LeafLens: Detect Plant Disease Before They Spread
Автор: Mrs. S.A. Vijayalakshmi, K.C. Akhila, Mr. S.P. Srikanth, A.C. Ananya, A. Jahnavi, Anjana Dinesh
Журнал: Science, Education and Innovations in the Context of Modern Problems @imcra
Статья в выпуске: 4 vol.7, 2024 года.
Бесплатный доступ
LeafLens is an intelligent plant disease detection system designed to assist farmers and agriculturists in identifying plant diseases efficiently and accurately. Leveraging the power of deep learning, the system employs a Convolutional Neural Network (CNN) architecture trained on a comprehensive dataset of diseased and healthy leaf images. The application streamlines the diagnosis process by analyzing images of plant leaves and providing precise predictions about the disease type. The backend is developed using Python and PyTorch, ensuring robust model performance and scalability. The system integrates image preprocessing techniques such as resizing, cropping, and normalization to ensure uniform input to the model. The frontend is designed to be user-friendly, allowing users to upload leaf images seamlessly. Real-time inference capabilities provide instant feedback, displaying the disease classification alongside recommendations for treatment. LeafLens also incorporates features like data visualization for training and validation losses, accuracy tracking, and detailed performance metrics, enabling continuous improvement of the model. The system's framework supports scalability, allowing integration into mobile and web platforms for broader accessibility. By offering a reliable, efficient, and cost-effective solution, LeafLens aims to revolutionize agricultural disease management, promoting healthier crops and sustainable farming practices.
Deep Learning, Convolutional Neural Network (CNN), Image Classification, Agricultural Technology
Короткий адрес: https://sciup.org/16010303
IDR: 16010303 | DOI: 10.56334/sei/7.4.10
Текст научной статьи LeafLens: Detect Plant Disease Before They Spread
Introduction.
LeafLens is an intelligent plant disease detection system aimed at addressing one of the critical challenges in agriculture—identifying plant diseases early to mitigate their impact on crop yield and quality. Plant diseases, caused by various pathogens, result in significant losses each year, emphasizing the need for efficient and scalable diagnostic solutions. Traditional methods for disease detection rely on manual inspection and expert knowledge, which are timeconsuming, resource-intensive, and often inaccessible to small-scale farmers.
Recent advancements in deep learning have revolutionized image-based disease detection, enabling automated and accurate identification of plant diseases. Leveraging Convolutional Neural Networks (CNNs), LeafLens analyzes images of plant leaves to classify them as healthy or diseased. Existing systems often focus on limited disease classes or require high-end infrastructure, making them impractical for widespread adoption. Additionally, they lack integrated recommendations, leaving farmers without actionable advice for managing plant health.
The proposed system overcomes these challenges by offering a comprehensive, scalable platform that combines disease detection with a built-in recommendation system. Leveraging sophisticated image preprocessing techniques and a pre- trained CNN model, LeafLens delivers accurate and consistent classification across diverse plant species and disease types. Additionally, it provides actionable insights, including tailored treatment suggestions and preventive strategies, enabling users to respond promptly and effectively.
By combining a user-friendly interface with a scalable deployment framework, LeafLens aims to democratize access to plant health management tools. This project contributes to sustainable agriculture by reducing crop losses, minimizing chemical overuse, and promoting ecofriendly farming practices. With its innovative approach, LeafLens represents a significant step toward enhancing global food security and supporting farmers worldwide.
Background
Plant disease detection models rely on datasets, which are captured under controlled conditions. However, real-world agricultural environments involve varied lighting, backgrounds, and leaf orientations, which can cause models trained on such controlled datasets to perform poorly in practical, dynamic settings. This lack of diversity in the training data means that models may struggle to generalize to real-world scenarios where variability in environmental conditions is common [1, 2, 4].
Another challenge is class imbalance. Rare plant diseases often have fewer examples in datasets, leading to an imbalance where the model becomes biased toward more common diseases. This reduces the model’s ability to accurately detect rarer diseases, which can still be harmful in agricultural contexts. Addressing this issue requires techniques such as data augmentation and synthetic image generation to balance the representation of both common and rare diseases in the dataset [6, 7].
Deep convolutional neural networks (CNNs) are powerful tools for plant disease detection, but their computational requirements can pose a challenge, particularly when deployed in resource-constrained environments. These models require significant processing power for both training and inference, making it difficult to implement them on edge devices like smartphones or low-cost computing systems typically used in agricultural settings. To mitigate this, lightweight architectures and optimization techniques are needed to reduce computational load while maintaining accuracy [4].
Hyperparameter tuning is another time-intensive challenge in CNN-based models. Optimizing hyperparameters such as learning rates, batch sizes, and network architecture can require extensive experimentation. This process is often complex and resource-draining, especially when working with large datasets. Automated techniques like grid search or Bayesian optimization can help streamline this process, making model tuning more efficient [6, 8].
Over fitting is a common issue in deep learning, particularly when models are trained on specific datasets. When this happens, the model may capture noise rather than general patterns, leading to poor performance when exposed to new, unseen data. This generalization issue can be mitigated through techniques like dropout, weight decay, and early stopping, which help the model generalize better and avoid over fitting [1, 2, 4].
Models also struggle with detecting multiple diseases on a single leaf. In reality, plants may suffer from more than one infection at the same time, and the ability to identify multiple diseases is essential for accurate diagnosis. Models need to incorporate multi-label classification capabilities to effectively handle such scenarios, improving their practical applicability in the field [3, 7].
Deep learning models are often referred to as "black boxes" because their decisionmaking process is not easily interpretable. This lack of transparency can make it difficult for users, such as farmers, to trust the model’s predictions. For models to be widely adopted, especially in agricultural settings, it is crucial to integrate explainable AI features, such as visualizations, to highlight the areas influencing the model’s predictions, helping to build trust and improve user acceptance [8].
Finally, data acquisition poses significant challenges, particularly when it comes to rare plant diseases. Obtaining a diverse and comprehensive dataset can be costly, especially for diseases with low incidence rates. Limited training data can restrict model performance, making it harder for the model to generalize across different disease types. Approaches such as crowdsourcing, data augmentation, and the use of publicly available datasets can help address this issue, expanding the dataset without incurring high costs [5, 6].
By focusing on these challenges—such as improving dataset diversity, reducing model complexity, optimizing hyperparameter tuning, enhancing model generalization, incorporating multi-label classification, ensuring interpretability, and addressing data acquisition barriers— future research can make deep learning models more effective and practical for plant disease detection in real- world agricultural environments.
Methodology
The methodology outlines the step-by-step processes used in the system for dataset augmentation, model training, and evaluation.
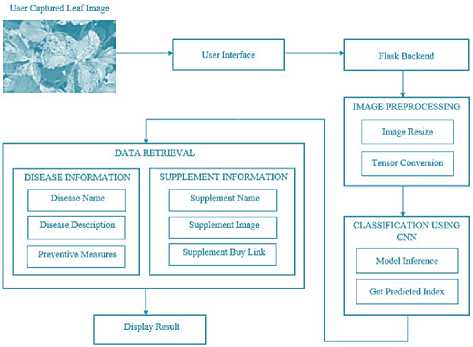
-
Fig. 1. System Architecture
-
a) Data Collection and Preparation
The development of LeafLens begins with the careful assembly of a robust dataset that forms the foundation for accurate plant disease detection. A variety of data sources, such as Kaggle and PlantVillage, are used to gather an extensive collection of plant leaf images. These images include both healthy and diseased leaves, ensuring the dataset captures a wide array of crop types and disease categories. The inclusion of diverse crop varieties and disease manifestations is critical to developing a generalizable model that can accurately diagnose various conditions across different environments.
Each image is meticulously labeled with its corresponding class—whether it's healthy or affected by a specific disease. This labeling ensures that the CNN model can correctly associate each image with its appropriate class, facilitating effective training. To ensure uniformity across the dataset, preprocessing techniques such as resizing, cropping, and noise reduction are applied.
These steps standardize the images, making them more compatible with the model and ensuring that the CNN focuses on the key features of the plant leaves while minimizing irrelevant information.
-
b) Model Training and Evaluation
Once the dataset is curated and preprocessed, a Convolutional Neural Network (CNN) is employed to train the model. CNNs are particularly well-suited for image-related tasks due to their ability to automatically learn hierarchical features from images. The model is fine-tuned to optimize its accuracy in classifying leaf diseases. Techniques like data augmentation are applied to artificially increase the dataset size by transforming existing images, such as rotating, flipping, and adjusting the brightness of images. This helps in improving the model’s robustness by simulating variations in real-world conditions.
Batch normalization is incorporated during the training process to stabilize the learning process and speed up convergence. This technique normalizes the input layer by adjusting and scaling activations, which helps prevent issues such as vanishing/exploding gradients. During the training, key performance metrics such as accuracy, precision, recall, and F1-score are continuously monitored to evaluate the model’s performance. Additionally, confusion matrices and loss curves are used as diagnostic tools to assess the model’s progress. These visualizations provide insight into the types of errors the model makes and help fine-tune the hyper parameters, improving overall performance.
-
c) Backend Implementation
The backend of LeafLens is developed using Flask, a lightweight and flexible web framework. Flask serves as the intermediary between the frontend interface and the trained CNN model, allowing for smooth communication and data flow. The backend is responsible for handling image uploads from users, preprocessing them into the appropriate format, and making inferences using the CNN model.
Uploaded images are resized to a standard dimension and converted into tensors— numerical representations of the image data—so they can be fed into the CNN model for prediction. After the image is processed, the CNN model predicts the disease class associated with the plant leaf. This prediction is then translated into a human-readable disease name, using a predefined dictionary that links disease labels to their corresponding names. The backend also handles any exceptions or errors during the image upload or inference process, ensuring that the system operates smoothly even under varied conditions.
-
d) Recommendation Engine Integration
To provide users with actionable insights, a recommendation engine is integrated into the LeafLens system. After the model classifies the disease, the recommendation engine retrieves relevant information from a comprehensive database, which includes details about the detected disease. This information might include the disease's name, symptoms, possible causes, prevention strategies, and treatment options. The engine pulls this data from a structured CSV file, which serves as the primary data source for the recommendations. This approach ensures that recommendations are dynamic and updated based on the disease classification results.
By incorporating this recommendation engine, LeafLens goes beyond simple disease detection to provide farmers with valuable, context-specific advice. This empowers users to take immediate actions in managing plant health, improving crop yield, and minimizing the impact of plant diseases on their farming operations. Also ensures that users, regardless of the device they are using, can access the system seamlessly.
Responsive design principles are applied, meaning that the user interface adjusts and reflows automatically to fit the screen size and resolution of the device. The interface includes clear and concise instructions, guiding users through each step of the process, from uploading an image of the leaf to viewing the predicted disease results. In addition to this, visual cues such as progress bars or loading spinners are integrated to keep the user informed about the system's processing status, enhancing the user experience.
-
e) Testing and Validation
The development of LeafLens includes a rigorous testing and validation phase to ensure the reliability, accuracy, and robustness of the system. Multiple layers of testing are performed at different levels:
Unit testing verifies the functionality of individual components such as image preprocessing, the CNN model inference, and data handling. Integration testing ensures thatall components, including the frontend, backend, and model, work together seamlessly without issues or failures. System testing evaluates the entire system to ensure a smooth user experience and accurate predictions under real-world conditions, including simulating typical user interactions and edge cases.
Through these rigorous testing processes, LeafLens is refined to meet high standards of performance and reliability, providing accurate results under varying conditions.
-
f) Scalability
LeafLens is designed with scalability in mind. The modular architecture allows for the easy addition of new crops, diseases, and features in the future. This flexibility ensures that as agricultural needs evolve, LeafLens can evolve alongside them. Future enhancements might include:
The system can be improved in several ways to enhance its functionality and accessibility. First, it could support additional crops and diseases to keep up with changing agricultural needs. It could also include multi-language support to reach a broader global audience. Additionally, LeafLens could be enhanced to process real-time images from mobile cameras, allowing farmers to use the system directly in the field for immediate disease detection.
This scalability ensures that LeafLens remains an evolving tool for farmers, capable of adapting to their changing needs and the advancing technological landscape.
Results and Discussions
The LeafLens system demonstrated remarkable results in addressing plant disease detection using a deep learning model based on Convolutional Neural Networks (CNN). The system successfully classified plant diseases with a high degree of accuracy, exceeding 85% on validation and testing datasets. This performance indicates the robustness of the model in identifying and categorizing diseases from leaf images, ensuring its reliability for real-world applications. The confusion matrix analysis further validated these results, showcasing strong recall and precision values for most disease categories. Misclassifications were minimal, suggesting that the model is well-trained and capable of accurately identifying diseases like Powdery Mildew and Cercospora Leaf Spot, even under diverse testing conditions.
The image displays a normalized confusion matrix for a classification task, showing the performance of a predictive model across multiple classes Each cell contains the proportion of instances for a specific true-predicted label combination, normalized between 0 and 1. A strong diagonal pattern, where most values are concentrated, indicates that the model correctly predicted most instances for each class. Off- diagonal values represent misclassifications, where the true label differs from the predicted label. The visualization helps assess model accuracy and identify patterns of misclassification across the 39 classes.
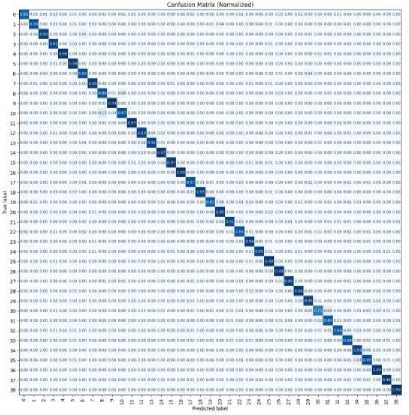
Fig. 2. Confusion matrix
The system's recommendation functionality provided practical and disease-specific solutions, offering clear treatment plans and prevention strategies. This feature not only bridged the gap in existing tools that lacked actionable outputs but also empowered users with actionable insights for managing plant health by providing users with supplement recommendations. By providing this level of detail, the system established itself as more than a diagnostic tool —it became a comprehensive assistant for plant care.
Visualization played a significant role in the project's results. Metrics tracking training and validation performance over epochs highlighted the consistency and reliability of the model's learning process. Additionally, data visualization techniques were employed to analyze the distribution of diseases across the dataset, identifying the most prevalent issues faced by farmers and agriculturists. These visual insights not only enhanced the interpretability of the results but also informed future improvements in the model and system functionality.
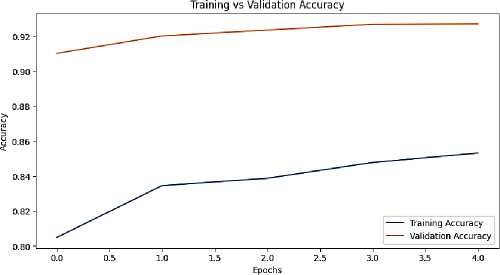
Fig. 3. Training and validation accuracy
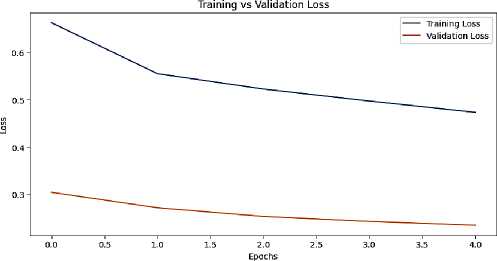
Fig. 4.Training and validation loss
While the results underscored the system's strengths, certain limitations were noted. The accuracy, though high, could benefit from further enhancement by incorporating additional real-world datasets. Such datasets would address rare and edge-case scenarios, improving the model's ability to generalize across diverse agricultural contexts. Moreover, the static nature of the input, limited to still images, constrained the system's capacity to track the progression of plant diseases over time. Addressing these limitations would significantly augment the system’s utility in dynamic farming environments.
Looking ahead, plans for improvement and expansion have been outlined to elevate the LeafLens system’s capabilities. Enhancing the dataset to include a wider range of plant species and disease variations will improve generalizability and ensure the tool remains relevant across different agricultural regions. Introducing features such as real-time plant health monitoring and sensor integration will provide users with dynamic insights, enabling them to track and address issues proactively. Furthermore, multi-language support is under consideration to make the platform globally accessible and inclusive for diverse user groups.
In conclusion, the LeafLens system has proven to be a powerful tool for plant disease detection, combining advanced deep learning techniques with user-friendly design and actionable recommendations. Its success in achieving high accuracy, providing tailored solutions, and presenting visually enriched insights underscores its potential to revolutionize plant health management. By addressing its current limitations and integrating future enhancements, LeafLens is poised to become an indispensable resource for promoting sustainable agriculture and ensuring healthier crops worldwide.


