Molecular-Phylogenetic Research of the Genus Hypericum L. in Flora of Azerbaijan
Автор: Fatdayeva A.
Журнал: Бюллетень науки и практики @bulletennauki
Рубрика: Биологические науки
Статья в выпуске: 11 т.7, 2021 года.
Бесплатный доступ
Hypericum is one of the 100 largest flowering plant genera forming the family Hypericaceae Juss., which belongs to the clusioid clade of the Malpighiales. Hypericum is represented in Azerbaijan flora by 19 native species and 1 subspecies belonging to 7 taxonomic sections. The chloroplast DNA of 8 species from the genus was studied by PCR-RFLP analysis. Total genomic DNA was extracted from leaf tissue using the DNeasyPlantMini kit. (Qiagen Inc.; Valencia, CA, USA) following the supplied protocol and quanti field using a Nanodrop (Nanodrop Technologies; Wilmington, DE, USA) spectrophotometer. The article is part of an experimental study that comprises molecular-phylogenetic research of this genus in the flora of Azerbaijan.
Hypericum, species, subspecies, molecular-phylogenetic research.
Короткий адрес: https://sciup.org/14121219
IDR: 14121219 | УДК: 582.824.3 | DOI: 10.33619/2414-2948/72/02
Текст научной статьи Molecular-Phylogenetic Research of the Genus Hypericum L. in Flora of Azerbaijan
Бюллетень науки и практики / Bulletin of Science and Practice
UDC 582.824.3
Hypericum is one of nine genera and represents approximately80%of the diversity of the family Hypericaceae Juss. This genus is originated from Eurasia and widely distributed in tropical and subtropical regions. Species of this genus grow on damp soils, meadows, and swamps.
Morphologically genera of Hypericum are characterized by the presence of different kinds of secretory glands and channels, including transparent, darkglands. The secretory structures are the accumulation of biologically active substances and their various configurations are important in the classification of the genus.
Species of this genus are annual and perennial herbaceous, semi-shrubs, and shrubs. Theleavesareopposite, rarely whorled, whole-edged, sessile, or with short petioles on the surface and at the edges often with transparent, sometimes with black point glands. Flowers are collected in the corymbose inflorescence.
Plant material and DNA extraction
Fieldwork was conducted during the expeditions (2015-2018) between May September, at different stages of plant development (Table 1). Dried herbarium specimens deposited in the Herbarium fond of the Institute of Botany ANAS (BAK) and were examined according to standard procedures.
Table 1.
PLACES OF COLLECTION OF MATERIAL
|
Species |
Collecting data |
|
AZ0001 Hypericum helianthemoides |
Shach-buz (Kuku village) 2260 m |
|
AZ0002 H. perforatum subsp. veronense. |
Gabala503 m |
|
AZ0003 H. elongatum |
Shach-buz(mountainofYellica ) 2300 m |
|
AZ0004 H. tetrapterum |
Lankaran (Dasdatuk village) 800 m; Gadabay(Soyudluvillage) 1473 m |
|
AZ0005 H. lydium |
Shach-buz (Kuku village, Safdara) 2270 m |
|
AZ0006 H. androsaemum |
Zagatala (Gabizdaravillage) 643 m, Gakh (Lakit village) 1571 m, Gabala (Vandam village) 563 m |
|
AZ0008 H. perforatum |
Zagatala (Gabizdara village) 562 m , Car666 m |
Also, the Herbarium specimens stored in the Herbarium fond of the Institute of Botany ANAS were used in this study. Classic comparative morphological and results of the molecular-phylogenetic analysis were used for the identification of species. From each sample of 100-200 mg of young leave plants put in 2 ml tubes. Leaf material was obtained from three individual plants per accession, flash-frozen in liquid nitrogen, and stored at —200 °C.
The buffer is added to the dried DNA and stored in the refrigerator for 1 night. Amplification reactions were shown in Table 2.
Table 2.
AMPLIFICATION REACTIONS
|
Components |
Stock Cons. |
Reac. Cons. |
|
PCR Buffer |
10X |
1X |
|
MgCl2 |
25mM |
1,5 mM |
|
dNTP mix |
20 mM |
0,2 mM |
|
F. Primer |
10 µM |
0,3 µM |
|
R. Primer |
10 µM |
0,3 µM |
|
Taq DNA Polymerase |
5U/ µl |
2 U |
|
DNA template |
3 µl |
|
|
PCR grade with H2O |
35 µl |
Amplifications were performed as follows: first denaturation (3 min at 940C), 35 cycles of denaturation (15 s at 94 °C), elongation (at 720C), and final extension of (5 min at 72 °C). The amplified products were precipitated with ice-cold ethanol, washedwith70% ethanol, and dissolved in water. PCR products were verified byelectrophoresison1.5% agarose gels containing ethidium bromide in this-acetate EDTA (TAE) buffer and detected under UV light (Table 2).
The restriction fragments with 100 bladders (Gene Ruler TM 100 bp ladder, Fermentas) as a size marker were separated on 1% agarose gels in Tris-acetate EDTA (TAE) buffer (Figure 1).
Table 2.
CYCLES AND THE DURATION OF THE POLYMERASE CHAIN REACTION
|
Temperature of PCR |
Duration |
Cycle |
|
940 C |
3 min |
1 |
|
940 C |
15 sec |
|
|
500 C |
15 sec 35 J |
|
|
720 C |
30 sec |
|
|
720 C |
5 min |
1 |
Checking the amount and purity of DNA
The amount of DNA is determined by a spectrophotometer (260 and 280 nm) (Nano Drop 2000C UV-VisSpectrophotometer-Thermo Scientific). The mixture was used to determine the amount of 20 µl extracted DNA and 1980 µl of DD H2O. The density of theDNAsolutioniscalculated as follows: DNA density (NG/µl) = (OS260H100 (dilution factor) x 50 ng/ml) / 1000. The optical densityratiobetween260and280 nm (OS26 0/OS280) shows the purity of sound acids. The optimum cleaning speed for the PCR is 8-2.0. After determining the amount of DNA, from each sample, are prepared 50 ng / μl of DNA for each PCR reaction.

100 bp DNA Ladder 1% agarose, lx TBE
Figure 1. 100bp_DNA_Ladder
PCR reaction with its primers
The total volume of reactionfor1sampleis20 µl (2 µl of DNA sample + 18 µl of the reaction mixture). The whole reaction should be arried outside the ice and the unit needs to be centrifuged. The tags added to theology measure action mixture, stirring, and the total reaction mix tourist thoroughly vortex. The amount of DNA is pre-placed in a tube or RDA (plate) and then stop resonance. Then, 18 µl is poured from the reaction mixture onto each sample and swirled again. After the reaction tube is placed in the apparatus PCR (Gene Amp System 2720, Applied Biosystems Foster City, CA and BigDyeTerminator v3.1 Cycle) and the programs are compiled in the following sequence.
Results
Nucleotide sequence reading (Sequencing)
Be for the sequencing of the resulting products PCRQIA quick Gel Extraction Kit was cleaned by using a kit (Qiagen, Germany). Then PCR sequence lingua automated ABI 3730 XL with the head edition of primers is placing rid and nucleotides equine cesarean.
Table 2.
THE (5-3) NUCLEOTIDES SEQUENCE OF IT SPRIMERS
|
DNA region |
Primer |
Primer sequence 5-3 |
Reference |
|
ITS |
ITS 1 |
TCC GTA GGT GAA CCT GCG G |
White and other, 1990 |
|
ITS 4 |
TCC TCC GCT TAT TGA TAT GC |
White, and other 1990 |
|
|
Trn L intron |
trnL C |
CGA AAT CGG TAG ACG CTA CG |
Taberlet et al., 1991 |
|
trnLD |
GGG GAT AGA GGG ACT TGA AC |
Taberlet et al., 1991 |
The obtained nucleotides equine care orted bytes of aware Clustal W [8]. Results of some kinds of America Gen Banker and data-fornication were taken. The nucleotide sequences were included in the Molecular Evolutionary Genetics Analysis program (MEGA 6.0). The phylogenetic tree according to the model of Tamura-Nei [9] was constructed with 500 bootstrap-sample using the Maximum Likelihood (ML) method (Figure 2).
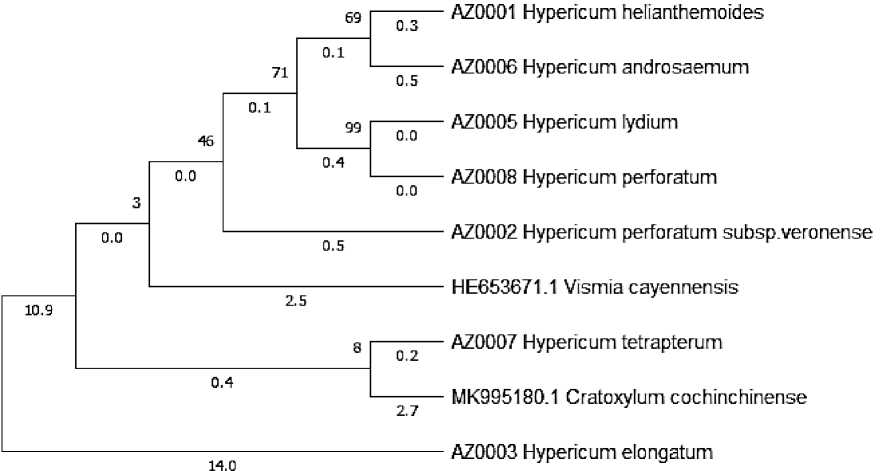
Figure 2. Phylogenetic tree of the genus Hypericum based on the Maximum Likelihood (ML) method
The obtained nucleotide sequences were compared with the data centers of the USA General Bank and the following results were obtained:
Sample AZ0001 with the data of Gen Bank with the type of Hypericum helianthemodes Spach.
Sample AZ0002 with the data of Gen Bank with the type of Hypericum perforatum subsp. veronense . Schrank.
Sample AZ0003 with the data of Gen Bank with the type of Hypericum elongatum Ledeb.
Sample AZ0005 with the data of Gen Bank with the type of Hypericum lydium Boiss.
Sample AZ0006 with the data of Gen Bank with the type of Hypericum androsaemum God.
Sample AZ0007 with the data of Gen Bank with the type of Hypericum tetrapterum Fries.
Sample AZ0008 with the data of Gen Bank with the type of Hypericum perforatum L.
Список литературы Molecular-Phylogenetic Research of the Genus Hypericum L. in Flora of Azerbaijan
- Clapham, A. R., Tutin, T. G., & Moore, D. M. (1990). Flora of the British isles. CUP Archive.
- Flora of Turkey (1967). Ed. by P. H. Davis. Edinburg, II, 355 401.
- Robson, N. B. (1987). Studies in the genus Hypericum L.(Guttiferae). VII: Section 29. Brathys (part 1). Bulletin of the British Museum. Natural History. Botany, 16(1), 1 106.
- Gorshkova, S. (1949). Rod Hypericum L. In Flora SSSR. Leningrad, 15, 203 258. (in Russian).
- Rzazade, R. (1955). Rod Hypericum L. In Flora Azerbaidzhana. Baku, 6, 248 259.
- Bondarenko, S. V. (2012). Konspekt flory Kavkaza. 3(2), Moscow. 308 314. (in Russian).
- Crockett, S. L., Douglas, A. W., Scheffler, B. E., & Khan, I. A. (2004). Genetic profiling of Hypericum (St. John’s Wort) species by nuclear ribosomal ITS sequence analysis. Planta medica, 70(10), 929 935. https://doi.org/10.1055/s 2004 832619
- Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic acids research, 22(22), 4673 4680. https://doi.org/10.1093/nar/22.22.4673
- Felsenstein, J. (1985). Phylogenies and the comparative method. The American Naturalist, 125(1), 1 15. https://doi.org/10.1086/284325


