Microarray gene retrieval system based on LFDA and SVM
Автор: Lt. Thomas Scaria, T. Christopher
Журнал: International Journal of Intelligent Systems and Applications @ijisa
Статья в выпуске: 1 vol.10, 2018 года.
Бесплатный доступ
The DNA microarray technology enables the biologists to observe the expressions of multiple thousands of genes in parallel fashion. However, processing and gaining knowledge from the voluminous microarray gene data is serious issue. It is necessary for the biologists to retrieve the required data in a reasonable time. In order to address this issue, this work presents a gene retrieval system, which is based on feature dimensionality minimization and classification of the microarray gene data. The feature dimensionality minimization is achieved by Local Fisher Discriminant Analysis (LFDA), which inherits the merits of both Fisher Discriminant Analysis (FDA) and Locality Preserving Projection (LPP). Support Vector Machine (SVM) is employed as the classifier to classify between the genes. The LFDA is chosen for reducing the dimensionality of the features, owing to its better performance on multimodal data. The SVM is trained with the feature dimensionality reduced microarray gene data, which improves the efficiency and overthrows the computational complexity. The performance of the proposed approach is compared with the LPP and FDA. Additionally, the performance of SVM is compared with the k-Nearest Neighbour (k-NN) classifier. The combination of LFDA and SVM serves better in terms of accuracy, sensitivity and specificity.
DNA microarray technology, feature reduction, SVM classification, LFDA
Короткий адрес: https://sciup.org/15016449
IDR: 15016449 | DOI: 10.5815/ijisa.2018.01.02
Текст научной статьи Microarray gene retrieval system based on LFDA and SVM
Published Online January 2018 in MECS
Data mining is the process of examining a vast amount of data to provide well-defined information. Hence, the process of transformation from raw data to clear-cut information is called as knowledge discovery process. Recognising the superiority of data mining, several applications utilize the technology. Some of the noteworthy applications are business platforms, finance, biomedicine, networks, education and so on [1].
Microarray technology enables the life scientists to study and analyse the genomic expression of a living organism [2, 3]. DNA microarray technology makes it possible to observe the expression level of several thousands of genes in a concurrent fashion [4,5]. Numerous research activities exploit the microarrays for definitive solution. For instance, toxicology, medical diagnosis, gene regulative studies depend on the microarrays [6-8].
Data mining techniques intend to gain knowledge from the microarray data by means of data clustering, classification and association mining. The overall process of microarray data analysis consists of data normalization, data processing and post-processing. Based on the operative nature of data mining techniques, the learning algorithms are classified into supervised and unsupervised learning.
Supervised learning is comprised of two phases and they are training and testing. In the training phase, the classifier is fed with knowledge of the subject samples of several categories. The testing stage aims to find the category of the test sample being passed. Unsupervised learning techniques do not require any prior knowledge about the processing data.
Irrespective of the presence of several data mining techniques, it is still difficult to gain knowledge from the voluminous microarray data [9]. It would be beneficial for the scientists to have a system that can fetch the data being searched and is popularly called as information retrieval system. The microarray information retrieval system strongly depends on the gene classification process. However the classification process is not simple, as the microarray data is quite voluminous and high dimensional [10, 11].
Understanding the necessity of the information retrieval from microarray gene data, this article proposes a microarray gene information retrieval system. The entire work is decomposed into two different phases, which are microarray feature reduction and classification. The initial phase aims to reduce the dimensionality of the microarray data and the next phase engages itself in gene classification. This work employs Local Fisher Discriminant Analysis (LFDA) to reduce the dimensions of the features. The reasons for the employment of LFDA are it preserves the local structure of the data and clearly separates different classes [12].
The Support Vector Machine (SVM) is trained with the dimensionality reduced feature set. SVM is employed owing to its margin optimization and exclusion of local maxima [13]. The key points of the proposed work are listed below.
-
• Incorporation of LFDA to minimize the dimensionality of the features, which makes the entire process simpler.
-
• The classifier SVM is trained with the dimensionality reduced set of features, which enhances the speed of learning.
-
• The information retrieval is faster, as LFDA reduces the feature dimension to train the SVM.
-
• The computational complexity is overthrown, as the process of classification is done after feature dimensionality minimization.
The rest of the paper is organized as follows. Section 2 presents the related review of literature with respect to the microarray gene data classification. The proposed information retrieval system for microarray gene data is presented in section 3. The performance of the proposed approach is analysed in section 4. The conclusions are drawn in section 5.
-
II. Related Works
This section reviews the related literature with respect to the classification of microarray gene data.
In living organisms, all the cells possess nucleus which in turn contains DNA. Every DNA contains the coding and decoding segments. These coding segments are called as genes and the genes describe the structure of proteins. The formation of proteins is achieved in two steps. Initially, the gene is converted to mRNA and mRNA is converted to proteins.
DNA microarray is the advanced technology that renders a provision to have a global insight of the cell and this paves way for measuring the expression level of several thousands of genes concurrently [14]. However, the serious challenge associated with the microarray gene data is the data dimensionality and this issue leads to misclassification [15, 16].
Effective gene selection is the remedy to this issue and the gene selection is the process of detecting and eliminating unwanted features during the training phase. As the knowledge is gained by the system without unwanted features, the knowledge discovery process is more effective [17]. Feature selection is the most important step, as it decides the efficiency of the work.
A perfect feature selection technique reduces the time consumption and computational complexity involved in the gene classification. However it is not an easy task, as it is difficult to determine which are relevant features and irrelevant features. On successful gene selection, the researchers can analyse the data to classify between the normal and the gene expression related to some diseases.
Understanding the significance of this issue, several researchers engage themselves in this research.
Globally, the feature selection techniques are classified into filter, wrapper and embedded techniques. Filter based approaches depend on the overall features of the training data for feature selection. Wrapper based approaches involve in the optimization process of the predictor for feature selection. Finally, the embedded techniques employ machine learning algorithms to extract the relevant set of features. However, it is reported that the wrapper and embedded approaches show computational burden [18].
The filter based approaches measure the relevancy of the genes by examining the fundamental features of the data, where a single gene or a subgroup of genes is analysed against the class labels. The advantage of filter based approach is its minimal time consumption and the drawback is that the classification results are not up to the mark.
Wrapper approaches failed to grab the attention of the researchers, as the computational overhead is high. Embedded techniques utilize machine learning for feature selection and the mostly used classifier is the Support Vector Machine (SVM). Besides these traditional feature selection techniques, there are several ensemble and hybrid approaches.
Fisher Discriminant Analysis (FDA) classifies the entities by organizing the entities in a single dimensional space and then the classification process is done by fixing threshold. The fisher criterion is utilized to reduce the dimensionality. However, the results produced by FCA are not convincing, when the samples in a class form multiple clusters [19, 20].
Local Preserving Projection (LPP) conserves the local structure of the data, while minimizing the data dimensionality [21]. The local structure is preserved, as the embedding transformation takes the neighbouring pair of data in the embedding space. However, the LPP doesn’t take the label information into account such that it does not work well in the supervised environment.
In [22], an associative classification algorithm is proposed for microarray gene classification. This work clubs both the association rule and classification mining. This work is based on four different phases, which are statistical gene filtering, discretization, class association rules and prediction. The initial phase is meant for distinguishing between the genes and to choose the important genes in the gene expression. The discretization phase is for converting the continuous values to discrete values. The next phase is responsible for producing a group of association rules by utilizing closed frequent itemset. Finally, the classification process is carried out by employing a scoring function. The performance of the proposed approach is tested against Linear Discriminant Analysis (LDA), SVM and decision tree.
In [23], a technique for gene selection and classification is proposed. This work is based on Random Forest Ranking (RFB) and Binary Balck Hole Algorithm (BBHA). The experimental results of this work are compared with several classification techniques and the proposed work is proved to be better.
Motivated by these works, this article intends to present an information retrieval system for microarray data with two stages, which are data dimensionality reduction and classification. This work produces expected results, as the dimensionality of the features are minimized prior to the classification stage, which is achieved by LFDA.
The dimensionality minimization stage reduces the computational complexity and time consumption. The SVM is trained with the obtained feature reduced microarray data, which improves the learning speed and performance. The following section presents the proposed approach in a detailed fashion.
-
III. Proposed Gene Retrieval System with Lfda and Svm
This section elaborates the phases involved in the proposed approach along with the overall working principle.
-
A. Overall Idea
The microarray gene expression data is complex to manage, owing to the nature and the high dimensionality of the data [24]. As the microarray gene data contains numerous expressions, it is difficult to manage the data. For this sake, this article proposes a gene retrieval system by utilizing the LFDA and SVM for the biomedical applications.
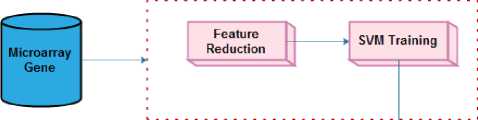
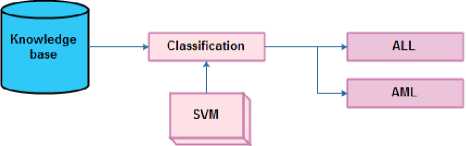
Fig.1. Overall flow of the proposed approach
To achieve the goal, the research is carried out in two steps, which are feature dimensionality minimization and classification. The initial step intends to reduce the data by means of LFDA. The SVM is then trained by the data with minimized dimensionality. The overall flow of the proposed work is presented in figure 1.
The LFDA is chosen by this work, as it clearly increases the class partitions and conserves the local structure of the data. Thus, LFDA works well both between-class and within-class scenarios. This dimensionality minimized data is utilized for the purpose of training the SVM. SVM is selected as the classifier, as the learning ability is better with few samples. The gene retrieval system yields better results, as the feature reduction and classification phases are employed together. The following subsections explain the phases involved in the proposed gene retrieval technique.
-
B. Feature Dimensionality Minimization by LFDA
The major goal of dimensionality minimization is to acquire a low dimensional depiction of high dimensional data along with the preservation of the local structure of the original data [25]. A perfect dimensionality minimization paves way for better classification. Recognizing the importance of feature dimensionality minimization, this work utilizes LFDA which inherits the ideas of both FDA and LPP. FDA is the traditional technique for dimensionality reduction but it cannot perform well with multimodal data.
LPP addresses this issue by minimizing the dimension of multimodal data while it preserves the local structure of the data also. Basically, LPP is an unsupervised dimensionality reduction technique and so it does not take the class labels into consideration. This implies that this technique does not suit supervised dimensionality reduction.
LFDA combines the idea of both FDA and LPP [26], such that it works well for multimodal data. LFDA can address the dimensionality minimization issue by solving the generalized eigenvalue issue. Let the microarray gene data possess к' labelled entities and is represented as {( at , b ,)} JL , , where b , E {1,2, ,„,c c} which is the class label related to the entity a^ and cc is the count of classes.
Consider k'n as the count of labelled entities which are in class n E {1,2, ... cc} and this can be represented as к' = E“ ik'(1)
The local between-class and local within-class matrices are denoted by SMlb et and SMlwlt respectively and are represented as follows.
SMlbet = 1^ ^-a^-rf(2)
SMiwit = 1E £=1^(а-ау)(а-ау)Т
Here, Pjb e 1 and Pj™11 are the к' x k' matrices and represented in the following equation.
(A-4-) if bt = b< plbet _ ) 1 ,j \m' kb. I 1 1 j
U if bt * bj
$if b=bj
(^0 if bt * bj
Where X i, j is the affinity value between ai and a,, subject to the local scaling heuristic. The local scaling is performed on class by class basis, such that the local structure of the microarray gene data is maintained. In eqns 4 and 5, Xi j I —--;- is negative. On the other
- ym kbJ hand, -4^ and 4 are positive. This means that the entities
, present in the same class are closer to each other and the entities of different class are farther. This local scaling operation reduces the computational cost as well. The transformation matrix for microarray gene data TMLFDA is formed as
TMLFDA
= ^^XZzttT MT S MlbetTM(TMTSM lwttTM )-4
The LFDA forms the transformation matrix of the genes, which maximizes the local between-class scatter (TMTSMLbetTM) and minimizes the local within-class scatter (7'M T SM lwit TM) in the embedding space. The generalized eigen value issue is solved by
C =SM(be 1 f D = SM(wlt (7)
Suppose if the value of X i, j = 1 V -,j , then the SM (1 be)> and SM (lwi)> are minimized to SM (be)> and SM (wit) respectively. Thus, the scatter matrices are formed for the microarray gene data and the dimensionality reduced data is obtained.
-
C. Gene Classification by SVM
As soon as the feature dimensionality reduction is achieved on microarray gene data, the SVM is trained with that data. The goal of SVM is to separate the entities by considering the hyperplane.
Let the training entities for SVM is denoted as ТЕ, with which the SVM is trained. The SVM gains knowledge from the training entities and classify the entities into either class A or class B, by means of hyperplane. The hyperplane is formed by solving the below given equation.
f (X) = ^! PiP (X t. x) +I (8)
Where is the lagrange multipler that aims to segregate the hyperplane ( . ) . is the threshold that determines the classification policy by the hyperplane. With respect to the threshold value, the entities are classified to be either in class A or B and this can be represented as
(f (x) > 0; Class A If (x) < 0; Class В
By this way, the SVM classifies between the entities present in the microarray gene data. The next section analyses the performance of the proposed gene retrieval system.
-
IV. Experimental Analysis
This section evaluates the performance of the proposed approach. Initially, a short description about the dataset is presented, followed by which the performance of the proposed work is analysed.
-
A. Dataset Descripton
The experiments are carried out on the Leukemia dataset, which is publicly available [27]. This dataset is formed by collecting the microarray data from Affymetrix chip, which contains 6817 genes. The genes are filtered after which the gene count is reduced to 3051. The training data contains 38 cases, which constitutes 27 Acute Lymphoblastic Leukemia (ALL) and 11 Acute Myeloid Leukemia (AML).
-
B. Results and Discussion
The proposed approach is implemented in Matlab environment (version 8.2). The feature dimensionality of the microarray gene data is minimized by LFDA. The obtained feature dimensionality minimized data is passed for SVM classification. The performance of the proposed technique is analysed by varying the feature reduction techniques and classification techniques in terms of accuracy, sensitivity, specificity and misclassification rates.
Accuracy is the most important measure of any classification technique, and is the ratio of the correctly classified samples to the total number of samples being involved in classification. Sensitivity rate is computed by the ratio of correctly classified samples to be in the correct class to the sum of correctly classified samples in the correct class and the samples which are misclassified to be the members of the wrong class.
Specificity is the rate of samples which are correctly classified as non-native samples of the class to the sum of samples that are wrongly classified as the member of a specific class and the samples that are correctly classified as non-native samples of the class. All the above stated performance measures are represented as follows.
True„+Truen . __ ....
ас cur асу =---------- -----------x 100 (10)
Truep+Truen+Falsep+Falsen
. True,,. _ _ s e ns 1t1 v i ty = x 100(11)
ТтиврА-Fais eu specific tty = —True"—
Fais врА-Ттиеи misclass ification = 100 - accuracy(13)
In the above equations, Truep, True^, aa lsep and Fa Is eH are the true positive, true negative, false positive and false negative rates. Initially, the feature reduction techniques are varied and the performance of the proposed approach is analysed.
The proposed work is compared by implementing LPP [21] and FDA [19] individually against LFDA. The second round of performance analysis deals with the classifiers. This work compares the performance of SVM against k-NN classifier. The experimental results are presented below. The experimental results present the accuracy, sensitivity, specificity and misclassification rates with respect to both ALL and AML.
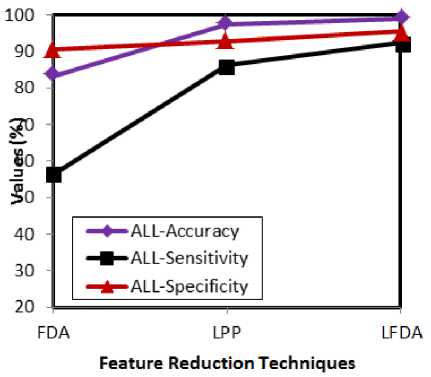
Fig.2. Comparative analysis w.r.t feature reduction techniques for ALL
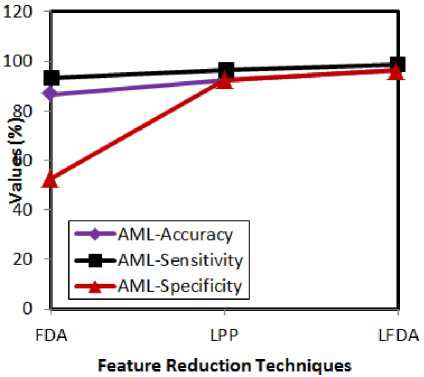
Fig.3. Comparative analysis w.r.t feature reduction techniques for AML
The above presented graphs show the experimental results of both ALL and AML cases with respect to the feature reduction techniques. The attained results prove that the accuracy, sensitivity and specificity rates of the LFDA are better than the FDA and LPP. LFDA achieves better results, as it works well for both between and within class relationships of entities.
Besides this, the LFDA addresses the dimensionality minimization issue by solving the generalized eigen value effectively. All these factors help the LFDA to perform better than the analogous techniques. FDA does not serve well for multimodal data and this issue is addressed by LPP.
However, LPP cannot perform well for supervised environment and thus, LFDA inherits the merits of both the techniques to render better results. The following graphs (fig 4 and 5) present the accuracy, sensitivity and specificity rates by varying the classifiers for ALL and AML.
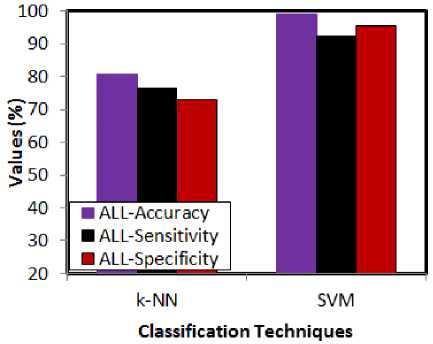
Fig.4. Comparative analysis w.r.t classification techniques for ALL
k-NN
SVM
Classification Techniques
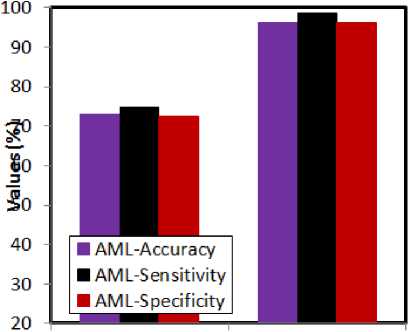
Fig.5. Comparative analysis w.r.t classification techniques for AML
The performance of the classification technique is needed to be justified. In order to achieve this, the feature dimensionality reduced microarray gene data is fed to k-NN and SVM for checking the performance. The learning ability of the k-NN classifier is not upto the mark, when compared to SVM.
Besides this, the learning speed of SVM is greater than k-NN. For these reasons, the choice of SVM is justified in the above presented experimental results. SVM shows the greatest accuracy rates in both ALL and AML cases. The maximum accuracy rate being shown by SVM is 98.9. The highest sensitivity and specificity rates being shown by SVM are 98.6 and 96.3 respectively. This shows the efficacy of the SVM over k-NN classifier. The following graphs (fig 6 and 7) present the misclassification rates by taking the feature reduction and classification techniques into account.
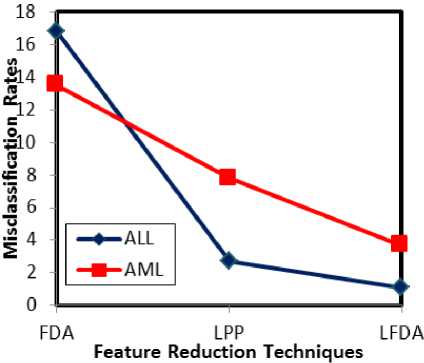
Fig.6. Misclassification rate analysis w.r.t feature reduction techniques
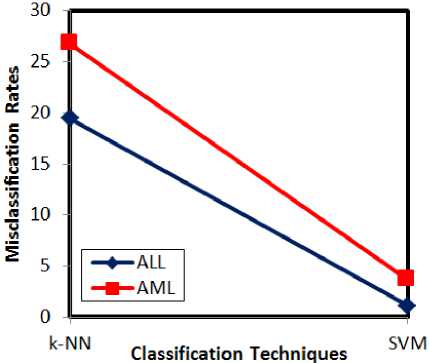
Fig.7. Misclassification rate analysis w.r.t classification techniques
Misclassification rate is indirectly proportional to the accuracy rate. Hence, it is obvious that the misclassification rate of LFDA is comparatively lower than the analogous techniques. LFDA shows the least misclassification rates for both ALL and AML, which are 1.1 and 3.7 respectively. This proves the efficacy of the LFDA over FDA and LPP.
On varying the classifiers, SVM shows the least misclassification rates, when compared to k-NN. From the experimental results, it is evident that the proposed approach serves better with LFDA for feature dimensionality reduction followed by SVM classification. This results in better retrieval rates for both ALL and AML cases.
-
V. Conclusion
This article presents an effective gene retrieval system, which is based on LFDA and SVM. LFDA is utilized for reducing the feature dimensionality of the microarray gene data. This activity of feature reduction helps in removing the unwanted data and thereby saves memory and reduces computational complexity of the forthcoming phase. The learning speed improves, as the SVM gains knowledge from the feature dimensionality reduced microarray data. The performance of the proposed approach is analysed by varying the feature reduction and classification techniques.
LFDA outperforms the LPP and FDA, as it inherits the benefits of both feature extraction techniques. The SVM proves its efficiency over k-NN in terms of classification accuracy. The experimental results prove the efficacy of the proposed approach in terms of accuracy, sensitivity and specificity rates. In future, we plan to continue the research by analysing several feature reduction and classification techniques for microarray gene data.
Список литературы Microarray gene retrieval system based on LFDA and SVM
- H. Chen, "Homeland Security Data Mining Using Social Network Analysis". In: Ortiz-Arroyo D., Larsen H.L., Zeng D.D., Hicks D., Wagner G. (eds) Intelligence and Security Informatics. Lecture Notes in Computer Science, Vol. 5376, pp. 4-4, 2008.
- M.M. Babu, "Introduction to microarray data analysis". Computational genomics: Theory and application. Vol. 17, No.6, pp.225-49, 2004.
- V. Filkov, S. Skiena and J. Zhi, ‘Analysis Techniques for Microarray Time Series Data’, Journal of Computational Biology, Vol. 9, No. 2, pp. 317-330, 2002.
- M. Schena, D. Shalon, R.W. Davis, and P. O. Brown, “Quantitative monitoring of gene expression patterns with a complementary DNA microarray,” Science, Vol. 270, No. 5235, pp. 467–470, 1995.
- J. L.DeRisi, V.R. Iyer, and P.O.Brown, “Exploring themetabolic and genetic control of gene expression on a genomic scale,” Science, vol. 278, no. 5338, pp. 680–686, 1997.
- “The chipping forecast I,” Supplement to Nature Genetics, vol. 21, no. 1, 1999.
- “The chipping forecast II,” Supplement to Nature Genetics, vol. 32, 2002.
- D.P. Berrar, W. Dubitzky, M. Granzow, editors. A practical approach to microarray data analysis. Boston, Mass, USA: Kluwer academic publishers; 2003.
- I. Slavkov, S. Džeroski, J. Struyf, S. Loskovska. "Constrained clustering of gene expression profiles". In : Proc. of the Conference on Data Mining and Data Warehouses at the Seventh International Multi-Conference on Information Society, pp. 212-215, 2005.
- Y.Y. Leung, Y.S. Hung, "An integrated approach to feature selection and classification for microarray data with outlier detection, In: Proc. of 8th Annual International Conference on Computational Systems Bioinformatics, Aug. 10-12, Stanford, CA, USA, pp. 1-4, 2009.
- A. Osareh and B. Shadgar, "Classification and Diagnostic Prediction of Cancers using Gene Microarray Data Analysis", Journal of Applied Sciences, Vol. 9, No. 3, 459-468, 2009.
- M. Sugiyama, "Dimensionality reduction of multimodal labeled data by local Fisher discriminant analysis". Journal of Machine Learning Research, Vol.8, pp.1027–1061, 2007.
- J. Shawe-Taylor, and Cristianini N. "Kernel methods for pattern analysis". Cambridge university press, 2004.
- G. Piatetsky-Shapiro, P. Tamayo, Microarray data mining: facing the challenges, ACM SIGKDD Explor. Newsl. Vol. 5, No. 2,pp. 1-5, 2003.
- T. Golub, D. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek, J. Mesirov, H. Coller, M. Loh, J. Downing, M. Caligiuri, et al, "Molecular classification of cancer: class discovery and class prediction by gene expression monitoring", Science, Vol. 286, No.5439, 531–537, 1999.
- A. Jain, D. Zongker, Feature selection: evaluation, application, and small sample performance, IEEE Trans. Pattern Anal. Mach. Intell. Vol.19, No. 2, pp.153–158, 1997.
- I. Guyon, S. Gunn, M. Nikravesh, L.A. Zadeh, editors. "Feature extraction: foundations and applications". Springer, Vol. 207, 2008.
- V. Bolón-Canedo, N. Sánchez-Maroño, A. Alonso-Betanzos, J.M. Benítez, F. Herrera, "A review of microarray datasets and applied feature selection methods", Information Sciences, vol.282, pp.111-135, 2014.
- R. A. Fisher. "The use of multiple measurements in taxonomic problems". Annals of Eugenics, Vol. 7, No. 2, pp. 179–188, 1936.
- K. Fukunaga. "Introduction to Statistical Pattern Recognition". Academic Press, Inc., Boston, second edition, 1990.
- X. He and P. Niyogi. "Locality preserving projections". In S. Thrun, L. Saul, and B. Sch ¨olkopf, editors, Advances in Neural Information Processing Systems 16. MIT Press, Cambridge, MA,2004.
- S Alagukumar, R Lawrance, "Classification of microarray gene expression data using associative classification", International Conference on Computing Technologies and Intelligent Data Engineering, 7-9 Jan, Kovilpatti, 2016.
- Elnaz Pashaei, Mustafa Ozen, Nizamettin Aydin, "Gene selection and classification approach for microarray data based on Random Forest Ranking and BBHA", IEEE-EMBS International Conference on Biomedical and Health Informatics, 24-27 Feb, Las Vegas, USA, 2016.
- S. Dehuri and S. Cho, “Multiobjective Classification Rule mining Using Gene Expression Programming”, In : Proc of the International Conference on convergence and Hybrid Information Technology, 11-13 November, Busan, Vol. 2, pp. 754-760, 2008.
- G.E. Hinton and R.R. Salakhutdinov, “Reducing the dimensionality of data with neural networks”, Science, Vol. 313, pp. 504–507, 2006.
- M. Sugiyama, M. Krauledat and K.R. Muller. “Covariate shift adaptation by importance weighted cross validation”. Journal of Machine Learning Research, Vol. 8, pp. 985–1005, 2007.
- http://ligarto.org/rdiaz/Papers/rfVS/.


