаз. В суперсемейство интеграз входят такие ре-золвазы как RuvC, CceI, Ydc2, а также РНКаза Н и фермент вируса коровьей оспы А22. К суперсемейству нуклеаз относятся Т7 эндонуклеаза I, резолва-зы архей, λ-экзонуклеаза, а также ферменты рестрикции [10, 19]. Большинство нуклеаз – сравнительно небольшие основные белки (обычно pI calc >8.5), связывающиеся со структурой Холлидея в строго определённых местах. При этом образуются комплексы димерного белка и ДНК-перекреста, сохраняющие стабильность в присутствии 1000-кратного избытка двухцепочечной ДНК [20, 21]. На данный момент механизмы разрешения структуры Холлидея изучены не полностью. Это обусловлено тем, что до настоящего времени не получены данные о кристаллической структуре комплекса ДНК-перекреста с присоединённой к нему эндонуклеазой.
К хорошо изученным резолвазам относится эндонуклеаза I бактериофага Т7 (T7EI). Этот фермент принадлежит к суперсемейству нуклеаз [10]. Благодаря широкой субстратной специфичности, данный фермент взаимодействует со множеством разветвлённых форм ДНК, включая линейные гетеро-дуплексные молекулы с неспаренными нуклеотидами [11]. Можно предположить, что перекрест-расщепляющие ферменты гидролизуют фосфодиэфирные связи, используя молекулу воды,
активированную ионами металлов. В пользу этого утверждения свидетельствует тот факт, что среди структур предполагаемых активных центров многих изученных ферментов встречаются кластеры кислых аминокислотных цепей, вероятно, отвечающих за связывание и координацию ионов металлов [20, 22, 23, 24, 25]. Кроме того, в активном сайте Т4 эндонуклеазы VII обнаружен ион кальция [26]. Позднее в активном сайте Т7 эндонуклеазы I, также были найдены два иона кальция [27].
Эндонуклеаза T7EI кроме структуры Холлидея взаимодействует также и с другими ДНК-структурами, вплоть до гетеродуплексов с однонуклеотидными ошибочными спариваниями – мисмэтчами [11]. Несмотря на это, механизм специфического узнавания субстрата до сих пор в деталях не изучен. В случае с ферментами, расщепляющими перекресты, определение молекулярных основ узнавания осложнено изменением конформации ДНК при прикреплении нуклеаз [5, 7, 18].
Для образования комплекса «фермент-субстрат» с Т7 эндонуклеазой I молекула ДНК должна войти в высокоэнергетическое конформационное состояние. Следовательно, из-за высокой энергии активации реакция протекает медленно, но для определённых последовательностей ДНК (например, для дуплексов с несовершенной комплементарной структурой) энергия активации может быть ниже, чем для других. По этой причине ДНК с ошибочно спаренными основаниями или с фосфодиэфирным разрывом в одной из цепей являются для T7EI наиболее предпочтительными субстратами. Такие участки с неполной комплементацией оснований по своей природе более гибкие – они являются своего рода горячими точками неспецифической нуклеазной активности генома.
Цель исследования состояла в клонировании гена эндонуклезы I бактериофага Т7 в экспрессирующей векторной конструкции, с последующим созданием штамма-продуцента данного фермента на основе штамма E. coli .
МАТЕРИАЛ И МЕТОДЫ
Объект исследования – фермент T7EI, кодируемый третьим геном кольцевой молекулы ДНК бактериофага Т7 [1, 13, 15]. Данный фермент представляет собой небольшой симметричный гомоди- мер с молекулярной массой 17 kDa, каждый домен которого состоит из 149 аминокислотных остатков.
Выделение и очистка ДНК бактериофага Т7. ДНК бактериофага Т7 выделяли из жидкой культуры бактериальных клеток E. coli , содержащих вирусные частицы Т7. Лизис проводили при помощи 2% SDS 10 мин при 4°С. К лизату добавляли 1М NaCl, после чего смесь центрифугировали в течение 5 мин при 14500 об./мин на микроцентрифуге Mini-Spin plus (Eppendorf, Германия). Далее фаговая ДНК, находящаяся в супернатанте, была выделена методом фенольно-хлороформной экстракции и в последующем осаждена 70% спиртом.
Выделение и очистка плазмидной ДНК . Плазмидную ДНК выделяли методом мягкого лизиса [2].
Полимеразная цепная реакция. Объем реакционных смесей для ПЦР составлял 30 мкл. Реакционная смесь содержала 67 мМ трис-HCl, 16,6 мМ (NH 4 ) 2 SO 4 , 1,5 мМ MgCl 2 , 0,01% Twin-20, 0,1 мкл геномной ДНК, по 10 пМ каждого праймера, по 200 мкм дАТФ, дЦТФ, дТТФ, дГТФ и 1 единицы ДНК-полимеразы. ПЦР проводили в амплификаторе МС2 «Терцик» компании "ДНК-технология" (Россия). Для получения полноразмерной последовательности гена Т7 эндонуклеазы I были подобраны праймеры: Т7Е1-U 5'-
AAGGAGGATC-CATGGCAGGTTACGGCGCTAAA-3', включаю щий область, соответствующую участку с 1 по 26 нуклеотид "+" цепи ДНК гена Т7Е1, T7E2-U 5'-AGGGAGGATC-CGATGGCAGGTTACGGCGCTAAA-3', включает область, комплементарную участку с 427 по 450 нуклеотиды "+" цепи ДНК гена Т7Е1, и Т7E1-L 5'-GTACAGGATCCATTATTTCTTTCCTCCTTTCCT-TTT-3'. Дополнительно в данные праймеры был введён сайт рестрикции для рестриктазы BamHI (последовательности подчёркнуты).
Расщепление ДНК рестрикционными эндонуклеазами проводили в буферах, рекомендованных фирмами-поставщиками. Время рестрикции варьировало от 1 ч до 1 сут в зависимости от цели эксперимента, а также от количества расщепляемого препарата ДНК и единиц активности взятого в реакцию фермента. В экспериментах по молекулярному клонированию рестрикционные эндонуклеазы инактивировали добавлением ЭДТА до конечной концентрации 20 мМ с последующей обработкой смесью фенола и хлороформа.
Определение нуклеотидных последовательностей амплифицированных фрагментов генома бактериофага Т7, содержащих ген эндонуклеазы I, проводили на автоматическом секвенаторе ABI PRISM 310 фирмы “Applied Biosystems” (США), используя наборы для секвенирования “Big Dye Terminator v.3.0”.
Экспрессия и очистка Т7 эндонуклеазы I. Штаммы бактерий E. coli BL21(DE3)pLysS, содержащие необходимую векторную молекулу со вставкой целевого гена, инкубировали до оптической плотности культуры 0,6 при 600 нм. Затем индуцировали бактериальную культуру внесением 0,5 мМ IPTG. Культивирование с IPTG продолжалось от 3 до 6 ч. Клетки осаждали при центрифугировании на центрифуге К-23 при 4ºС 3000 об/мин в течение 10 мин и суспедировали в лизирующем буфере (Na-фосфатный буфер 100 мМ; NaCl 300 мМ; Тритон X-100 1%; глицерин 20%; pH 8,0). Далее клетки подвергали УЗ-дезинтеграции 5-6 раз по 15 сек при 4ºС на УЗДН-2Т (СССР), после чего клеточный дебрис был удалён центрифугированием на ультрацентрифуге Avanti® J-E в бакет-роторе JA-20.1 (Beckman, США) при 4ºС 20000 об/мин в течение 30 мин. После осветлённый белковый экстракт загружали в Ni-сефарозные колонки Pierce® Centrifuge Columns (Thermo Scientific, США). Целевой белковый продукт выделяли градиентом концентрации имидазола (от 50 мМ до 400 мМ) по методике, предоставляемой производителем. Полученные фракции диализовали против градиента 20 мМ Tris-HCl (pH 8.0), 1 мМ EDTA, 1мМ дитиотри-тола, 50% глицерина 2 ч. При 4ºС. Очищенный белковый продукт анализировался в SDS-полиакриламидном геле.
РЕЗУЛЬТАТЫ И ИХ ОБСУЖДЕНИЕ
По завершении ПЦР геномной ДНК фага Т7 был амплифицирован фрагмент, размер которого совпал с длиной гена эндонуклеазы I бактериофага Т7 и составил 474 п.н. Полученный амплификат был клонирован в плазмидный вектор pKRX (National BioResource Project: E. coli Strain, Япония).
Далее был проведён рестрикционный анализ продукта амплификации плазмидной ДНК клонов pKRX, содержащих вставку, по уникальным сайтам рестрикции гена Т7Е1 ( Csp 61, Hae III). При рестрикционном анализе полноразмерный ген Т7Е1 расщепляется на два фрагмента с ожидаемыми для данных сайтов рестрикции размерами ( Csp 61 – 293 и 157 п.н.; Hae III – 397 и 53 п.н.). Проведённый рестрикционый анализ дал фрагменты, соответствующие ожидаемым (рис. 1).
После необходимой наработки плазмиды ген Т7Е1 был амплифицирован и клонирован в экспрессирующие векторные конструкции по сайтам рестрикции Bam HI в вектора рЕТ-3а и PinPointXA-2, и Bam HI, Hin dIII в рЕТ-22b. Полученные штаммы анализировались методом автоматического секвенирования. В результате были отобраны штаммы, содержащие векторные молекулы без изменения рамки считывания, с сохранением всех необходимых регуляторных областей гена Т7Е1 и плазмиды для экспресии данного гена (pET-22bT7cl2, pET-22bT7cl7, pET-22bT7cl14, PinPointXA-2cl42, PinPointXA-2cl78, PinPointXA-2cl83).
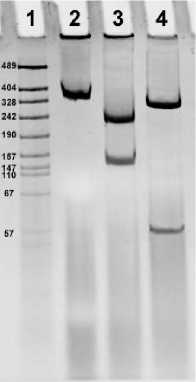
Рис. 1. Рестрикционный анализ фрагмента плазмиды pKRXT7-E1, гомологичного гену Т7Е1. 1 – маркер; 2 – амплифицированный фрагмент плазмиды pKRXT7-E1; 3 – результат рестрикции амплификата Csp 61; 4 – результат рестрикции амплификата Hae III.
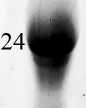
После индукции с различными условиями среди отобранных клонов был выделен один (pET-22bT7cl2), который по результатам электрофореграмм клеточного лизата нарабатывал белковый продукт сходный по размерам с эндонуклеазой I бактериофага Т7. Далее белковый препарат клона pET-22bT7cl2 был очищен методом аффинной хроматографии и сконцентрирован при помощи диализа (рис. 2).
45 *
18^
М 7 8 9 D
Рис. 2. Электрофореграмма очищенного методом аффинной хроматографии белкового препарата pET-22bT7cl2. М - белковый маркер (цифровые обозначения слева - молекулярный вес фрагмента в кДа); 7, 8, 9 - фракции белкового препарата, выделенные после аффинной хроматографии pET-22bT7cl2; D - фракции белкового препарата pET-22bT7cl2 после процедуры диализа.
Необходимо отметить, что на стадии индукции бактериальной культуры и последующей очистки фермента мы столкнулись с трудностью наработки достаточного для идентификации количества Т7 эндонуклеазы I.
Было опробовано несколько вариантов культивирования штаммов E. coli , которые по предварительным данным могли синтезировать целевой фермент.
Из всех условий наиболее удовлетворительной для наработки T7EI оказалась индукция 0,5 мМ
IPTG при 32º С в течение 6 ч с последующей УЗ-дезинтеграцией клеток на снегу клона pET-22bT7cl2. Наиболее вероятным объяснением наработки фермента при данных условиях может являться тот факт, что помимо основной своей функции (которая заключается в разрезании структур Холлидея, образующихся при рекомбинации во время литического цикла бактериофага Т7) T7EI при высокой концентрации в клетке расщепляет ДНК самой бактерии [15]. При пониженной температуре культивирования в отдельно взятой клетке E. coli фермента T7ЕI нарабатывается меньше, его растворимость повышается. Однако изначально высокая численность клеток непосредственно перед индукцией позволяет выделить достаточное для детекции гель-электрофорезом количество Т7EI.
В рамках данного исследования нами был создан ряд клонов, содержащих вставку гена эндонуклеазы I бактериофага Т7 в экспрессирующих векторных системах рЕТ и PinPoint. Данный факт подтверждается наработкой в результате ПЦР с праймерами к гену Т7ЕI фрагмента ДНК одинаковой длины с амплифицированным ранее геном Т7ЕI из ДНК фага Т7, рестрикционным анализом и автоматическим секвенированием плазмидной ДНК трансформированных клеток E. coli . Однако только в одном штамме (pET-22bT7cl2) была отмечена возможность стабильной наработки целевого фермента. Поскольку экспрессия вставки в используемой векторной системе рЕТ-22b приводит к образованию белкового продукта с гистидиновым «хвостом» на С-конце, фермент Т7ЕI был специфично очищен из клеточного лизата методом аффинной хроматографии. Выделенная и очищенная фракция клеточного лизата pET-22bT7cl2 по своей электрофоретической подвижности аналогична Т7ЕI. Опираясь на эти результаты, мы можем предположить, что созданный штамм E. coli BL21(DE3)pLysS, содержащий плазмиду pET-22bT7cl2, является штаммом-продуцентом эндонуклеазы I бактериофага Т7.
Список литературы Клонирование и экспрессия гена эндонуклеазы I бактериофага Т7
- Center M.S., Richardson C.C. An endonuclease induced after infection of Escherichia coli with bacteriophage T7. I. Purification and properties of the enzyme//J. Biol. Chem. 1970. V. 245. Р. 6285-6291.
- Clewell D.B., Helinski D.R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form//Proc. Natl. Acad. Sci. USA. 1969. V. 62. N 4. P. 1159-1166.
- Cox M.M., Goodman M.F., Kreuzer K.N. et al. The importance of repairing stalled replication forks//Nature. 2000. V. 404. Р. 37-41.
- Flores-Rozas, H., Kolodner R.D. Links between replication, recombination and genome instability in eukaryotes//Trends Biochem. Sci. 2000. V. 292. Р. 196-200.
- Fogg J.M., Kvaratskhelia M., White M.F., Lilley D.M.J. Distortion of DNA junctions imposed by the binding of resolving enzymes: a fluorescence study//J. Mol. Biol. 2001. V. 313. P. 751-764.
- Giraud-Panis M.-J.E., Lilley D.M.J. Structural recognition and distortion by the DNA junction-resolving enzyme RusA//J. Mol. Biol. 1998. V. 278. P. 117-133.
- Holliday R. A mechanism for gene conversion in fungi//Genet. Res. 1964. V. 5. P. 282-304.
- Holliday R. Molecular aspects of genetic exchange and gene conversion//Genetics. 1974. V. 78. Р. 273-287.
- Kerr C., Sadowski P. D. The involvement of genes 3, 4, 5 and 6 in genetic recombination in bacteriophage T7//Virology. 1975. V. 65. P. 281-285.
- Lilley D.M.J., White M.F. Resolving the relationships of resolving enzymes//Proc. Natl. Acad. Sci. USA. 2000. V. 97. Р. 9351-9353.
- Mashal R.D., Koontz J., Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases//Nat. Genet. 1995. V. 9. P. 177-183.
- Powling A., Knippers R. Recombination of bacteriophage T7 in vivo//Mol. Gen. Genet. 1976. V. 149. Р. 63-71.
- Sadowski P. D. Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B//J. Biol. Chem. 1971. V. 246. P. 209-216.
- Smith K.C. Recombinational DNA repair: the ignored repair systems//Bioessays. 2004. V. 26 (12). Р. 1322-1326.
- Studier F.W. The genetics and physiology of bacteriophage T7//J. Virol. 1969. V. 39. P. 562-574.
- Subramaniam S., Tewari A.K., Nunes-Duby S.E., Foster M.P. Dynamics and DNA substrate recognition by the catalytic domain of lambda integrase//J. Mol. Biol. 2003. V. 329. N 3. Р. 423-439.
- Tsujimoto Y., Ogawa H. Intermediates in genetic recombination of bacteriophage T7 DNA. Biological activity and the roles of gene 3 and gene 5//J. Mol. Biol. 1978. V. 125. Р. 255-273.
- White M.F., Lilley D.M.J. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction//J. Mol. Biol. 1997. V. 266. P. 122-134.
- Makarova K.S., Aravind L., Koonin E.V. Holliday junction resolvases and related nucleases: identifcation of new families, phyletic distribution and evolutionary trajectories//Nucleic Acids Res. 2000. V. 28. P. 3417-3432.
- Duckett D.R., Panis M.E.G., Lilley D.M.J. Binding of the junction-resolving enzyme bacteriophage T7 endonuclease I to DNA: separation of binding and catalysis by mutation//J. Mol. Biol. 1995. V. 246. Р. 95-107.
- White M.F., Lilley D.M.J. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae//J. Mol. Biol. 1996. V. 257. Р. 330-341.
- Saito A., Iwasaki H., Ariyoshi M. et al. Identification of four acidic amino acids that constitute the catalytic centre of the RuvC Holliday junction resolvase//Proc. Natl Acad. Sci. USA. 1995. V. 92. P. 7470-7474.
- Giraud-Panis M.-J.E., Lilley D.M.J. Structural recognition and distortion by the DNA junction-resolving enzyme RusA.//J. Mol. Biol. 1998. V. 278. P. 117-133.
- Parkinson M.J., Pöhler J.R.G., Lilley D.M.J. Catalytic and binding mutants of the junction-resolving enzyme endonuclease I of bacteriophage T7: role of acidic residues//Nucleic Acids Res. 1999. V. 27. P. 682-689.
- Wardleworth B.N., Kvaratskhelia M., White M.F. Sitedirected mutagenesis of the yeast resolving enzyme Cce1 reveals catalytic residues and relationship with the intronsplicing factor Mrs1.//J. Biol. Chem. 2000. V. 275. P. 23725-23728.
- Raaijmakers H., Vix O., Toro I. et al. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture//EMBO J. 1999. V. 18. P. 1447-1458.
- Hadden J.M., Déclais A.C., Carr S.B. et al. The structural basis of Holliday junction resolution by T7 endonuclease I.//Nature. 2007. V. 449. N 4. P. 621-625.


